Abstract
Acyl-homoserine lactones (AHLs) are employed by several Proteobacteria as quorum-sensing signals. Past studies have established that these compounds are subject to biochemical decay and can be used as growth nutrients. Here we describe the isolation of a soil bacterium, Pseudomonas strain PAI-A, that degrades 3-oxododecanoyl-homoserine lactone (3OC12HSL) and other long-acyl, but not short-acyl, AHLs as sole energy sources for growth. The small-subunit rRNA gene from strain PAI-A was 98.4% identical to that of Pseudomonas aeruginosa, but the soil isolate did not produce obvious pigments or AHLs or grow under denitrifying conditions or at 42°C. The quorum-sensing bacterium P. aeruginosa, which produces both 3OC12HSL and C4HSL, was examined for the ability to utilize AHLs for growth. It did so with a specificity similar to that of strain PAI-A, i.e., degrading long-acyl but not short-acyl AHLs. In contrast to the growth observed with strain PAI-A, P. aeruginosa strain PAO1 growth on AHLs commenced only after extremely long lag phases. Liquid-chromatography-atmospheric pressure chemical ionization-mass spectrometry analyses indicate that strain PAO1 degrades long-acyl AHLs via an AHL acylase and a homoserine-generating HSL lactonase. A P. aeruginosa gene, pvdQ (PA2385), has previously been identified as being a homologue of the AHL acylase described as occurring in a Ralstonia species. Escherichia coli expressing pvdQ catalyzed the rapid inactivation of long-acyl AHLs and the release of HSL. P. aeruginosa engineered to constitutively express pvdQ did not accumulate its 3OC12HSL quorum signal when grown in rich media. However, pvdQ knockout mutants of P. aeruginosa were still able to grow by utilizing 3OC12HSL. To our knowledge, this is the first report of the degradation of AHLs by pseudomonads or other γ-Proteobacteria, of AHL acylase activity in a quorum-sensing bacterium, of HSL lactonase activity in any bacterium, and of AHL degradation with specificity only towards AHLs with long side chains.
Many bacterial species control and modulate their physiology in response to increases in their population densities in a process known as quorum sensing (reviewed in references 11 and 25). Several dozen species of Proteobacteria use acyl-homoserine lactones (AHLs) as dedicated signal molecules in this process. A diversity of AHL structures and the enzymes and proteins involved in their synthesis and recognition have been elucidated (12, 27, 31, 35). One of the best studied quorum-sensing species is the opportunistic pathogen Pseudomonas aeruginosa, which makes and responds to two distinct AHLs: 3-oxododecanoyl-homoserine lactone (3OC12HSL, also known as PAI, the signal component of the Pseudomonas las quorum-sensing system), and butanoyl-HSL (C4HSL, also known as PAI-2, the autoinducer of the rhl quorum-sensing system). The two quorum circuits are known to control several physiologies and virulence factors associated with the infection of immunocompromised individuals and those with cystic fibrosis (40). Recently, the influence of AHLs on the global regulation of gene expression by P. aeruginosa has been examined and found to be vast (37, 42). Clearly, AHL-mediated signaling and signal dynamics are very important to the biology of this species, and it is important to understand issues related to signal stability.
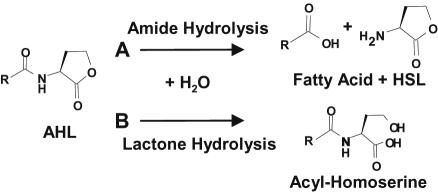
AHLs are known to be chemically inactivated via alkaline hydrolysis, yielding the cognate acyl-homoserine (41), but are stable for weeks or months at pH values of 5 to 6 (34). AHLs are also subject to biological inactivation (Fig. 1). In a process similar to abiotic ring hydrolysis, acyl-homoserine can be generated by acyl-homoserine lactonase encoded by Bacillus cereus (and its close relatives) and by Agrobacterium tumefaciens (6, 8, 22, 32, 46). None of these strains have been found to further degrade the molecule, and no net oxidation occurs during this inactivation reaction. A more extensive degradation of acyl-homoserine can occur, as evidenced by an Arthrobacter soil isolate that utilizes oxohexanoyl-homoserine as an energy source (9). In another mechanism of AHL inactivation, the amide bond of AHL is cleaved by AHL acylases during the utilization of quorum signals as growth nutrients by Variovorax and Ralstonia species (21, 23). HSL is released as a product of these reactions, and the acyl moiety is further metabolized as an energy substrate (19, 21). When a gene encoding an AHL acylase from Ralstonia, aiiD, was expressed in both Escherichia coli and P. aeruginosa, it effectively inactivated endogenously produced AHL quorum signals and quenched quorum sensing in the latter strain (23).
FIG. 1.
Two mechanisms by which AHLs can be inactivated. A, cleavage of the amide bond by bacterial AHL acylase yields HSL and the corresponding fatty acid (21, 23). The AHL amide bond is chemically stable under conditions of nonextreme temperature and pH. B, cleavage of the lactone ring by bacterial AHL lactonase yields the corresponding acyl-homoserine (7, 46). The lactone ring is also subject to chemical hydrolysis; the chemical half-life of the ring is ca. 10[7−pH] days, i.e., it is less stable with increased alkalinity. The acyl side chain diversity of known, naturally occurring AHLs has been reviewed (11).
A close homologue of the Ralstonia AHL acylase has been identified in P. aeruginosa strain PAO1 and in the genomes of several other sequenced pseudomonads (23). This finding raised the possibility that members of this genus, which often produce their own AHLs and engage in quorum sensing, may also be capable of degrading these signals. In this study we detail our isolation of a soil pseudomonad capable of utilizing 3OC12HSL as a sole energy source, our subsequent discovery that P. aeruginosa also exhibits growth on long-acyl AHLs, and our examination of pvdQ, an aiiD homologue in P. aeruginosa, for AHL acylase activity.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used were as follows: Pseudomonas strain PAI-A (isolation described below); P. aeruginosa strain PA14, obtained from Dianne Newman of Caltech; Pseudomonas aeruginosa strains PAO1 and QSC112a, obtained from E. Peter Greenberg of the University of Iowa (43); P. aeruginosa strain PAO1 and a mutant with an in-frame pvdQ deletion and Genr cassette replacement of this PAO1 Denver strain, both obtained from Michael Vasil of the University of Colorado Health Sciences Center (29); P. aeruginosa PAO1 containing pPvdQ-Nde (see below), a constitutive PvdQ expression vector derived from pUCP-Nde (4); E. coli DH5α, carrying pUCP18-Nde obtained from Ciaran Cronin of the University of California, San Francisco; E. coli BL21PRO containing pPvdQ-PROTet, a pPROTet.E133-derived Tet-inducible PvdQ expression vector encoding tetracycline and spectinomycin resistance (see below); and E. coli BL21PRO containing the autonomously replicating plasmid, pPROTet.E133 (Clontech).
Media and growth conditions.
Lysogen broth (LB) medium, amended with antibiotics when appropriate, was used for growth and maintenance of all strains unless otherwise stated. For the 3OC12HSL-dependent enrichment of strain PAI-A and other growth experiments performed with this strain, we modified MES 5.5, the defined medium used to enrich and study Variovorax paradoxus VAI-C (21). These modifications included buffering with 5 mM MOPS [3-(N-morpholino)-propanesulfonic acid] at pH 7.2 and the addition of sodium sulfate (14 g · liter−1) and magnesium chloride (4 g · liter−1). For growth experiments with P. aeruginosa, MES 5.5 defined medium was used as described previously (21), with the exception that it contained sodium sulfate, not sulfite (a typographical error in the reported recipe), as a source of sulfur. Unless otherwise noted, the medium was buffered to a pH of 5.5 with 5 mM 2-(N-morpholino)-ethanesulfonic acid (MES). Ammonium-free MES 5.5 basal medium was used to examine the utilization of AHLs and HSL as potential nitrogen sources. Stock solutions (100 mM) of AHLs were prepared by dissolving AHLs in ethyl acetate that had been acidified with glacial acetic acid (0.01% [vol/vol]), and stocks were stored at −20°C. The AHLs used in these studies were as follows: N-3-oxododecanoyl-l-homoserine lactone (3OC12HSL; Quorum Sciences Inc., Iowa City, Iowa); N-3-oxohexanoyl-l-homoserine lactone (3OC6HSL; Sigma); and, from Fluka, N-3-butanoyl-dl-homoserine lactone (C4HSL), N-3-hexanoyl-dl-homoserine lactone (C6HSL), N-3-heptanoyl-dl-homoserine lactone (C7HSL), N-3-octanoyl-dl-homoserine lactone (C8HSL), N-3-dodecanoyl-dl-homoserine lactone (C10HSL), N-3-dodecanoyl-dl-homoserine lactone (C12HSL), and N-3-tetradecanoyl-dl-homoserine lactone (C14HSL). For growth experiments, the AHL was dispensed into sterile tubes, the ethyl acetate was removed by evaporation under a stream of sterile air, and sterile medium was added to the dried AHL that remained. Stocks of l-HSL (Sigma) were prepared just prior to their use from well-desiccated reagent stored at −20°C. Thin-layer chromatography (TLC) with subsequent ninhydrin staining was used to confirm the purity of HSL stock solutions (15). Cells were grown in 5 ml of medium in 18-mm-diameter tubes with shaking at 37°C unless otherwise noted. AHL molecules are stable for 30 days under the conditions of low pH in our defined medium (34; A. Eberhard, personal communication). Unless noted, all other reagents were of reagent grade.
Enrichment and isolation procedures.
Turf soil was collected in May 2000 at the University of Iowa. The soil was disrupted and dispersed with a metal spatula, and remaining large particles were removed. One hundred milligrams of the soil was added to 5 ml of the vitamin-replete, ammonium-replete enrichment medium containing 1 mM 3OC12HSL as a sole energy source (see above). After 2 days of incubation with shaking at 37°C, sequential 1% (vol/vol) transfers were made into like medium before being streaked for isolation on rich media. Because 3OC12HSL is not soluble at the concentrations employed for growth, isolation was on LB agar with subsequent verification of the AHL degradation phenotype in the defined liquid medium.
Growth studies.
Measurements of optical density at 600 nm (OD600) were performed by using a Spectronic 20 spectrophotometer. AHLs with side chains of more than six carbons in length were poorly soluble, so the ethyl acetate carrier was evaporated in the glass tube so that a uniform coating of AHL was beneath the spectrophotometer's light path. When care was taken to vortex the tubes gently, the changes in optical density reflecting growth could be monitored accurately. Molar growth yields were determined in the defined media containing the indicated substrate at a final concentration of 1 or 2 mM. For both Pseudomonas strain PAI-A and P. aeruginosa PAO1, the factors for converting optical density to cell dry mass were determined by growing cells in media containing succinate as the energy source and NH4Cl as the nitrogen source, washing the cells with 50 mM ammonium acetate buffer (pH 5.5), and drying cell samples to a constant weight. Such determinations were made in quadruplicate.
Other analyses.
Strain PAI-A was examined for several traits exhibited by P. aeruginosa, which was used as a positive control. Fluorescent pigment production was examined by using Wood lamp illumination of colonies grown on LB agar. Pyocyanin production in glycerol-alanine medium was examined (10). Production of acyl-HSLs in both LB and defined media was examined by using previously described radioassay methods (33). A Beckman System Gold high-performance liquid chromatography (HPLC) running a methanol gradient was used in the chromatographic analysis of ethyl acetate extracts as previously described (34). Radioactivity was monitored via online, solid scintillation counting using an in-line HPLC β-particle detector (Model 3; IN/US Systems, Tampa, Fla.). Microscopic examinations were performed by using a Zeiss Stemmi 2000 stereomicroscope (low magnification) and a Zeiss Axioplan research microscope (higher magnification; phase-contrast and dark-field microscopy). Nitrate-dependent anaerobic growth was tested by using both MOPS-buffered defined media or LB medium amended to contain 10 mM potassium nitrate dispensed under a 100% N2 headspace in Bellco (Vineland, N.J.) 18-mm-diameter butyl, serum-stoppered Balch tubes.
Analysis of cell-free culture and reaction fluids.
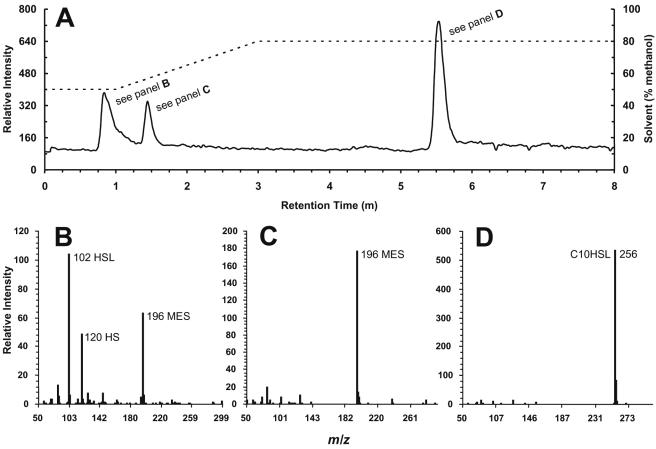
For the initial characterization of the intermediates in AHL degradation, a TLC method was used (15). For a refined analysis, a liquid-chromatography-atmospheric pressure chemical ionization-mass spectrometry (LC/APCI-MS) technique was developed to monitor and quantify the disappearance of AHL and appearance of a number of AHL degradation products. For this analysis, 50-μl samples of culture fluids were taken in triplicate from AHL-grown cultures and were centrifuged at 15,800 × g for 10 min. The cell-free culture fluids were stored at −20°C until all samples had been collected for analysis. For LC/MS analysis, samples were mixed 1 to 1 with acetic acid-acidified methanol (1% [vol/vol]). Dilutions were made with MES 5.5 medium. A C18 ultra-aqueous reverse-phase column (5-μm-diameter bead size; 50 by 3.2 mm; Restek catalogue no. 9178553) was used. The initial mobile phase was 50:50:1 (methanol-water-acetic acid) running at 0.5 ml · min−1 isocratically over the first minute after injection and increased (via a linear gradient) to 80:20:1 (methanol-water-acetic acid) over the following 2 min (see Fig. 3). By this method, a diversity of AHLs, their corresponding acyl-homoserines, HSL, and homoserine in samples could be quantified from cultures growing in the MES 5.5 defined medium. These analyses were performed at Caltech's Environmental Analysis Center by using a Hewlett-Packard 1100 Series LC/APCI-MS instrument.
FIG. 3.
LC/APCI-MS analysis of a cell-free fluid sampled from a P. aeruginosa culture utilizing C10HSL as a sole energy source in MES 5.5 medium. The details for resolving AHLs from their degradation products and other media components are described in Materials and Methods. (A) Chromatogram showing the separation of homoserine and/or HSL, MES buffer, and decanoyl-HSL (left axis). The hatch marks correspond to changes in methanol/water solvent ratios during the course of the run (right axis). (B) The mass spectrum of the first peak resolves homoserine from homoserine lactone; note that the peak tail can overlap with, but can be resolved from, the component in the second peak. (C) Mass spectrum of the second peak, morpholinoethane sulfonic acid (MES buffer). (D) Mass spectrum of the third peak, decanoyl-HSL. This method can be applied to separate and determine the concentrations of a number of other AHLs and any acyl-homoserine degradation products (not shown).
Standards over a range of concentrations (125 nM to 1 mM) were prepared by using either water or MES 5.5 basal media, depending on the origin of the sample, and were diluted 1 to 1 with acetic acid-acidified methanol (1% [vol/vol]). For a 20-μl sample injection, the detection limits for standards prepared in MES 5.5 media were as follows: 2.5 pmol for 3OC12HSL and its corresponding acyl-homoserine, 10 pmol for C10HSL and its corresponding acyl-homoserine, and 100 pmol for HSL. The limit of detection for standards prepared in water was lower than for those prepared in medium, but the former were used only to quantify AHLs recovered from evaporated ethyl acetate extracts. Ethyl acetate extraction did not recover HSL or homoserine. The accurate quantitation of homoserine in standards and samples was complicated by its partial lactonization into HSL, a chemical reaction that occurred after injection into the LC/APCI-MS instrument. Thus, while homoserine plus HSL pool sizes could accurately be quantified, homoserine itself was determined with less precision by using the LC/APCI-MS method and was usually a slight underestimate.
Nucleotide sequence analysis of the small-subunit (SSU) rDNA strain PAI-A.
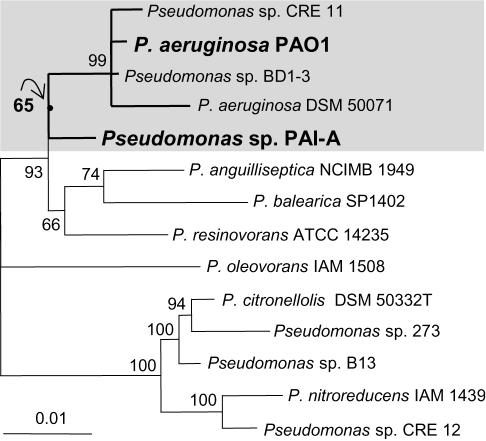
The 16S ribosomal DNA (rDNA) of strain PAI-A was PCR amplified by using standard 27-Forward and 1492-Reverse primers (18); the PCR product was cloned, sequenced, and analyzed by using previously described procedures (20, 21). Sequence reads were assembled and edited by using Sequencher (Genecodes, Ann Arbor, Mich.). Multiple sequence alignments, translations, and phylogenetic analyses were performed by using the Linux ARB freeware package (www.arb-home.de/). Phylograms were constructed via Puzzle-Map 5.0 maximum-likelihood analyses (36). The tree layout was performed by using Treeview 1.6.6 for Windows (30). The 1,401-bp sequence for strain PAI-A has been submitted to GenBank (see below). The GenBank accession numbers for the other sequences presented in Fig. 2 are as follows: Pseudomonas strain CRE 11, U37338 (28); P. aeruginosa PAO1, AE004949 (38); Pseudomonas strain BD1-3, AB015516; P. anguilliseptica, X99540 (5); P. balearica U26418 (3); P. resinovorans, AB021373 (1); P. oleovorans, D84018 (2); P. citronellolis, Z76659 (26); Pseudomonas strain 273, AF039488 (44); Pseudomonas strain B13, AJ272544 (24); P. nitroreducens, D84021 (2); and Pseudomonas strain CRE 12, U37339 (28).
FIG. 2.
rRNA-based phylogeny of strain PAI-A. Construction of the phylogram used 1,120 unambiguously aligned nucleotide positions in a 10,000-step Tree-Puzzle 5.0 maximum-likelihood analysis (36, 39). The bar represents evolutionary distance as 0.01 changes per nucleotide position, determined by measuring the lengths of the horizontal lines connecting the species. The numbers provide support for the robustness of the adjacent nodes. The arrow points to the short node from which the five strains within the shaded box radiate. See Materials and Methods for GenBank accession numbers.
Cloning and expression of pvdQ (PA2385) encoding a putative P. aeruginosa AHL acylase.
Genomic DNA was isolated from P. aeruginosa PAO1 by using the DNeasy tissue kit (Qiagen) and used as a template for the PCR. The deduced coding region for PvdQ (gene PA2385; http://www.pseudomonas.com) was amplified from the genomic DNA by using primers 5′-AGGCCAAGCTTATGGGGGATGCGTACCGTACTG-3′ and 5′-GTTATATAGCGGCCGCTAGGCATTGCTTATCATTCG-3′ (underlining indicates HindIII and NotI restriction sites in the former and latter primers, respectively), cloned into the appropriately digested expression vector, pPROTet.E133 (Clontech), and transformed into E. coli BL21PRO. Recombinant AHL acylase activity was examined as follows. After growth in LB medium containing spectinomycin (50 μg · ml−1) and chloramphenicol (34 μg · ml−1) and after gene induction by the addition of anhydrotetracycline (100 ng · ml−1) at 18°C, cells were pelleted and resuspended to a final optical density of 1.2 in MOPS-buffered media (pH 7.2) containing 10 μM 3OC12HSL. Recombinant cells that had not been induced with anhydrotetracycline were used as a negative control. Reaction mixtures were incubated at 30°C. Samples of 150 μl were removed at 0, 15, 30, and 60 min and analyzed for AHL disappearance and product appearance via LC/APCI-MS (see above).
Constitutive overexpression of pvdQ in P. aeruginosa.
For the constitutive expression of pvdQ in strain PAO1, the coding sequence was PCR amplified with the following primers: 5′-AAGAGGACATATGGGGGATGCGTACCGTACTG-3′ and 5′-CTAAAGCTTGGCTGTGGGCCGCCTCTATGG-3′ (underlining indicates NdeI and HindIII restriction sites in the former and latter primers, respectively). The PCR product was cloned into the E. coli-Pseudomonas shuttle expression vector pUCP-Nde digested with NdeI and HindIII (4). The resulting construct, pPvdQ-Nde, was transformed into P. aeruginosa PAO1 via electroporation. Since the repression of gene expression from this vector requires LacI, and since wild-type P. aeruginosa PAO1 does not encode this repressor, the probable acylase was expected to be constitutively expressed, a prediction which was borne out after the examination of total cell proteins via polyacrylamide gel electrophoresis.
Nucleotide sequence accession number.
The 1,401-bp sequence for strain PAI-A has been submitted to GenBank under accession no. AY288072.
RESULTS
Enrichment and isolation of a bacterium that utilizes 3OC12HSL as a sole energy source.
An enrichment culture using a 3OC12HSL-containing minerals and vitamins medium became turbid within 48 h after inoculation with turf soil. No growth was evident in a control lacking energy nutrient. The cells were rods of uniform morphology and were well dispersed in the medium. They did not form clumps or a pellicle, nor did they attach to the glass at the air-medium interface. When the culture was streaked on LB agar medium for isolation, a single, uniform colony morphotype was observed. Pure cultures were obtained after several successive streaks from single colony picks. Growth of a representative isolate, designated strain PAI-A, was confirmed in the 3OC12HSL-containing liquid medium.
Phylogenetic analysis of strain PAI-A.
A nearly complete sequence of the SSU rDNA was obtained. Web-based similarity searches against rDNA in the RDP-II and GenBank databases suggested that strain PAI-A was most closely related to P. aeruginosa and several other pseudomonads. The SSU rDNA shared 98.4 and 98.1% sequence identity with P. aeruginosa PAO1 and Pseudomonas resinovorans, respectively. By any of the distance (not shown), parsimony (not shown), and maximum-likelihood methods employed (Fig. 2), the SSU rDNA from strain PAI-A clustered most closely with those from P. aeruginosa and its close relatives.
Properties of Pseudomonas strain PAI-A.
Strain PAI-A grew aerobically in both defined media and LB at 30 and 37°C, but not at 42°C. Cultures doubled every 35 min in defined medium with succinate as the sole carbon source at 37°C. The isolate grew on a number of tested substrates at both pH 7.2 and pH 5.5; however, cultures did not grow in AHL-containing media at the latter pH. Cells did not grow anaerobically in either succinate-defined or LB media amended with nitrate. Exponentially growing cells sampled from AHL-containing media were vigorously motile rods, 2.5 by 0.8 μm in dimension. The isolate formed creamy white colonies with spreading edges. After several days, colonies become smooth, nonsticky, leathery, and extremely recalcitrant to disruption with an inoculating loop.
Strain PAI-A did not produce colored or fluorescent pigments in or on LB or glycerol-alanine (pyocyanin production) media. The cultures did not have any aroma of note. Cells did not grow in media containing 30 μg of nalidixate · ml−1. To examine Pseudomonas strain PAI-A for the production of AHL quorum signals, cultures grown in both defined and LB media were incubated with [14C]carboxyl methionine. Since no radioactive peaks were evident after chromatography of the ethyl acetate extract fraction, this isolate does not appear to accumulate AHLs under the conditions examined.
Examination of P. aeruginosa strains for the ability to utilize 3OC12HSL.
Two clinical strains of P. aeruginosa, PAO1 and PA14, were examined for the ability to utilize 3OC12HSL in defined, ammonia-replete media at both pH 5.5 and pH 7.2. Both strains grew rapidly at both pHs with succinate as a sole energy source. Although initially it appeared as if neither would utilize the quorum signal as an energy nutrient, the strains began to grow exponentially with a doubling time ranging from 11 to 25 days after several weeks of incubation. The length of the initial lag phase in cultures inoculated by using naïve cells (those not previously grown on AHL) was highly variable, ranging from 10 to 30 days. Curiously, AHL-grown cells that were transferred directly into AHL-containing media did not show significant lags in growth, but those transferred and grown in media containing a different energy substrate followed by reintroduction into AHL broth reexhibited long lag phases. The issues underlying the long lags exhibited by naïve cells and their subsequent adaptation to growth on AHLs have not been further clarified.
Pseudomonas strain PAI-A and P. aeruginosa PAO1 degrade and utilize long-acyl AHLs.
Strains PAI-A and PAO1 grew on a number of AHLs, but no growth was observed with AHLs with acyl side chains shorter than eight carbons (Table 1). When provided with 1 mM C4HSL as a cosubstrate in 3OC12HSL-containing media, PAI-A and PAO1 did not degrade detectable amounts of the short-chain AHL or exhibit any C4HSL-dependent stimulation of their growth yields. Optical density to dry weight biomass conversion factors were determined for an OD600 of 1.0 as 346 ± 7 μg · ml−1 for strain PAO1 and 337 ± 8 μg · ml−1 for strain PAI-A. The doubling times of strains PAI-A and PAO1 were comparable for many substrates (Table 1). P. aeruginosa PAO1 utilized both the d and l forms of AHLs, as determined by substrate disappearance and comparison of the molar yields on l and dl forms. Curiously, no increase in molar growth yield was observed as a function of AHL acyl lengths, i.e., when comparing growth on C10HSL, C12HSL, and C14HSL (see Fig. 5 of reference 21). The AHL molar growth yields for strain PAO1 were poor, e.g., only 49 to 74% of that achieved during its growth on the corresponding fatty acids (Table 1). Growth on fatty acids revealed the expected incremental increase in molar yield as a function of increased acyl length. Neither strain PAI-A nor strain PAO1 used HSL as a sole or supplementary energy source.
TABLE 1.
Growth of P. aeruginosa PAO1 and Pseudomonas strain PAI-A on acyl-homoserine lactones and other energy sourcesa
| Substrate |
P. aeruginosa PAO1
|
Pseudomonas strain PAI-A
|
||
|---|---|---|---|---|
| Yield (g · mol−1) | Doubling time (h) | Yield (g · mol−1) | Doubling time (h) | |
| C8-dl-HSL | 95 ± 4 | 15.0 ± 2 | +, NDb | ND |
| C10-dl-HSL | 97 ± 3 | 14.0 ± 3.8 | +, ND | ND |
| 3OC12-l-HSL | 76 ± 10 | 25.0 ± 3 | 84 ± .2 | 25.0 ± 3.5 |
| C12-dl-HSL | 84 ± 7 | 14.9 ± 7.5 | 80 ± 18 | 16.5 ± 3 |
| C14-dl-HSL | 84 ± 12 | 21.0 ± 5.3 | +, ND | ND |
| Succinate | 43 ± 3 | 0.6 ± .09 | 49 ± 3.8 | 0.6 ± .02 |
| Decanoate | 130 ± 3 | ND | 126 ± .5 | ND |
| Dodecanoate | 141 ± 4 | ND | 155 ± 8.4 | ND |
| Tetradecanoate | 177 ± 16 | ND | 198 ± 3 | ND |
Values represent the averages ± standard errors from at least duplicate cultures. Studies were performed with minimal salts media buffered at pH 5.5 for strain PAO1 and pH 7.2 for strain PAI-A. Neither strain utilized HSL, homoserine, C4HSL, 3OC6HSL, C6HSL, or C7HSL as an energy source.
+, positive yield; ND, not determined.
FIG. 5.
E. coli cells expressing recombinant pvdQ degrade 3OC12HSL and generate stoichiometric amounts of HSL. Substrate disappearance and product accumulation were determined by using the LC/APCI-MS method (for an example, see Fig. 3.). Induced cells containing plasmid pPvdQ-PROTet were washed and suspended in MOPS (pH 7.2) buffered medium to a final OD600 of 1.2. AHL degradation and HSL accumulation were not observed over the duration of the experiment in either heat-killed suspensions of the same cells, or in no-cell controls (not shown).
Strains PAI-A and PAO1 release HSL as an initial product of AHL degradation.
TLC of clarified reaction fluids, harvested from dense cell suspensions of strains PAI-A and PAO1 incubated with 25 mM C12HSL, revealed the AHL-dependent release of ninhydrin-reactive materials. These had the same yellow and purple staining characteristics and migration characteristics as authentic HSL and homoserine, respectively (data not presented). In contrast, cell-free, AHL-free, and cell-and AHL-free controls did not produce ninhydrin-reactive materials after similar incubation periods. The TLC data suggest that both strains catalyze the initial step of AHL degradation via an HSL-releasing acylase. Because analyses of biological AHL degradations are less ambiguous at pH 5.5 than they are at pH 7.2, and because strain PAI-A does not grow on AHLs at pH 5.5, P. aeruginosa was chosen for further experiments.
LC/APCI-MS analyses confirmed that P. aeruginosa PAO1 releases HSL and homoserine as AHL degradation products. A representative chromatogram of cell-free fluid from a C10HSL-grown culture is shown in Fig. 3A. Although HSL and HS elute at similar times, both compounds were resolved by extracting the M + 1 molecular ions 102 and 120, respectively, from the raw chromatogram (a standard MS practice).
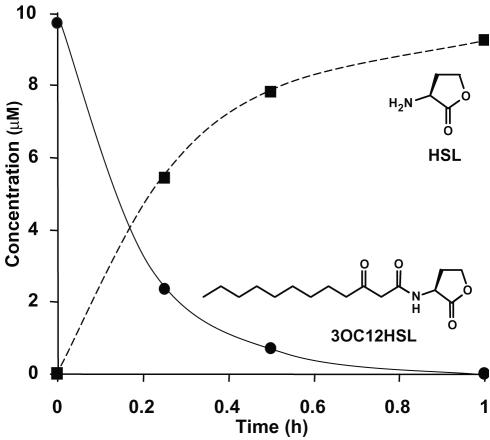
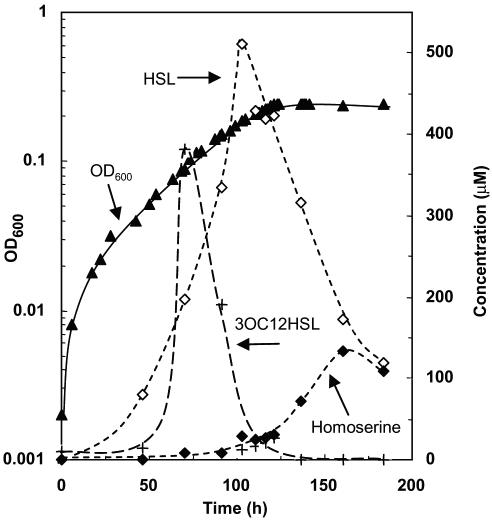
P. aeruginosa PAO1 growth and utilization of 3OC12HSL as the sole energy source in MES 5.5 medium is shown in Fig. 4; the cells used as the inoculum had been grown out in 3OC12HSL-containing MES 5.5 medium, so the long lag phase was not evident. Because 1 mM 3OC12HSL is essentially insoluble, the concentration of the substrate in the medium at time zero appeared low; a transient peak of the substrate appeared during the growth phase, possibly reflecting the dissolution of the particles via surfactant production. By early stationary phase, all of the white, nearly insoluble 3OC12HSL substrate was consumed such that concentrations of less than 125 nM remained. HSL accumulated throughout the growth phase and reached a maximum of ca. 500 μM just before the onset of stationary phase, after which it decreased to less than 80 μM by 100 h into stationary phase. Concomitant with the disappearance of HSL, the amino acid homoserine accumulated and then decreased to concentrations below 80 μM by 100 h into stationary phase (Fig. 4). Since the culture pH was well controlled at pH 5.5, and since the half-life decay of HSL into homoserine at this pH is on the order of weeks, an enzymatic HSL lactonase, not abiotic alkaline hydrolysis, is most likely responsible for the evolution of homoserine. P. aeruginosa did not grow with either HSL or homoserine as a sole energy source in MES 5.5 media. When provided with long-chain AHLs as sole sources of carbon and nitrogen, strain PAO1 grew about twice as slowly as cultures utilizing AHL plus ammonium (not shown). Cells did not use either homoserine or HSL as sole sources of energy or nitrogen.
FIG. 4.
Growth of P. aeruginosa PAO1 in ammonia-replete MES 5.5 media containing 1 mM 3OC12HSL as the sole energy source. Substrate consumption and product accumulation were determined via LC/APCI-MS. Note that since the 3OC12HSL substrate was poorly soluble at the initial concentrations employed, virtually no AHL was observed in the culture fluid at the time of inoculation. As growth progressed, a transient spike of AHL in solution was observed. HSL accumulated throughout the growth phase but was degraded upon entry into stationary phase yielding a transient intermediate, homoserine. 3OC12-homoserine concentrations remained static throughout the course of the experiment and never exceeded 0.1% of the initial AHL concentration (not plotted). Culture pH was examined and found to be well controlled throughout.
Analysis and expression of P. aeruginosa pvdQ, which encodes a candidate AHL acylase.
We explored whether the P. aeruginosa gene PA2385 (recently named pvdQ [17]), which was identified as a close homologue to a gene encoding an HSL-releasing AHL acylase from Ralstonia XJ12B (23), might encode a protein with AHL acylase activity and confer the AHL-dependent growth of P. aeruginosa. The coding region of this gene was amplified from the genomic DNA, cloned into an expression vector, and expressed in E. coli. The polypeptide encoded by the gene was predicted to be posttranslationally cleaved into two distinct subunits (discussed in reference 23). Polyacrylamide gel electrophoresis analysis of the total protein fractions from E. coli cells expressing recombinant pvdQ revealed small amounts of the two expected subunits (data not shown). The majority of the recombinant protein was recovered as the unprocessed 80-kDa propeptide. This observation is similar to that noted by Lin et al. for recombinant aiiD, the Ralstonia AHL acylase gene (23).
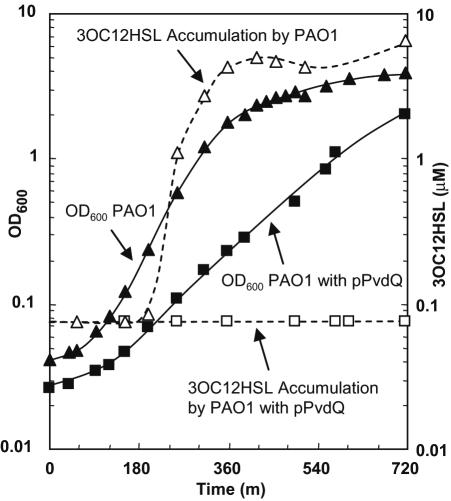
Resting E. coli cell suspensions expressing pvdQ were incubated with 10 μM 3OC12HSL, which is a concentration relevant to the quorum-sensing physiology of P. aeruginosa. AHL disappearance and the appearance of HSL and 3OC12-homoserine were evaluated by using the LC/APCI-MS analysis of cleared reaction fluids (Fig. 5). Within an hour, the AHL disappeared concurrently with the accumulation of stoichiometric amounts of HSL as product. No 3OC12-homoserine accumulation was observed. Cell-free and uninduced cell controls did not catalyze HSL release or the degradation of the AHL over the same time period. In a pattern similar to the AHL utilization data (Table 1), cells of E. coli expressing the recombinant acylase catalyzed the HSL-releasing degradation of C14HSL, C12HSL, C10HSL, and C8HSL, but not 3OC6HSL or C6HSL. The effects of the constitutive expression of pvdQ in P. aeruginosa PAO1 were also examined. In comparison to the wild type, which accumulated 3OC12HSL to concentrations in excess of 6 μM during growth in LB at 30°C (Fig. 6), cultures expressing the acylase did not accumulate any of this quorum signal above the threshold of detection.
FIG. 6.
Growth and accumulation of endogenous 3OC12HSL by P. aeruginosa PAO1 wild type (▴, ▵), and a recombinant derivative constitutively expressing pvdQ (▪, □). Because of the organic complexities of LB, sampled cell-free culture fluids were extracted with ethyl acetate before LC/APCI-MS analysis; the limit of detection for 3OC12HSL was 75 nM and was plotted in place of zero. Cultures were grown at 30°C in LB. Under similar culture conditions, a pvdQ knockout mutant grew and accumulated 3OC12HSL in parallel with the wild type (not shown).
A pvdQ deletion replacement mutant and the strain QSC112a, which carries a Tn5 insertion into pvdQ, were also examined for growth in defined media with 3OC12HSL as the sole energy source. Surprisingly, both mutants remained capable of growing on long-acyl AHL signals. Accumulations of endogenously produced 3OC12HSL by LB-grown cultures of the pvdQ deletion mutant were identical to that of the wild type grown under the same conditions. Evidently, although pvdQ encodes an enzyme with an HSL-releasing AHL acylase activity specific towards long-chain AHLs, another enzyme must be a significant contributor to the growth phenotype on AHL.
DISCUSSION
We have enriched and isolated a novel soil pseudomonad, strain PAI-A, based on its ability to degrade 3OC12HSL, a known virulence factor produced and used as a dedicated signal in the quorum-sensing physiology of the opportunistic pathogen P. aeruginosa. Subsequently, we found that two clinical strains of P. aeruginosa were capable of degrading and growing on 3OC12HSL and on other long-acyl AHLs. None of the pseudomonads examined degraded either butanoyl-HSL, the other distinct AHL quorum signal produced by P. aeruginosa, or other short-acyl AHLs tested. Although closely related to P. aeruginosa, strain PAI-A is not a member of that species as it did not produce pigments or AHLs or grow at 42°C or anaerobically with nitrate as a terminal electron acceptor.
By use of a newly refined LC/APCI-MS technique, the soil and clinical pseudomonads were shown to degrade AHLs via an HSL-releasing activity. These results suggest that these pseudomonads use an AHL acylase in the initial step of AHL degradation, a mechanism previously described in Variovorax and Ralstonia isolates (21, 23). P. aeruginosa accumulated HSL as a transient intermediate during degradation of long-acyl AHLs. The HSL was subsequently delactonized to form homoserine, which was then consumed (Fig. 4). Since the culture pH was well controlled in order to preclude chemical hydrolysis of the lactone ring, the observed HSL degradation was due to a biological, and not an abiotic, hydrolysis event. Enzymes with HSL lactonase activity have been found in fungal and mammalian biota (14, 16). To our knowledge, this is the first reported demonstration of an HSL lactonase activity in bacteria. Curiously, neither HSL nor homoserine was used as an energy or nitrogen source by P. aeruginosa, as they are by Variovorax and Arthrobacter species (9, 21). It is possible that HSL lactonase- and homoserine-degrading activities serve as detoxification mechanisms, since both of these compounds are known to be toxic to diverse biota (9, 13, 14, 45).
There are notable differences between AHL utilization by the pseudomonads and V. paradoxus and Ralstonia strain XJ12B. Ralstonia was reported to degrade and grow equally rapidly with short- and long-acyl AHLs, exhibiting doubling times of 8.5 to 10.5 h on 3OC12HSL and C4HSL, respectively (23). V. paradoxus was reported to utilize the entire spectrum of short- and long-acyl AHLs tested, growing most rapidly by using 3OC6HSL with a doubling time of 3.5 h and with molar growth yields that corresponded well with the acyl length of the given AHL (21). The pseudomonads examined in this study, however, did not degrade AHLs with acyl side chains shorter than eight carbons, and no correspondence between molar growth yields and AHL acyl side chain lengths was observed (Table 1). Curiously, such a correspondence was observed when the cells were grown with long-chain fatty acids.
AHL utilization by P. aeruginosa exhibited another key difference from Variovorax, Ralstonia, and even Pseudomonas strain PAI-A. When cultures not previously grown on AHL were inoculated into media containing long-acyl AHL, it generally took 1 to 3 weeks before logarithmic growth commenced. No such lag was observed when AHL-grown cells were subcultured into such media. This adaptation does not appear to reflect a stable mutation, as long lags returned if the subculturing process was punctuated with a transfer into or onto media containing a different energy substrate. The long initial lag time suggests that AHL degradation by P. aeruginosa is not immediately induced by the quorum signal and is not controlled as a function of the catabolic needs of the cell or by cell starvation.
Recently, an acylase from a Ralstonia sp., AiiD, that inactivates both long- and short-chain AHLs was described (23). Heterologous expression in E. coli of pvdQ, the closest homologue of the Ralstonia acylase gene carried by P. aeruginosa, conferred to E. coli AHL acylase activity specific towards long-acyl-chain but not short-acyl-chain AHLs (Fig. 5). Expression of pvdQ in P. aeruginosa is known to be well regulated. Under the monikers of QSC 112a and QSC 112b, pvdQ was identified as being a late responder to the 3OC12HSL quorum-sensing circuit (43), although gene microarray studies have not provided further support for this observation (37, 42). pvdQ has also been found to be iron regulated (Fur repressed) and appears to be involved in pyoverdine biosynthesis based on evidence from both microarray and mutagenesis studies (17, 29). Because of its complicated control, we wished to examine the effects of the constitutive expression of plasmid-encoded pvdQ in strain PAO1. Remarkably, in striking contrast to the wild type, which produced micromolar amounts of this quorum signal 3OC12HSL when grown in rich medium, P. aeruginosa constitutively expressing pvdQ did not accumulate 3OC12HSL (Fig. 6).
Surprisingly, two pvdQ knockout mutants were able to grow with 3OC12HSL as a sole energy source. This suggests that another enzyme must confer the AHL growth phenotype in strain PAO1. Although some contribution of pvdQ to AHL utilization cannot be ruled out, it seems more likely that this protein is, as has previously been suggested, involved in a yet to be fully described editing reaction during the maturation of the pyoverdine siderophore (17, 29). It remains possible, however, that 3OC12HSL is subject to inadvertent biochemical degradation by pvdQ during times of pyoverdine expression. In addition to PvdQ, the genome of P. aeruginosa encodes three other N-terminal nucleophile hydrolase homologues. It remains to be determined whether the AHL growth phenotype is conferred by one of these or another protein that is not homologous with them.
A. tumefaciens has been shown to degrade 3OC8HSL, its AHL quorum signal, during early stationary phase (46). That P. aeruginosa can degrade one, but not the other, of its two AHL quorum signals has revealed a novel AHL degradation apparatus. Signal decay, in addition to providing utilizable nutrients, may play a role in the regulation of the LasR/LasI/3OC12HSL-controlled quorum-sensing regulon. It has been noted that the two principal AHL quorum signals of P. aeruginosa, C4HSL and 3OC12HSL, are present in the sputum of cystic fibrosis patients and in laboratory biofilms at ratios quite different from those encountered in planktonic, liquid-grown cultures. Sputum and biofilm samples were found to contain significantly higher levels of C4HSL with respect to 3OC12HSL, and this could be a reflection of the biochemical turnover of the latter. In addition to examining this possibility, we hope to identify the loci encoding the AHL acylase and HSL lactonase enzymes involved during AHL-dependent growth and to reveal the regulatory details controlling signal decay by Pseudomonas strain PAI-A and P. aeruginosa PAO1 under diverse cultivation conditions.
Acknowledgments
This research was supported by the National Science Foundation (DBI-0107908), the Department of Agriculture (CSREES 2001-01242), the Defense Advanced Research Projects Agency (DARPA; no. N66001-02-1-8929), the W. M. Keck Foundation Fund for Discovery in Basic Medical Research at the California Institute of Technology, and an NIH Training Grant for Biology (5T32GM07616).
Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the DARPA.
We thank Bob Becker and Nathan Dalleska for technical discussions and for help with performing LC/APCI-MS analyses and C. Saltikov, D. Newman, and E. R. Leadbetter for their helpful comments.
REFERENCES
- 1.Anzai, Y., H. Kim, J. Y. Park, H. Wakabayashi, and H. Oyaizu. 2000. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50:1563-1589. [DOI] [PubMed] [Google Scholar]
- 2.Anzai, Y., Y. Kudo, and H. Oyaizu. 1997. The phylogeny of the genera Chryseomonas, Flavimonas, and Pseudomonas supports synonymy of these three genera. Int. J. Syst. Bacteriol. 47:249-251. [DOI] [PubMed] [Google Scholar]
- 3.Bennasar, A., R. Rossello-Mora, J. Lalucat, and E. R. Moore. 1996. 16S rRNA gene sequence analysis relative to genomovars of Pseudomonas stutzeri and proposal of Pseudomonas balearica sp. nov. Int. J. Syst. Bacteriol. 46:200-205. [DOI] [PubMed] [Google Scholar]
- 4.Cronin, C. N., and W. S. McIntire. 1999. pUCP-Nco and pUCP-Nde: Escherichia-Pseudomonas shuttle vectors for recombinant protein expression in Pseudomonas. Anal. Biochem. 272:112-115. [DOI] [PubMed] [Google Scholar]
- 5.Domenech Fernandez-Garayzabal, J. F., P. Lawson, J. A. Garcia, M. T. Cutuli, M. Blanco, A. Gibello, M. A. Moreno, M. D. Collins, and L. Dominguez. 1997. Winter disease outbreak in sea-bream (Sparus aurata) associated with Pseudomonas anguilliseptica infection. Aquaculture 156:317-326. [Google Scholar]
- 6.Dong, Y. H., A. R. Gusti, Q. Zhang, J. L. Xu, and L. H. Zhang. 2002. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 68:1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong, Y. H., L. H. Wang, J. L. Xu, H. B. Zhang, X. F. Zhang, and L. H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 8.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flagan, S., W.-K. Ching, and J. R. Leadbetter. 2003. Arthrobacter strain VAI-A utilizes acyl-homoserine lactone inactivation products and stimulates quorum signal biodegradation by Variovorax paradoxus. Appl. Environ. Microbiol. 69:909-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank, L., and R. DeMoss. 1959. On the synthesis of pyocyanine. J. Bacteriol. 77:776-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 12.Hanzelka, B., M. R. Parsek, D. L. Val, P. V. Dunlap, J. J. E. Cronan, and E. P. Greenberg. 1999. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J. Bacteriol. 181:5766-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakubowski, H. 1997. Aminoacyl thioester chemistry of class II aminoacyl-tRNA synthetases. Biochemistry 36:11077-11085. [DOI] [PubMed] [Google Scholar]
- 14.Jakubowski, H. 2000. Calcium-dependent human serum homocysteine thiolactone hydrolase. A protective mechanism against protein N-homocysteinylation. J. Biol. Chem. 275:3957-3962. [DOI] [PubMed] [Google Scholar]
- 15.Jakubowski, H. 1995. Proofreading in vivo. Editing of homocysteine by aminoacyl-tRNA synthetases in Escherichia coli. J. Biol. Chem. 270:17672-17673. [PubMed] [Google Scholar]
- 16.Kobayashi, M., M. Shinohara, C. Sakoh, M. Kataoka, and S. Shimizu. 1998. Lactone-ring-cleaving enzyme: genetic analysis, novel RNA editing, and evolutionary implications. Proc. Natl. Acad. Sci. USA 95:12787-12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamont, I. L., and L. W. Martin. 2003. Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology 149:833-842. [DOI] [PubMed] [Google Scholar]
- 18.Lane, D. J. 2001. 16S/23S rRNA sequencing. In E. Stackebrandt (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 19.Leadbetter, J. R. 2001. Plant microbiology. Quieting the raucous crowd. Nature 411:748-749. [DOI] [PubMed] [Google Scholar]
- 20.Leadbetter, J. R., and J. A. Breznak. 1996. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl. Environ. Microbiol. 62:3620-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, S. J., S. Y. Park, J. J. Lee, D. Y. Yum, B. T. Koo, and J. K. Lee. 2002. Genes encoding the N-acyl homoserine lactone-degrading enzyme are widespread in many subspecies of Bacillus thuringiensis. Appl. Environ. Microbiol. 68:3919-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, Y.-H., J.-L. Xu, J. Hu, L.-H. Wang, S. L. Ong, J. R. Leadbetter, and L.-H. Zhang. 2003. Acyl-homoserine lactone acylase from Ralstonia str. XJ12B represents a novel and potent class of quorum quenching enzymes. Mol. Microbiol. 47:849-860. [DOI] [PubMed] [Google Scholar]
- 24.Mikkat, S., E. A. Galinski, G. Berg, A. Minkwitz, and A. Schoor. 2000. Salt adaptation in pseudomonads: characterization of glucosylglycerol-synthesizing isolates from brackish coastal waters and the rhizosphere. Syst. Appl. Microbiol. 23:31-40. [DOI] [PubMed] [Google Scholar]
- 25.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 26.Moore, E. R. B., M. Mau, A. Arnscheidt, E. C. Boettger, R. A. Hutson, M. D. Collins, Y. Van de Peer, R. De Wachter, and K. N. Timmis. 1996. The determination and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships. Syst. Appl. Microbiol. 19:478-492. [Google Scholar]
- 27.Moré, M. I., D. Finger, J. L. Stryker, C. Fuqua, A. Eberhard, and S. C. Winans. 1996. Enzymatic synthesis of a quorum-sensing autoinducer through the use of defined substrates. Science 272:1655-1658. [DOI] [PubMed] [Google Scholar]
- 28.Mueller, J. G., R. Devereux, D. L. Santavy, S. E. Lantz, S. G. Willis, and P. H. Pritchard. 1997. Phylogenetic and physiological comparisons of PAH-degrading bacteria from geographically diverse soils. Antonie Leeuwenhoek 71:329-343. [DOI] [PubMed] [Google Scholar]
- 29.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 30.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 31.Parsek, M. R., D. L. Val, B. L. Hanzelka, J. E. Cronan, and E. P. Greenberg. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 96:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reimmann, C., N. Ginet, L. Michel, C. Keel, P. Michaux, V. Krishnapillai, M. Zala, K. Heurlier, K. Triandafillu, H. Harms, G. Defago, and D. Haas. 2002. Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology 148:923-932. [DOI] [PubMed] [Google Scholar]
- 33.Schaefer, A. L., E. P. Greenberg, and M. R. Parsek. 2001. Acylated homoserine lactone detection in Pseudomonas aeruginosa biofilms by radiolabel assay. Methods Enzymol. 336:41-47. [DOI] [PubMed] [Google Scholar]
- 34.Schaefer, A. L., B. L. Hanzelka, M. R. Parsek, and E. P. Greenberg. 2000. Detection, purification and structural elucidation of acylhomoserine lactone inducer of Vibrio fischeri luminescence and other related molecules. Methods Enzymol. 305:288-301. [DOI] [PubMed] [Google Scholar]
- 35.Schaefer, A. L., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1996. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc. Natl. Acad. Sci. USA 93:9505-9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 37.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 39.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 40.Van Delden, C., and B. H. Iglewski. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voelkert, E., and D. R. Grant. 1970. Determination of homoserine as the lactone. Anal. Biochem. 34:131-137. [DOI] [PubMed] [Google Scholar]
- 42.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wischnak, C., F. E. Loffler, J. Li, J. W. Urbance, and R. Muller. 1998. Pseudomonas sp. strain 273, an aerobic α, ω-dichloroalkane degrading bacterium. Appl. Environ. Microbiol. 64:3507-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zakataeva, N. P., V. V. Aleshin, I. L. Tokmakova, P. V. Troshin, and V. A. Livshits. 1999. The novel transmembrane Escherichia coli proteins involved in the amino acid efflux. FEBS Lett. 452:228-232. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, H. B., L. H. Wang, and L. H. Zhang. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]