Abstract
Vibrio vulnificus is an estuarine bacterium capable of causing rapidly fatal infections through both ingestion and wound infection. Like other opportunistic pathogens, V. vulnificus must adapt to potentially stressful environmental changes while living freely in seawater, upon colonization of the oyster gut, and upon infection of such diverse hosts as humans and eels. In order to begin to understand the ability of V. vulnificus to respond to such stresses, we examined the role of the alternate sigma factor RpoS, which is important in stress response and virulence in many pathogens. An rpoS mutant of V. vulnificus strain C7184o was constructed by homologous recombination. The mutant strain exhibited a decreased ability to survive diverse environmental stresses, including exposure to hydrogen peroxide, hyperosmolarity, and acidic conditions. The most striking difference was a high sensitivity of the mutant to hydrogen peroxide. Albuminase, caseinase, and elastase activity were detected in the wild type but not in the mutant strain, and an additional two hydrolytic activities (collagenase and gelatinase) were reduced in the mutant strain compared to the wild type. Additionally, the motility of the rpoS mutant was severely diminished. Overall, these studies suggest that rpoS in V. vulnificus is important for adaptation to environmental changes and may have a role in virulence.
The gram-negative bacterium Vibrio vulnificus is one of the most invasive and rapidly fatal human pathogens, causing three distinct syndromes of infection: primary septicemia through ingestion of raw or undercooked shellfish (primarily oysters), wound infection by exposure of a preexisting wound to seawater or shellfish, and gastroenteritis (35). Primary septicemia results in a high fatality rate of ca. 60% but occurs primarily in individuals who are immunocompromised or have a chronic liver disease (for recent reviews, see references 20 and 46). V. vulnificus is widely distributed in aquatic environments and has been isolated from seawater, sediment, plankton, animals, fish, and bivalves primarily along the eastern and Gulf coasts of the United States. This bacterium has also been found in various other countries, including Brazil, Korea, and several European countries (35).
V. vulnificus encounters numerous stresses in its natural environment and upon entry into the human host. The stresses include both high and low temperatures, periods of nutrient starvation, osmotic changes, and oxidative stress. Bacteria react to such environmental perturbations by the production of stress proteins which guarantee the continued viability of the bacteria under otherwise deleterious conditions. In Escherichia coli, the rpoS-encoded sigma factor, RpoS (σs), is a central regulator of many stationary-phase-responsive genes, as well as genes involved in adaptation to diverse types of stresses. These stresses include starvation, osmotic stress, low pH, nonoptimal high or low temperatures, and oxidative damage (9). In general, RpoS appears to be present in the gamma branch of proteobacteria and functions as a general stress regulator (9, 49). In Vibrio cholerae, RpoS plays a crucial role in survival under a variety of stressful situations, including exposure to H2O2, hyperosmotic stress, and nutrient deprivation (50). In contrast, RpoS in Vibrio harveyi does not appear to have a role in surviving oxidative or hyperosmotic challenge, but it does function in surviving ethanol stress and persisting during the stationary phase of growth (19). In addition to its role in stress adaptation, RpoS is known to control the expression of various virulence genes (4, 5, 8, 17) and is involved in colonization and adhesion to host tissue (24, 37, 40). A recent publication reported increased mRNA levels of Pseudomonas aeruginosa rpoS in chronically infected cystic fibrosis patients (6), suggesting an upregulation of rpoS upon entry into the human host and therefore a role during infection.
With V. vulnificus, an indication that adaptation to environmental stress may be important was our finding that cells encountering an osmotic upshift respond by becoming resistant to potentially lethal temperatures (both high and low), to osmotic and ionic shock, and to oxidative stress (44). Furthermore, osmotic shock- or nutrient-stressed cells of V. vulnificus resist the bactericidal effects of human serum (34). Thus, the regular environmental stresses that V. vulnificus encounters appear to result in cells which are more able to survive these adverse environments and may also make the cells more able to initiate human infections.
V. vulnificus has been shown to produce many extracellular products which could help in adaptation to environmental changes. V. vulnificus expresses capsule, produces both hydroxamate and phenolate siderophores, and secretes macromolecule-degrading enzymes, such as hemolysin, protease (vvp), hyaluronidase, elastase (vvpE), chondroitin sulfatase, phospholipase, and mucinase (20). These proteolytic activities have been shown to increase vascular permeability, cause hemorrhagic damage, and facilitate the acquisition of iron by digesting various proteins, such as heme proteins (30, 31, 33). Interestingly, the expression of vvp seems to be controlled by SmcR, the V. harveyi LuxR homologue in V. vulnificus, and is thereby linked to the quorum-sensing (cell-density-dependent) regulatory system, although the regulatory mechanism (positively or negatively regulated) is still unclear (23, 43). Similarly, the expression of the hemagglutinin (HA)/protease gene hap in V. cholerae is positively controlled by the LuxR homologue, HapR (14). In addition, the expression of hap and/or its secretion is controlled by RpoS, as an rpoS-deficient mutant lacks HA/protease activity in its supernatant (49). Furthermore, in V. vulnificus, expression of the elastase gene, vvpE, is decreased 10-fold in an rpoS mutant compared to the parent strain (12).
In order to begin to study the role of rpoS in the adaptation of V. vulnificus to environmental changes, we constructed an rpoS mutant strain by homologous recombination. Phenotypic evaluation indicated that the rpoS mutant is more susceptible than the parent strain to many stressful environmental changes. Moreover, activity of five of the nine exoenzymes tested was abolished or reduced in the mutant. Finally, the motility of the rpoS mutant was dramatically reduced. Taken together, our data indicate that the alternate sigma factor, RpoS, is important for the adaptation of V. vulnificus to the highly variable environmental conditions that the bacterium is likely to face in seawater, oyster gut, eels, and humans.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are described in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| V. vulnificus | ||
| C7184o | Human wound isolate | CDCa |
| AH1 | C7184o rpoS::pHA4; rpoS deficient | This study |
| AH1(pJAK13) | AH1 with vector control | This study |
| AH1(pTR3) | AH1 with rpoS on pTR3; rpoS positive | This study |
| E. coli | ||
| SM10λpir | thi thr tonA lacY supE recA::RP4-2-Te::Mu Kmr | 44 |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80 lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(araA-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| Plasmids | ||
| pCRII Topo | PCR product cloning vector, Apr Kmr | Invitrogen |
| pNQ705 | R6K γ ori, oriT of RP4, Cmr | 29 |
| pHA3 | 550-bp rpoS fragment on pCRII Topo | This study |
| pHA4 | 600-bp pHA3 fragment as XbaI/SstI on pNQ705 | This study |
| pJAK13 | IncQ, LacI+, ptac, mobilized by RP4, Smr | D. Figurski |
| pTR1 | pCRII Topo with promoterless rpoS | This study |
| pTR3 | pJAK13-rpoS, inducible RpoS expression | This study |
CDC, Centers for Disease Control and Prevention.
Media and growth conditions.
V. vulnificus cells were grown in heart infusion (HI) broth (Difco, Detroit, Mich.) (pH 7.0) at 22°C with agitation. For cultivation of the rpoS mutant (AH1), chloramphenicol was added to the growth medium at 15 μg/ml. E. coli cells were typically grown in Luria-Bertani (LB) broth (26) at 22°C with shaking. The semisolid motility agar plates (M agar) contained 10 g of tryptone per liter, 20 g of NaCl per liter, and 3.25 g of agar per liter (16). If required for plasmid selection, kanamycin (50 μg/ml), ampicillin (75 μg/ml), chloramphenicol (15 μg/ml), or streptomycin (50 μg/ml) was added. In order to induce ptac expression, plates were supplemented with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
Standard DNA methods.
Genomic DNA from V. vulnificus was isolated using the Qiagen Genomic-tip system (Qiagen, Valencia, Calif.). Plasmids were prepared from E. coli using the Qiagen Plasmid Mini kit. Digestion by endonucleases, ligation reactions, and PCR were performed by standard procedures (41).
Gene cloning.
A 550-bp PCR fragment of the rpoS-coding region was generated using V. vulnificus C7184o cells as a template, Taq polymerase (Promega, Madison, Wis.), and oligonucleotides (AH03 [5′ TGC CGA AGA AGA AGT] and AH04 [5′ TCT TTG ACG ACA TGA AT]) (Bio-Synthesis, Inc., Lewisville, Tex.) derived from the conserved regions of RpoS proteins in other bacteria. This rpoS fragment lacks ca. 220 and 264 bp at the 5′ and 3′ ends, respectively, of the putative open reading frame. Amplification of part of the V. vulnificus rpoS gene was verified by sequencing (Retrogene, San Diego, Calif.). Protein homology searches were done with BLAST (7), and multiple alignments were performed using CLUSTAL X (11). The amplified fragment was ligated into cloning vector pCRII Topo using the Topo TA cloning kit (Invitrogen, Carlsbad, Calif.). Cells of E. coli strain Top10 were transformed with the ligation mixture, and ampicillin-resistant transformants were screened for the presence of an rpoS fragment, yielding plasmid pHA3.
Construction of an rpoS mutant.
An rpoS mutant of V. vulnificus strain C7184o was constructed by inserting a suicide vector into the chromosome as originally described by Miller and Mekalanos (27). Briefly, plasmid pHA3 was digested with the restriction endonucleases XbaI and SstI, and the rpoS fragment was ligated to the suicide vector pNQ705 (28) that had been linearized with the same enzymes and dephosphorylated. E. coli SM10λpir cells were transformed with the ligation mixture, and chloramphenicol-resistant transformants were tested for the presence of the correct fragment by restriction analysis, resulting in plasmid pHA4. E. coli SM10λpir (donor) was used for mobilizing the suicide construct pHA4 into V. vulnificus C7184o (recipient). Cultures of donor and recipient grown overnight were washed in LB broth. Recipient cells were incubated at 45°C for 15 min and mixed with donor cells at a 1:3 ratio (vol/vol). Aliquots (100 μl) were spotted onto a nitrocellulose filter (0.45-μm pore size; Millipore, Bedford, Mass.) on an LB plate and incubated overnight at 22°C. The filter was transferred to a microcentrifuge tube containing LB broth to wash the cells. Chloramphenicol-resistant transconjugants carrying the mobilized plasmid integrated into the chromosome by homologous recombination were selected on LB agar plates containing chloramphenicol at 2 μg/ml and polymyxin B at 100 U/ml. The integral mutation was subsequently verified by Southern blot analysis, yielding mutant strain AH1. For Southern blot hybridization, the 550-bp rpoS fragment amplified with oligonucleotides AH03 and AH04 was labeled using a digoxigenin-dUTP DNA label and detection kit (Roche Diagnostics GmbH, Mannheim, Germany) and was subsequently used as a probe in Southern hybridization. Hybridization was performed according to the manufacturer's manual.
Construction of RpoS-complementing plasmid and strain.
An approximately 1-kb amplicon containing a promoterless copy of the V. vulnificus rpoS gene was generated using V. vulnificus strain C7184o genomic DNA as a template, Ex-Taq DNA polymerase (PanVera, Madison, Wis.) and the oligonucleotide primers Rpos up (5′-TGC TAA CTC GCC ATG GGG AG-3′) and Rpos dn (5′-GCC AAG CGT TAG AAG CCC TA-3′) (Bio-Synthesis, Inc.). The genomic sequence of V. vulnificus strain CMCP6 (GenBank accession no. AE016795) was used to generate the primers. The nucleotide sequence of the 550-bp fragment of rpoS used in the mutant matched exactly that segment of the rpoS gene from CMCP6 (data not shown). This amplicon was ligated into the cloning vector pCRII Topo. Cells of the E. coli strain TOP10 were transformed with the ligation mixture, and kanamycin-resistant transformants were screened for the presence of the rpoS gene, yielding plasmid pTR1. The rpoS gene was cloned into the IncQ, broad-host-range expression vector pJAK13 (a gift from D. Figurski), allowing inducible expression of the gene from the ptac promoter. A fragment containing the rpoS gene was obtained by digestion of pTR1 with KpnI and XbaI. This fragment was ligated into XbaI-KpnI-digested pJAK13. E. coli strain SM10λpir was transformed with the ligation mixture, and streptomycin-resistant transformants were screened for the presence of the rpoS gene, yielding pTR3. E. coli SM10λpir was used as the donor to transfer pTR3 into the V. vulnificus rpoS mutant strain AH1 as described above, with selection of the transconjugants on media containing chloramphenicol (2 μg/ml), streptomycin (50 μg/ml), and polymyxin B (100 U/ml). Transconjugants were confirmed to harbor pTR3 using restriction digestion, yielding the RpoS-complementing strain, V. vulnificus AH1(pTR3).
Catalase assays.
Dialyzed cell extracts were prepared as previously described (32). Protein concentrations were determined by the method of Lowry et al. (21) using bovine serum albumin as a standard. Catalase activity in cell extracts were determined by the method of Beers et al. (3).
Survival assays.
To test cells in the stationary growth phase, cultures were grown overnight in HI broth, centrifuged, washed in artificial seawater (ASW), and resuspended in ASW to provide ca. 108 CFU/ml. An aliquot (100 μl) was taken, added to an appropriate medium (900 μl) to give the test conditions described below, and incubated at 22°C. At indicated time points, an aliquot was withdrawn to determine the survival rate as indicated by plate counts.
Test conditions were oxidative challenge (2 mM H2O2 in ASW), osmotic challenge (2.7 M NaCl in ASW), ethanol challenge (10 and 12.5% ethanol in ASW), heat challenge (42°C in HI broth), acidic challenge (HI broth adjusted with HCl to pH 4), and starvation (ASW). In most experiments, ASW was used as a diluent, resulting in an osmotic upshift and nutrient deprivation. To ensure that this challenging condition was not responsible for the differences observed during the various tests, we observed survival in ASW over a time period of 60 min. No statistical difference was observed between the mutant and parent strains; the average survival rate was 70% (data not shown).
Values are the means of duplicate samples from a typical experiment, and each experiment was performed at least twice. Error bars represent standard errors of the means.
Exoenzyme expression.
Cultures were grown overnight in HI broth, and the presence of several exoenzymes was examined as described previously (36). Briefly, the activities of various enzymes were determined following inoculation of cultures onto HI agar to which the following substrates had been added: 0.2% hide powder azure (Sigma, St. Louis, Mo.) for collagenase, 1% gelatin for gelatinase, 1% Tween 80 for lipase, 1% porcine stomach mucin type II (Sigma) for mucinase, 5% sheep red blood cells for hemolysin, and 10 mg of bovine serum albumin (Sigma) per ml for albuminase. In addition, caseinase activity was observed on tryptic soy agar containing 1% skim milk, and phospholipase (lecithinase) on McClung Toabe agar (Difco). A spectrophotometric assay, monitoring release of dye from elastin-Congo red (Sigma) following incubation with test cells, was employed to assay elastase activity (36). In the plate assays, clearing zones (if appropriate, following addition of coagulants) were measured and compared with appropriate controls. All experiments were performed in duplicate and repeated at least once.
To determine exoprotease activity as a function of growth, cells were cultured in HI broth at 37°C and samples were taken at different time points. Cell-free supernatants were prepared and stored at −80°C. Elastase activity was determined as described above using elastin-Congo red (Sigma). Collagenase activity (1) was assayed using hide powder azure (Sigma). For caseinase activity, azocasein (Sigma) was used as described by Swift et al. (48). All experiments were performed in duplicate and repeated at least once.
Complementation.
The rpoS mutant was complemented in trans using the caseinase assay described above. Cultures of V. vulnificus strains C7184o, AH1, AH1(pJAK13), and AH1(pTR3) were grown overnight to saturation in HI broth at 22°C with antibiotics supplemented as appropriate. Ten-microliter aliquots of the culture were added to tryptic soy agar containing 1% skim milk (with 1 mM IPTG added to induce rpoS expression) to measure caseinase activity. Plates were grown over several days at either 22 or 37°C. Caseinase activity was determined by measuring the clearing zones formed around the colony.
Motility.
Cultures of V. vulnificus strains C7184o, AH1, AH1(pJAK13), and AH1(pTR3) were grown to stationary phase in HI broth at 22°C with antibiotics if appropriate. Two-microliter aliquots of the culture were added to M agar containing 1 mM IPTG. Plates were incubated for 20 h at 22°C. Motility was measured by the size of the growth ring.
Nucleotide sequence accession number.
The nucleotide sequence of the portion of the rpoS gene determined in this study has been deposited in the GenBank database under accession no. AY163815.
RESULTS
Construction of an rpoS mutant.
A 550-bp fragment of the rpoS gene in V. vulnificus was amplified and sequenced. The deduced amino acid sequence had 98, 98, 93, and 82% sequence identity values to the corresponding sequence of homologous proteins in V. harveyi (accession no. AF321124), Vibrio parahaemolyticus (accession no. AF144608), V. cholerae (accession no. AF000945), and E. coli (accession no. AF275947), respectively. The rpoS fragment was cloned into plasmid pCRII, yielding pHA3. Using the restriction sites within the polylinker of this vector, the insert was cut out of the vector and cloned into the suicide vector pNQ705, yielding pHA4. A V. vulnificus rpoS mutant strain was constructed by inserting the mobilized vector pHA4 into the chromosomal copy of rpoS by homologous recombination. Southern blot analysis was performed to verify the correct insertion (data not shown). Complementation of the rpoS mutation was seen when an RpoS expression plasmid was present in trans (see Fig. 3 and the caseinase activity described below under “Exoenzyme activity”; also data not shown). These findings, coupled with the observation that the next predicted gene after rpoS in the V. vulnificus genome is mutS, which is transcribed in the opposite direction, indicates that the insertion within the rpoS gene did not have a polar effect.
FIG. 3.
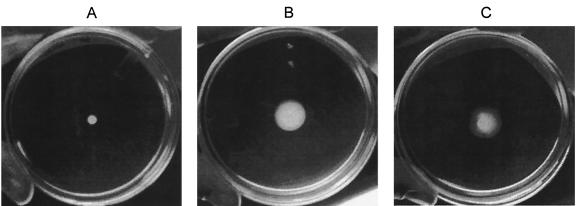
Motility of V. vulnificus wild-type and mutant strains. Cultures were grown to stationary phase at 22°C. Aliquots were plated on M agar plates and incubated for 20 h at 22°C. V. vulnificus AH1(pJAK13) (A), C7184o (B), and AH1(pTR3) (C) are shown.
Function of RpoS in stress survival.
Other investigators have shown that rpoS mutants of various enteric bacteria and various Pseudomonas species are more susceptible to a variety of challenging conditions that are believed to represent stressful environmental changes (2, 15, 18, 19, 22, 38, 49). According to these studies, we would expect the V. vulnificus mutant to be more sensitive to acidic, hyperosmotic, and oxidative conditions, among others. To test the overall fitness of the mutant in comparison with the parent strain, we cultivated both strains in HI broth at 22°C. Similar growth characteristics were observed with generation times of 43 and 48 min for V. vulnificus C7184o and AH1, respectively (data not shown).
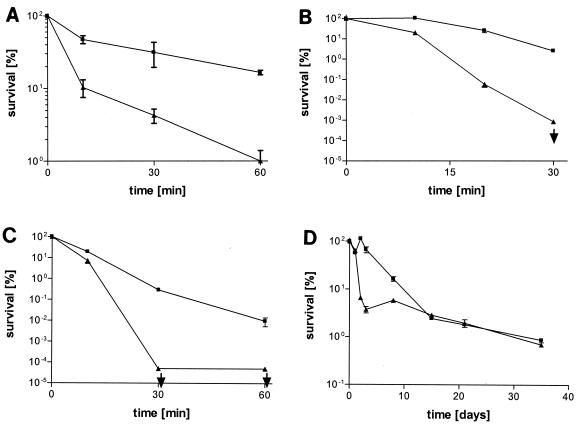
To study the role of RpoS in surviving diverse environmental stresses, V. vulnificus C7184o and AH1 cells from stationary-phase cultures were exposed to various challenges, and the survival rates were determined by plate counts. Over a time period of 60 min, AH1 cells were significantly less resistant to 2.7 M NaCl in ASW than were C7184o cells (Fig. 1A). In acidic conditions (HI broth, pH 4.0), RpoS enhanced survival markedly, as the ability to culture AH1 cells rapidly dropped below the detection limit, while C7184o cells remained at high levels (Fig. 1B). AH1 cells were much more sensitive to H2O2 than their wild-type counterparts (Fig. 1C). To mimic nutrient deprivation in a natural estuarine environment, microcosms of both strains in nutrient-free ASW were prepared. During the initial phases of starvation, the survival of AH1 cells was greatly reduced compared to the survival of C7184o. However, at later time points and throughout the duration of the study, the remaining population was not dramatically different from the parent strain population (Fig. 1D). When exposed to 12.5% ethanol, the mutant seemed to be only slightly more susceptible than the parent strain (data not shown). Both the parent strain and rpoS mutant survived 10% ethanol equally well but did not survive concentrations higher than 12.5% (data not shown). Both strains showed a similar decline in plate counts when exposed to a classic heat shock procedure (42°C after growth at 30°C [data not shown]).
FIG. 1.
Stress survival of stationary-phase V. vulnificus cells. Bacteria of the wild-type strain C7184o (▪) and rpoS::pHA4 strain AH1 (▴) were tested by incubating the cultures in 2.7 M NaCl in ASW (A), at pH 4 in HI broth (B), in 2 mM H2O2 in ASW (C), and under long-term starvation conditions in ASW (D). All experiments were conducted at 22°C. The detection limits of the ability to culture the bacteria are indicated by the arrows. Experiments were done in duplicate (at least) and repeated at least once. Values show the results of a typical experiment done in duplicate, and bars represent standard errors of the means.
Catalase activity.
As RpoS in E. coli is known to function in the oxidative stress response and our survival experiments suggested a major role for RpoS during H2O2 exposure, we monitored catalase activity in V. vulnificus cells. Results are shown in Table 2. Both the parent strain and the rpoS mutant strain possess an insignificant amount of catalase activity (0.2 and 0.3 U/mg of cell mass, respectively) during logarithmic phase. The catalase activity for wild-type cells in stationary phase and in exponentially growing cells that were exposed to 50 μM H2O2 increased to 4.1 and 5.8 U/mg, respectively. Catalase activity in the rpoS mutant was increased during stationary growth and after induction of logarithmic cells but was 37% and 28% less than that of the parent strain. The loss of catalase activity in the rpoS mutant is not nearly as dramatic as the decline in survivability when exposed to 2 mM H2O2 (Fig. 1C).
TABLE 2.
Catalase activity in V. vulnificus wild-type strain C7184o and rpoS mutant strain AH1
| Strain | Growth phase | Catalase activity after H2O2 treatmenta | Catalase activity (U/mg of protein in cells) |
|---|---|---|---|
| C7184o | Logarithmic | − | 0.2 |
| Logarithmic | + | 5.8 | |
| Stationary | − | 4.1 | |
| AH1 | Logarithmic | − | 0.3 |
| Logarithmic | + | 4.2 | |
| Stationary | − | 2.6 |
50 μM H2O2 was added to the cells at early logarithmic growth phase, and the culture was incubated for an additional 1 h. The presence (+) or absence (−) of catalase activity is shown.
Exoenzyme activity.
V. vulnificus produces and secretes a variety of macromolecule-degrading proteins that likely contribute to the survival of this organism in a variety of environmental conditions. Such enzymes include a metalloprotease, hemolysin, collagenase, and lipase. We investigated the ability of the V. vulnificus wild-type strain C7184o and the rpoS mutant strain to degrade nine different macromolecules. After the bacteria were grown overnight at 37°C, the wild-type strain showed a clearing zone around the colony of 3.0 ± 0.4 mm, indicative of caseinase activity. Neither the rpoS mutant nor the mutant with a vector control showed activity (0.2 ± 0.3 and 0.4 ± 0.6 mm, respectively). With the RpoS expression plasmid in trans, the clearing zone was comparable to that of the wild type (5.5 ± 0.1 mm), indicating a restoration of caseinase activity. The results from the remaining exoenzyme activity tests are summarized in Table 3. The wild-type strain gave positive results for all nine exoenzyme activities. The mutant was negative for three enzymes (albuminase, caseinase, and elastase). Additionally, clearing zones produced by collagenase and gelatinase activity were 35 and 40% smaller, respectively, than those of the wild type.
TABLE 3.
Expression of virulence factors in V. vulnificus wild-type strain C7184o and rpoS mutant strain AH1
| Virulence factor | Expressiona of virulence factor by strain:
|
|
|---|---|---|
| C7184o | AH1 | |
| Albuminase | + | − |
| Caseinase | + | − |
| Collagenase | + | +/− |
| Elastase | + | − |
| Gelatinase | + | +/− |
| Hemolysin | + (α) | + (α) |
| Lipase | + | + |
| Mucinase | + | + |
| Phospholipase | + | + |
Symbols: +, expressed; −, not expressed; +/−, activity was 35 to 40% less than the activity in the wild type on the basis of clearing zone size; + (α), alpha hemolysis.
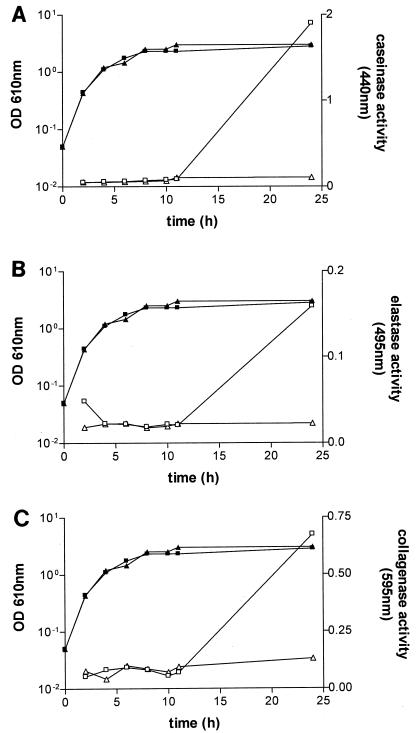
To investigate these differences in more detail, cultures of V. vulnificus C7184o and AH1 were grown and samples were taken periodically to determine cell density and caseinase, elastase, and collagenase activity of cell-free supernatants. These results are shown in Fig. 2. Exoenzyme activities in the wild type increased dramatically in the late stationary phase of growth. In fact, at earlier time points, enzyme activities were at the lower limit of detection and increased only slightly within the first 11 h. Similar results were observed for albuminase activity (data not shown). Almost no substrate degradation was detected for strain AH1, especially for azocasein (Fig. 2A) and elastin-Congo red (Fig. 2B). Only during incubation of cell-free supernatants of the mutant with hide powder azure (collagenase activity [Fig. 2C]) was some degradation measured (optical density at 610 nm of 0.13 for late-stationary-phase cells). Virtually no degradation was observed for azoalbumin (data not shown). These data confirm the plate results described above and show that RpoS is required for these exoenzyme activities.
FIG. 2.
Exoprotease activity of wild-type strain C7184o and rpoS::pHA4 strain AH1 of V. vulnificus. Cultures of C7184o (▪) and AH1 (▴) were grown in HI broth at 37°C with shaking. At various time points, aliquots were taken, and cell-free supernatants were prepared. Exoprotease activity in C7184o (□) and AH1 (▵) supernatants were determined. Graphs show degradation of azocasein (A), elastin-Congo red (B), and hide powder azure (C) at 37°C. Experiments were repeated at least once; shown is a typical result.
Motility.
The role of RpoS in the motility of V. vulnificus is demonstrated in Fig. 3. Aliquots of stationary-phase cultures were plated on M agar and incubated for 20 h at 22°C. The rpoS mutant strain did not move from the original inoculum spot (Fig. 3A). The wild-type strain C7184o showed typical movement after 20 h (Fig. 3B). The mutant containing the complementing plasmid, pTR3, appeared to be nearly as motile as the wild-type strain (Fig. 3C). These results indicate that RpoS is required for motility of stationary-phase V. vulnificus cells.
DISCUSSION
When grown to stationary phase, V. vulnificus lacking rpoS is impaired in its ability to survive osmotic, acidic, or oxidative challenge and the initial phase of long-term starvation. These results indicate that RpoS is necessary for the ability of V. vulnificus to adapt to many of the environmental stresses that it is likely to encounter. In addition, RpoS was required for full activity of five of the nine exoenzymes studied. These findings further indicate a role for RpoS in the adaptation of V. vulnificus to a dynamic environment. Finally, the rpoS mutant strain was shown to be significantly less motile than the wild type. Together these results demonstrate that RpoS is part of a regulatory network in V. vulnificus that helps the bacterium to thrive in diverse niches. These results are similar to those reported for Pseudomonas aeruginosa (15, 47), where an rpoS mutant was found to be more sensitive to carbon starvation, hydrogen peroxide, heat, elevated osmotic levels, acid pH, and ethanol. Altered motility and significant reduction in the level of exotoxin A, but not phospholipase C or elastase, have also been reported in P. aeruginosa mutants (47). Similarly, a V. cholerae rpoS mutant was found to be impaired in survival against hydrogen peroxide, hyperosmotic conditions, and carbon starvation and to exhibit reduced protease production (50). The regulation of rpoS transcription and the phenotypic consequences of RpoS loss in E. coli and other closely related bacteria have recently been reviewed by Venturi (49) and Hengge-Aronis (9, 10).
Most striking was the high sensitivity of V. vulnificus AH1 to oxidative stress (nearly a 6,000-fold decrease in viability). Reactive oxygen species may result from the oxidative burst of host macrophages during infection or from UV irradiation of water. Therefore, oxidative resistance plays a role in the persistence of pathogens in the human host and in survival in aquatic environments. In E. coli, this RpoS regulon includes a variety of genes involved in the oxidative stress response (for a recent review on oxidative stress, see reference 45). Among these genes are xthA (exonuclease III), dps (nonspecific DNA binding protein), sodC (periplasmic superoxide dismutase), and both katG and katE (hydroperoxidase I and II, respectively). The expression of some of these genes (e.g., katG and dps) is also under the control of the oxidative stress response regulator, OxyR. The relationship between both response regulators, OxyR and RpoS, is even more complex, as data suggest that that oxyR expression itself is under the control of RpoS (25). Additionally, the expression of rpoS is controlled by the OxyS RNA whose expression is activated by OxyR (51). We are currently examining the role of OxyR in the oxidative stress response of V. vulnificus.
It was reported earlier that V. vulnificus strain C7184o possesses a large variety of macromolecule-degrading enzymes (36). The rpoS-deficient mutant was impaired in albuminase, caseinase, and elastase activity. Additionally, gelatinase and collagenase activities were greatly reduced in this strain. At least two proteins could be responsible for these results. The purified metalloprotease (encoded by vvp) is reported to have caseinolytic, elastolytic, and collagenolytic activities (29). Additionally, the gene product of vvpE, an elastolytic metalloprotease (elastase), is known to be partially regulated by RpoS (12, 13). It is not clear whether the remaining collagenase and gelatinase activities observed in the mutant on plate assays are due to a third, still unknown collagenase. Taken together, our data suggest that RpoS regulates the expression of genes encoding possible virulence determinants that are responsible for the enormous tissue damage which occurs during infection (30, 31, 33). As knockout mutations in either vvpE or vvp do not affect the ability to infect mice (13, 42), one would assume that the sum of all factors involved rather than one single virulence determinant results in the highly invasive character of this human pathogen. As an alternative sigma factor, RpoS is involved in a variety of processes, and its deficiency likely has a pleiotropic effect under a variety of stressful situations that exceeds its influence on a single factor, e.g., the metalloprotease or elastase. Furthermore, we have shown that the rpoS mutant strain is substantially less motile than the wild-type strain. A recent report has shown that due to an insertion in flgC, nonmotile cells of V. vulnificus exhibit decreased virulence in mice (39). We are currently investigating the virulence of the rpoS mutant strain in the mouse model.
The studies reported here indicate that an RpoS-deficient mutant of V. vulnificus is impaired in surviving various stressful conditions. Moreover, it is shown that the rpoS mutant lacks proteolytic activities that are potentially important for virulence. More systematic work must be undertaken to understand the role RpoS plays in surviving host-induced stress and virulence of pathogenic bacteria. Our experimental results are supported by data published by others and suggest a larger function for this alternative sigma factor in infection and pathogenesis than what we previously understood.
Acknowledgments
We thank Lee A. Lewis for help in statistical analysis and David Figurski for supplying pJAK13.
This work was supported in part by the U.S. Department of Commerce, National Marine Fisheries Service grant NA17FD2364. We also acknowledge the Graduate School of the University of North Carolina at Charlotte for support of publication costs.
REFERENCES
- 1.Albertson, N. H., T. Nyström, and S. Kjelleberg. 1990. Exoprotease activity of two marine bacteria during starvation. Appl. Environ. Microbiol. 56:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 3.Beers, R. F., Jr., and I. W. Sizer. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195:133-140. [PubMed] [Google Scholar]
- 4.Beltrametti, F., A. U. Kresse, and C. A. Guzmán. 1999. Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J. Bacteriol. 181:3409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang, F. C., S. J. Libbey, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative σ factor KatF (RpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foley, I., P. Marsch, E. M. H. Wellington, A. W. Smith, and W. R. M. Brown. 1999. General stress response master regulator rpoS is expressed in human infection: a possible role in chronicity. J. Antimicrob. Chemother. 43:164-165. [DOI] [PubMed] [Google Scholar]
- 7.Gish, W., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nature 385:556-559. [DOI] [PubMed] [Google Scholar]
- 8.Hengge-Aronis, R. 2000. A role for the σs subunit of RNA polymerase in the regulation of bacterial virulence. Adv. Exp. Med. Biol. 485:85-93. [DOI] [PubMed] [Google Scholar]
- 9.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress response. American Society for Microbiology, Washington, D.C.
- 10.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σs (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 12.Jeong, H. S., K. C. Jeong, H. K. Choi, K.-J. Park, K.-H. Lee, J. H. Rhee, and S. H. Choi. 2001. Differential expression of Vibrio vulnificus elastase gene in a growth phase-dependent manner by two different types of promoters. J. Biol. Chem. 276:13875-13880. [DOI] [PubMed] [Google Scholar]
- 13.Jeong, K. C., H. S. Jeong, J. H. Rhee, S. E. Lee, S. S. Chung, A. M. Starks, G. M. Escudero, P. A. Gulig, and S. H. Choi. 2000. Construction and phenotypic evaluation of a Vibrio vulnificus vvpE mutant for elastolytic protease. Infect. Immun. 68:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 15.Jørgensen, F., M. Bally, V. Chapon-Herve, G. Michel, A. Lazdunski, P. Willams, and G. S. A. B. Stewart. 1999. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 145:835-844. [DOI] [PubMed] [Google Scholar]
- 16.Kim, Y., and L. L. McCarter. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 182:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowarz, L., C. Coynault, V. Robbe-Saule, and F. Norel. 1994. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvV and spvABCD virulence plasmid genes. J. Bacteriol. 176:6852-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 19.Lin, Y. H., C. Miyamoto, and E. A. Meighen. 2002. Cloning, sequencing, and functional studies of the rpoS gene from Vibrio harveyi. Biochem. Biophys. Res. Commun. 293:456-462. [DOI] [PubMed] [Google Scholar]
- 20.Linkous, D. A., and J. D. Oliver. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174:207-214. [DOI] [PubMed] [Google Scholar]
- 21.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 22.Martínez-García, E., A. Tormo, and J. M. Navarro-Llorens. 2001. Further studies on RpoS in enterobacteria: identification of rpoS in Enterobacter cloacae and Kluyvera cryocrescens. Arch. Microbiol. 175:395-404. [DOI] [PubMed] [Google Scholar]
- 23.McDougald, D., S. A. Scott, and S. Kjelleberg. 2001. SmcR-dependent regulation of adaptive phenotypes in Vibrio vulnificus. J. Bacteriol. 183:758-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merrell, D. S., A. D. Tischler, S. H. Lee, and A. Camilli. 2000. Vibrio cholerae requires rpoS for efficient intestinal colonization. Infect. Immun. 68:6691-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michán, C., M. Manchado, G. Dorado, and C. Pueyo. 1999. In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J. Bacteriol. 181:2759-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1992. Cloning a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174:7235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyoshi, N., C. Shimizu, S. Miyoshi, and S. Shinoda. 1987. Purification and characterization of Vibrio vulnificus protease. Microbiol. Immunol. 31:13-25. [DOI] [PubMed] [Google Scholar]
- 30.Miyoshi, S., and S. Shinoda. 1988. Role of the protease in permeability enhancement by Vibrio vulnificus. Microbiol. Immunol. 32:1025-1032. [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi, S., H. Nakazawa, K. Kawata, K. Tomochika, K. Tobe, and S. Shinoda. 1998. Characterization of the hemorrhagic reaction caused by Vibrio vulnificus metalloprotease, a member of the thermolysin family. Infect. Immun. 66:4851-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moody, C. S., and H. M. Hassan. 1984. Anaerobic biosynthesis of the manganese-containing superoxide dismutase in Escherichia coli. J. Biol. Chem. 259:12821-12825. [PubMed] [Google Scholar]
- 33.Okujo, N., T. Akiyama, S. Miyoshi, S. Shinoda, and S. Yamamoto. 1996. Involvement of vulnibactin and exocellular protease in utilization of transferrin- and lactoferrin-bound iron by Vibrio vulnificus. Microbiol. Immunol. 40:595-598. [DOI] [PubMed] [Google Scholar]
- 34.Oliver, J. D. 1997. Interaction of Vibrio vulnificus and the human host, p. 393-399. In M. T. Martins (ed.), Progress in microbial ecology. Proceedings of the Seventh International Symposium on Microbial Ecology. Brazilian Society for Microbiology, São Paulo, Brazil.
- 35.Oliver, J. D., and J. B. Kaper. 2001. Vibrio species, p. 263-300. In M. P Doyle (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. American Society for Microbiology, Washington, D.C.
- 36.Oliver, J. D., J. E. Wear, M. B. Thomas, M. Warner, and K. Linder. 1986. Production of extracellular enzymes and cytotoxicity by Vibrio vulnificus. Diagn. Microbiol. Infect. Dis. 5:99-111. [DOI] [PubMed] [Google Scholar]
- 37.Olsén, A., A. Arnqvist, M. Hammar, S. Sukupolvi, and S. Normark. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin binding curli in Escherichia coli. Mol. Microbiol. 7:523-536. [DOI] [PubMed] [Google Scholar]
- 38.Ramos-González, M. I., and S. Molin. 1998. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT22440. J. Bacteriol. 180:3421-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ran Kim, Y., and J. Haeng Rhee. 2003. Flagellar basal body flg operon as a virulence determinant of Vibrio vulnificus. Biochem. Biophys. Res. Commun. 304:405-410. [DOI] [PubMed] [Google Scholar]
- 40.Römling, U., K. Livers, T. Austin, R. Kaiser, D. Jensen, R. Burgess, and P. Blum. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Shao, C. P., and L. I. Hor. 2000. Metalloprotease is not essential for Vibrio vulnificus virulence in mice. Infect. Immun. 68:3569-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao, C. P., and L. I. Hor. 2001. Regulation of metalloprotease gene expression in Vibrio vulnificus by a Vibrio harveyi LuxR homologue. J. Bacteriol. 183:1369-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, D. J. 1999. M.S. thesis. University of North Carolina at Charlotte.
- 45.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-59. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress response. American Society for Microbiology, Washington, D.C.
- 46.Strom, M. S., and R. N. Paranjpye. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2:177-199. [DOI] [PubMed] [Google Scholar]
- 47.Suh, S.-J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. H. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swift, S., M. J. Lynch, L. Fish, D. F. Kirke, J. M. Tomás, G. S. B. Stewart, and P. Williams. 1999. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect. Immun. 67:5192-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venturi, V. 2003. Control of rpoS transcription in Escherichia coli and Pseudomonas: why so different? Mol. Microbiol. 49:1-9. [DOI] [PubMed] [Google Scholar]
- 50.Yildiz, F. H., and G. K. Schoolnik. 1998. Role of rpoS in stress survival and virulence of Vibrio cholerae. J. Bacteriol. 180:773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, A., S. Altuvia, A. Tiwari, L. Argaman, R. Hengge-Aronis, and G. Storz. 1998. The OxyS regulatory RNA represses rpoS translation and binds Hfq (HF-I) protein. EMBO J. 17:6061-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]