Abstract
Oligonucleotides can be used to direct site-specific changes in genomic DNA through a process in which mismatched base pairs in the oligonucleotide and the target DNA are created. The mechanism by which these complexes are developed and resolved is being studied by using Saccharomyces cerevisiae as a model system. Genetic analyses have revealed that in all likelihood the reaction occurs in two phases: DNA pairing and DNA repair. While the former phase involves strand assimilation, the latter phase likely involves an endonucleolytic processing step that leads to joint resolution. In this study, we established the importance of a functioning MRE11 gene in the overall reaction, as yeast strains deficient in MRE11 exhibited severely reduced activity. The activity could be rescued by complementation with wild-type MRE11 genes but not with MRE11 alleles lacking the nuclease function. Taken together, the data suggest that Mre11 provides nuclease activity for targeted nucleotide exchange, a process that could be used to reengineer yeast genes.
Genetic reengineering in lower eukaryotes holds promise for the creation of host strains that can produce useful metabolites or express desirable phenotypes. The majority of efforts in yeasts have focused on the development of knock-out strategies, where the aim is to completely disable the gene but in many cases the most desirable outcome is a (subtle) modulation of gene function. Thus, new approaches that are simple and straightforward and do not require multiple cloning steps must be developed to meet this goal. A direct approach centers around alteration of single bases at specific sites in chromosomal genes without the need for integration or extensive genetic rearrangement.
Single-stranded DNA molecules can be used to direct the alteration or exchange of specific nucleotides in the genome of Saccharomyces cerevisiae. Pioneering work by Moerschell and colleagues (16) demonstrated the feasibility of base repair in the CYC1 gene, a gene encoding cytochrome c isoform 1. A variety of point and frameshift mutations within CYC1 were reversed by the transformation of single strands of DNA in approximately 0.0003 to 0.002% of the cells (28). The mechanism of conversion, however, was not fully elucidated in these studies, although the reaction was found to be dose dependent and exhibited an interesting strand bias. In a similar line of experiments, we have begun to examine the potential for using modified single-stranded DNA oligonucleotides (MSOs) to repair point and frameshift mutations in the yeast chromosome, and our long-term goal is to apply the technique to mammalian cells (22; see reference 2 for a review). To this end, we focused on examining the mechanism by which MSOs direct nucleotide alteration in yeast cells.
S. cerevisiae has already served as a useful and genetically tractable model for identifying genes involved in the process which we term targeted nucleotide exchange (TNE) (3). The majority of the work has employed MSOs ranging in length from 25 to 100 nucleotides and containing three phosphorothioate linkages at the 3′ and 5′ termini (14). These modified linkages confer resistance to nuclease digestion and, as a result, increase the specific activity three- to fourfold (9). MSOs that are 70 to 80 bases long appear to be optimal for TNE on both episomes and chromosomes (12). Alternative modifications, such as locked nucleic acid residues, can also protect against nuclease digestion while high levels of activity are maintained (20). Locked nucleic acid residues may eventually prove to be useful since they have low levels of cellular toxicity, especially in mammalian cells (27). Each of these molecules has been shown to repair a mutated hygromycin-enhanced green fluorescent protein (eGFP) fusion gene, generating converted cells resistant to hygromycin and exhibiting green fluorescence (13). This dual genetic readout has enabled measurement of TNE frequency under selectable and nonselectable conditions. Molecules with similar designs were used to correct single-base mutations in the CYC1 gene by using an assay system in which only converted clones can grow in the presence of a nonfermentable carbon source (glycerol) (3).
An examination of the TNE activity in strains of yeast containing mutations in DNA repair genes provided insight into the mechanism of action by highlighting genes that may function in this pathway. For example, experimental data have implicated ScRad51 and ScRad54 in the initial phase of TNE, target localization and DNA pairing (6). In this phase, ScRad51, acting as a DNA recombinase, aligns the MSO in homologous register at the designated target sequence, while ScRad54 helps unwind the target helix to enable complementary base pairing. The other strand of the duplex is displaced, and a complex known as a displacement or D loop is formed. We have strengthened the argument that DNA pairing is a critical phase of this reaction by performing biochemical studies in which preformed D loops were shown to enhance the TNE reaction. Elegant and groundbreaking studies of Biet et al. (1) established the importance of D-loop formation and suggested potential strategies for improving the frequency of this reaction. During the second (repair) phase, mismatch repair proteins MSH2, MSH3, and MSH6 initialize the removal of a mismatched base (4, 21) and set the stage for nucleotide replacement or exchange directed by the oligonucleotide. The final step in the DNA repair phase of the reaction is predicted to involve resolution of the D-loop structure by the action of a site-specific or structure-specific endonuclease. This resolution may lead to oligonucleotide dissociation or directed integration, as described by Storici and colleagues in the delitto perfecto model (24).
The yeast protein Mre11 has enzymatic characteristics that might allow it to act on this type of DNA structure, perhaps to resolve the joint molecule. MRE11 is a member of the RAD52 epistasis group and functions in double-strand break repair (18). MRE11-deficient cells exhibit an increase in radiation sensitivity and a reduction in nonhomologous end joining (11), and mutations in the N-terminal nuclease domain result in severe reductions in the capacity to repair double-stranded breaks (8). Recently, this enzyme was also shown to cleave secondary structures, such as hairpin loops created during a strand separation event (25), and it can act on inverted repeats assembled into cruciforms (15). Thus, the specificity of Mre11 activity could be regulated or influenced by the structure of its substrate, and the enzyme could prefer at some point to cleave unusual conformations of DNA, such as the D loop. Based on the structural nature of the preferred substrates and the endonucleolytic properties, we examined the possibility that Mre11 functions in the TNE reaction pathway. We took a fundamentally genetic approach by evaluating TNE in a strain lacking functional Mre11 activity and then restored the activity by complementing a Δmre11 strain with an exogenously added wild-type MRE11 gene. In addition, we created several new strains in which the endogenous MRE11 gene was replaced with mutant MRE11 alleles encoding proteins that lack nuclease function. The results of these studies suggest that Mre11 is involved in targeted nucleotide alteration, perhaps providing a nuclease activity that can act either in the resolution phase of the reaction or in the disjunction of paired substrates.
MATERIALS AND METHODS
Plasmids and oligonucleotide vectors.
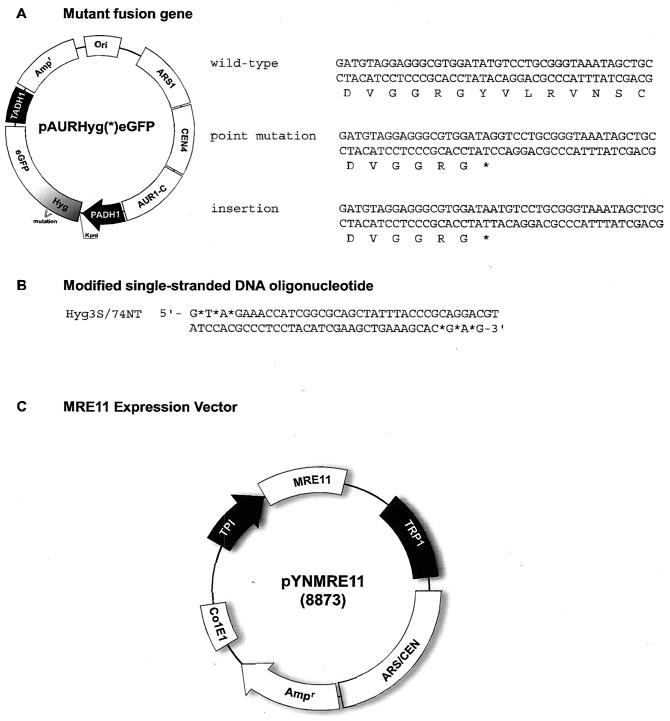
Plasmids pAURHyg(rep)eGFP and pAURHyg(ins)eGFP were constructed by inserting a cassette containing a mutant hygromycin gene and a fused eGFP gene into the pAUR123 shuttle vector, producing a stable episomal plasmid. This fusion cassette was inserted into pAUR101 to form plasmid pAUR101Hyg(rep)eGFP and was integrated into strain LSY678 to generate LSY678(Int)Hyg(rep) by electroporation; the integrated transformants were confirmed by Southern blot analyses. Plasmid pYNMRE11 was constructed by inserting MRE11 into the expression vector pYN132. The S. cerevisiae MRE11 cDNA was amplified from LSY678 genomic DNA by using the following set of primers: MRE11F (5′ CAGCATATG GACTATCCTGATCCAGACACA) and MRE11B (5′ GATCTCGAGCTATTTTCTTTCTTAGCAAGGAGACTTCCAAG). The PCR products were cut with NdeI and XhoI (New England BioLabs, Beverly, Mass.) and ligated into pYN132. All the plasmids mentioned above were confirmed by DNA sequencing. Plasmids pSM258:mre11(wt), pSM304:mre11-H125N, and pSM312:mre11-D56N are Mre11 nuclease wild-type or defective plasmids derived from pRS414 containing a CEN fragment and a TRP1 selective marker. In these plasmids, Mre11 is transcribed under control of its endogenous promoter. Most pSM438:mre11-H125N and pSM444:mre11-D56N integrative plasmids were also derived from pRS414; the exceptions were the plasmids carrying a Ura3-selective marker. The modified single-stranded DNA oligonucleotide vector Hyg3S/74NT, which is 74 bases long and contains three phosphorothioate linkages at the 3′ and 5′ termini, was also used in this study. This vector was designed to hybridize to the nontranscribed strand of the episomal or integrated hygromycin-eGFP fusion gene.
Transformation of S. cerevisiae and uptake of 32P-labeled oligonucleotides.
Plasmids and oligonucleotides were transformed into yeast cells by electroporation. Briefly, yeast cells were grown in YPD at 30°C to a density of approximately 2 × 107 cells/ml; then they were harvested and washed with distilled H2O twice and with 1 M sorbitol once. The cells were resuspended in 120 μl of 1 M sorbitol, and aliquots (40 μl; 2 × 108 cells) were electroporated by using a Bio-Rad gene pulser apparatus at the following settings: 1.5 kV, 25 μF, 200 Ω, one pulse, and 5-s pulse length. The cells were then allowed to recover in 3 ml of YPD supplemented with 1 M sorbitol for 16 h and plated (200 μl) on YPD-hygromycin (300 μg/ml) plates, YPD-aureobasidin A (0.5 μg/ml) plates, or SC agar plates lacking tryptophan or uracil. Colony counts were determined by using an AccuCount 1000 (Biologics, Inc.), and standard deviations were determined from the results of five independent experiments done in triplicate. To measure uptake efficiency, strains LSY678(mata) and LSY568(Δmre11) were grown in the same way, electroporated with 5 μl of 4 μM 32P-labeled Hyg3S/74NT, and recovered in 1 ml of YPD supplemented with 1 M sorbitol for 1 h. Cells were washed with 1 M sorbitol twice, and the radioactivity remaining in each cell pellet was determined with an LS6500 scintillation counter (Beckman, Fullerton, Calif.).
Establishment of a nuclease-defective Mre11 yeast strain.
Integrative plasmids pSM438:mre11-H125N and pSM444:mre11-D56N were linearized with BstEII (NEB) and electroporated into either LSY678(Int)Hyg(rep) or LSY678(Int)Hyg(ins). After a recovery time of 1 h, the cells were plated on SC agar lacking uracil and grown at 30°C for 48 h. The Ura+ transformants were then cultured in YPD until a cell density of 2 × 107/ml was reached, and 103 cells were spread on YPD plates containing 5′-fluororotic acid (5′-FOA) (1.25 g/liter). The 5′-FOA-resistant colonies, as well as LSY678(matα) and LSY568(Δmre11), were cultured in 5 ml of YPD overnight and then diluted to an optical density at 600 nm of 0.2. Ten-microliter portions of cells were spread on YPD plates containing 0.025% methyl methanesulfonate (MMS). Colonies showing MMS sensitivity were picked and used for DNA sequence analyses.
Targeted nucleotide alteration and gene complementation.
LSY678 (MATα leu2-3 112 trp1-1 ura3-1 his3-11 15 ade2-1 can1-100), LSY568(Δmre11) (LSY578 is LSY678 mre11::LEU2), and YGR258C(Δrad2) (YGR258C is MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rad2::Kanr) were electroporated with plasmid pAURHyg(rep)eGFP or pAURHyg(ins)eGFP, and this was followed by selection on YPD-aureobasidin A plates. After the presence of the plasmids was confirmed by colony PCR, experiments were begun by electroporation of the strains with 5 μg of Hyg3S/74NT, followed by a recovery period (16 h) in 3 ml of YPD supplemented with sorbitol. Cells (200 μl) were plated on YPD-hygromycin (300 μg/ml) plates and YPD-aureobasidin A (0.5 μg/ml) plates. Hygromycin-resistant colonies were quantified 48 h later. In another experimental protocol, plasmids pYN132 and pYNMRE11 were electroporated into yeast strains LSY678 and LSY568(Δmre11), respectively, which already contained an episome [pAURHyg(rep)eGFP or pAURHyg(ins)eGFP]. The cells were selected on SC agar plates lacking tryptophan but containing aureobasidin (0.5 μg/ml). The expression of Mre11 was confirmed by reverse transcription (RT)-PCR and by the development of MMS resistance. Yeast cells were grown to a density of 2 × 107 cells/ml and then targeted with 5 μg of Hyg3S/74NT, recovered for 16 h, and plated on YPD-hygromycin plates and YPD-aureobasidin A plates as described above. Hygromycin-resistant and aureobasidin-resistant colonies were counted, the values for the experiments performed were averaged, and the standard deviations were calculated by analysis of variance (SPSS6.14).
RT-PCR.
LSY678 and LSY568(Δmre11) bearing either pSM258:mre11(wt), pSM302:mre11-H125N, or pSM312:mre11-D56N were grown to a density of 2 × 107 cells/ml, and 1 × 107 to 5 × 107 cells were prepared for RNA isolation by using an RNeasy mini-kit (Qiagen) in which treatment of RNase-free DNase (Qiagen) was included. The first cDNA strand was synthesized by using a Superscript II transcription kit (Invitrogen) with primer MRE11B5 (5′-CTTACTGCTTTCCGCTTGAC-3′). PCR with the first cDNA strand was completed by using primers MRE11F3 (5′-CACAAGTTTTCACTACGATG-3′) and MRE11B5; the products were electrophoresed through 1% agarose by using a 1-kb ladder as a standard.
Southern blotting.
Genomic DNA was isolated from LSY678(Int)Hyg(rep) by phenol-chloroform extraction. Five micrograms of DNA was digested with MscI/AfeI at 37°C overnight and electrophoresed in a 0.7% agarose gel. The DNA was then transferred to an N+ nylon membrane in 20× SSC overnight (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and immobilized with a UV Stratalinker 2400 (Stratagene, La Jolla, Calif.). The membrane was hybridized to a [32P]dATP-labeled DNA probe made by performing PCR with two primers, pAUR743F (5′-CTATCAGAGAGACCTCCAAACTG-3′) and pAUR976B (5′-ATGTTAGGATGGGCAAGGCATTG-3′). After exposure for 16 h, the image was analyzed with a Molecular Dynamics Typhoon PhosphorImager, and the relative intensities of bands were quantified.
RESULTS
Assay systems.
Targeted nucleotide alteration or exchange (TNE) was measured by two assays; in both of these assays a mutation in the hygromycin resistance gene was used as the target for repair. In the first system, the mutant gene was in plasmid pAURHyg(rep)eGFP, which harbored a point mutation at nucleotide position 138 that rendered the plasmid unable to confer hygromycin resistance. The second system was similar to the first, except that plasmid pAURHyg(ins)eGFP, a plasmid containing an insertion mutation in the hygromycin gene, was used as the target (Fig. 1A). Repair of either hygromycin mutation resulted in expression of the gene and a hygromycin resistance phenotype. Each plasmid also contained an aureobasidin resistance gene, which could be used to normalize plating efficiency. Hence, dividing the number of hygromycin-resistant colonies by the number of aureobasidin-resistant colonies generated a value known as the correction efficiency. The values representing Hygr colonies were determined by averaging the numbers from at least three independent experiments, most often performed in duplicate, and standard deviations were calculated by using the Excel program and the raw data. The correction efficiency was another measure of the repair activity, but it took into account any differences in survival among host strains electroporated with oligonucleotides. In all previous cases, this value has reflected the hygromycin-resistant/aureobasidin-resistant colony counts, and the same was true here. Standard deviations for correction efficiency values were based on the Hygr/Aurr ratios generated directly from the colony counts.
FIG. 1.
Model systems for targeted nucleotide alteration in which plasmids pAURHyg(rep)eGFP and pAURHyg(ins)eGFP and oligonucleotides are utilized. (A) Plasmid pAURHyg(*)eGFP contains a synthetic expression cassette, which includes a hygromycin B gene, an eGFP gene fused to the alcohol dehydrogenase (ADH1) constitutive promoter, and a selective marker, AUR1-C. Plasmid pAURHyg(rep)eGFP contains a point mutation at nucleotide 138 of the hygromycin B coding sequence, which creates a replacement mutation, TAT (tyrosine) to TAG (stop codon) (asterisk). Plasmid pAURHyg(ins)eGFP contains an A residue at position 138, which creates a frameshift mutation. Correction of pAURHyg(rep)eGFP requires replacement of the mutant G residue with a C residue, while correction of pAURHyg(ins)eGFP requires removal of an A or T and replacement with a C residue. (B) Hyg3S/74NT, the oligonucleotide used in this study to direct nucleotide exchange. Asterisks indicate the phosphorothioate linkages in the oligonucleotide, located at both the 3′ and 5′ termini. (C) Plasmid pYN132 contains a constitutive TPI promoter, a TRP1 marker, and ARS/CEN elements. MRE11 was amplified by PCR and inserted into the expression vector pYN132 at the NdeI/XhoI sites to create pYNMRE11.
Low copy numbers of both plasmids were present, as the plasmids each contained a centromere and autonomously replicating sequence (CEN/ARS) element. In addition, the same single-stranded DNA molecule could be used to correct either mutation, and the sequence of this oligonucleotide is shown in Fig. 1B. It was a 74-mer modified at each terminus with three phosphorothioate linkages and was designed to hybridize to the nontranscribed strand of the target; it was designated Hyg3S/74NT to reflect all of these properties. Hyg3S/74NT has previously exhibited the capacity to direct significant levels of TNE on these episomal or chromosomal targets. The expression vector pYN132, a low-copy-number plasmid, was utilized for MRE11 expression under control of the triose phosphate isomerase (TPI) promoter. The selectable marker, TRP1, was used to maintain this plasmid in the cells (Fig. 1C).
Since the uptake of the oligonucleotide vector is of paramount importance for this type of study, we measured the transformation efficiencies of wild-type strain LSY678 and a strain lacking MRE11 (LSY568). Cells were grown to the same cell density (2 × 107 cells/ml), and 32P-labeled Hyg3S/74NT was electroporated into them; the cell density was determined, and the number of counts per minute per 108 cells was then calculated. The electrocompetencies of the strains were approximately equal, and the number of 32P counts per minute was measured relative to a cell density of 108 cells/ml (Table 1).
TABLE 1.
Electrocompetency of LSY678 and LSY568a
| Strain | Cell density (108 cells/ml) | 32P-labeled Hyg3S/74NT | Radioactivity
|
Relative radio- activity | |
|---|---|---|---|---|---|
| cpm | cpm/108 cells | ||||
| LSY678 | 1.49 | − | 24 | 16 | |
| LSY678 | 1.49 | + | 59,062 | 39,639 | 1 |
| LSY568(Δmre11) | 1.48 | − | 32 | 21 | |
| LSY568(Δmre11) | 1.48 | + | 69,384 | 46,881 | 1.18 |
Strains LSY678 and LSY568(Δmre11) were electroporated with 4 μM 32P-labeled Hyg/74NT oligonucleotide, and the intracellular radioactivity was detected after 1 h, as described in Materials and Methods. The uptake efficiency was calculated by determining the number of counts per minute per 108 cells, and the relative radioactivity was determined based on a value for LSY678 of 1.00. The data are averages of three experiments with a variance of ±15% (see Materials and Methods).
MRE11 is involved in targeted nucleotide alteration of a mutation in the episomal hygromycin-eGFP fusion gene.
The role of MRE11 in the TNE process was investigated by using plasmids pAURHyg(rep)eGFP and pAURHyg(ins)eGFP. These constructs were introduced into LSY678 and LSY568, respectively, and were maintained under aureobasidin selection. The cultures for targeting experiments were started from a single clone and propagated to obtain a density of 2 × 107 cells/ml, and then Hyg3S/74NT was introduced by electroporation. Repair of the mutation was then measured after 48 h (see Materials and Methods). Wild-type strain LSY678 supported high levels of TNE, while deletion strain LSY568 supported low levels, indicating that MRE11 plays a role in the correction process (Table 2). When the same experiment was carried out with pAURHyg(ins)eGFP as the plasmid target, the overall TNE frequency was again reduced. In fact, the reduction was amplified as the amount of repair of a frameshift mutation was less than that of a replacement mutation, even in LSY568, as previously reported.
TABLE 2.
Repair of an episomal targeta
| Strain | Mutation | Construct | No. of Hygr colonies | Correction efficiency (105) |
|---|---|---|---|---|
| LSY678 | pAURHyg(rep)eGFP | 932 ± 124 | 4.83 ± 0.77 | |
| LSY568 | Δmre11 | pAURHyg(rep)eGFP | 12 ± 6 | 0.74 ± 0.38 |
| LSY678 | pAURHyg(ins)eGFP | 619 ± 40 | 3.89 ± 1.02 | |
| LSY568 | Δmre11 | pAURHyg(ins)eGFP | 1.00 ± 0.82 | 0.06 ± 0.04 |
| YGR258C | Δrad2 | pAURHyg(rep)eGFP | 844 ± 96 | 4.37 ± 0.63 |
| YGR258C | Δrad2 | pAURHyg(ins)eGFP | 579 ± 20 | 3.61 ± 1.00 |
Strains LSY678 and LSY568 bearing either plasmid pAURHyg(rep)eGFP or plasmid pAURHyg(ins)eGFP were electroporated with 5 μg of Hyg3S/74NT and then spread on YPD plates supplemented with hygromycin. The average numbers of hygromycin-resistant colonies from these experiments and the standard deviations are shown along with the correction efficiencies calculated by determining the number of hygromycin-resistant colonies per aureobasidin-resistant colony for three experiments done in duplicate.
To demonstrate that these plasmids can be corrected in a strain other than LSY678, we introduced each of them into YGR258C(Δrad2) and maintained them under aureobasidin selection. Hyg3S/74NT was electroporated into each of the strains, and TNE activity was measured by quantifying the appearance of Hygr colonies. Both replacement and frameshift mutations were repaired in YGR258C(Δrad2) at a frequency comparable to that observed in the wild-type strain (Table 2).
The lower level of TNE observed in strain LSY568(Δmre11) provided evidence that MRE11 is involved in the TNE process. If this is true, then complementing the mutant strain with an expression construct containing wild-type MRE11 should have rescued the activity. To test this possibility, plasmid pYNMRE11 was constructed with the wild-type MRE11 gene under control of the constitutive promoter TPI. pYNMRE11 was introduced into LSY568 containing either pAURHyg(rep)eGFP or pAURHyg(ins)eGFP and maintained under TRP1 selection. Each of the strains was then electroporated with Hyg3S/74NT, and gene repair was assayed by examining colony growth on hygromycin plates. Overexpression of MRE11 rescued TNE activity in both cases, restoring the repair activity to approximately the wild-type levels for repair of replacement or frameshift mutations (Table 3).
TABLE 3.
Overexpression of MRE11 rescues targeted nucleotide alteration activity in LSY568(Δmre11)a
| Strain | Target | Construct | No. of Hygr colonies | Correction efficiency (105) |
|---|---|---|---|---|
| LSY678 | pAURHyg(rep)eGFP | pYN132 | 792 ± 63 | 4.26 ± 0.51 |
| LSY568(Δmre11) | pAURHyg(rep)eGFP | pYN132 | 23 ± 2 | 0.56 ± 0.18 |
| LSY568(Δmre11) | pAURHyg(rep)eGFP | pYNMRE11 | 209 ± 43 | 3.96 ± 0.84 |
| LSY678 | pAURHyg(ins)eGFP | pYN132 | 521 ± 125 | 3.12 ± 0.42 |
| LSY568(Δmre11) | pAURHyg(ins)eGFP | pYN132 | 2 ± 0.35 | 0.08 ± 0.03 |
| LSY568(Δmre11) | pAURHyg(ins)eGFP | pYNMRE11 | 244 ± 51 | 4.14 ± 0.76 |
Strain LSY678 or LSY568 containing either plasmid pAURHyg(rep)eGFP or plasmid pAURHyg(ins)eGFP and pYN132 or pYNMRE11 was electroporated with 5 μg of Hyg3S/74NT and then quantified by determining colony growth on YPD plates containing hygromycin. The values are the averages ± standard deviations for four experiments for each strain.
Repair of a chromosomal mutation.
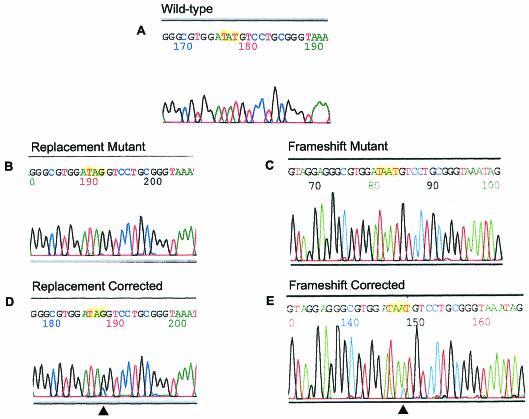
Previously, we modified strain LSY678 by integrating the hygromycin gene bearing a point mutation into the chromosome to create strains LSY678(Int)Hyg(rep) and LSY678(Int)Hyg(ins). Since the aureobasidin c gene is cointegrated, these cell lines could be propagated in the presence of aureobasidin. The integrated strains LSY678(Int)Hyg(rep) and LSY678(Int)Hyg(ins) contained three or four copies of the hygromycin cassette, as judged by Southern blotting (data not shown). An evaluation of how the overexpression of MRE11 influenced targeted nucleotide alteration of a chromosomal mutation in a wild-type strain could be performed by using this strain. Cultures were grown to a density of 2 × 107 cells/ml, and the oligonucleotide Hyg3S/74NT vector was electroporated into the cells. Chromosomal repair of the integrated hygromycin point mutation was assessed 48 h later (Table 4). Individually, overexpression of MRE11 stimulated the correction process approximately four- to fivefold. When we isolated clones from Hygr colonies and carried out DNA sequence analyses, a mixed peak was seen at the target site due, in all likelihood, to the fact that only one or two of the integrated mutant genes were corrected (Fig. 2). The presence of a black peak (G) coincident with a blue peak (C) illustrates this result. As in all cases, the targeted conversion was from TAG to TAC and not to the wild-type TAT in order to avoid concerns of cell contamination. In the case of the frameshift mutation (Fig. 2E), a C residue (blue peak) was introduced into the sequence, restoring the wild-type phenotype.
TABLE 4.
Targeted nucleotide alteration activity on a chromosomal mutation is increased by expression of MRE11a
| Strain | Construct | No. of Hygr colonies | Correction efficiency (105) |
|---|---|---|---|
| LSY678(Int)Hyg(rep) | pYN132 | 930 ± 130 | 3.91 ± 0.66 |
| LSY678(Int)Hyg(rep) | pYNMRE11 | 4,636 ± 241 | 18.68 ± 1.83 |
| LSY678(Int)Hyg(ins) | pYN132 | 697 ± 97 | 3.02 ± 0.28 |
| LSY678(Int)Hyg(ins) | pYNMRE11 | 3,755 ± 405 | 15.88 ± 1.91 |
LSY678(Int)Hyg(rep) and LSY678(Int)Hyg(ins) containing plasmid pYN132 or pYNMRE11 were targeted with 5 μg of Hyg3S/74NT, and the correction efficiency was assessed by determining colony growth on YPD plates supplemented with hygromycin as described in the text. The values are averages ± standard deviations for three experiments performed in duplicate.
FIG. 2.
DNA sequence analyses of mutant and repaired target genes. Single hygromycin-resistant colonies were picked for colony PCR, and the products were subjected to DNA sequence analyses. (A) DNA sequence of the wild type (TAT). (B) Hygromycin resistance gene sequence of replacement mutant (TAT → TAG). (C) Sequence of frameshift mutant (TAT → TAAT). (D) Sequence of DNA isolated from a hygromycin-resistant clone originally bearing the replacement mutation. (E) Same as panel D, except that the sequence illustrates correction of a frameshift mutation.
The amino acid sequence of MRE11 is partially homologous to the sequence of SbcC nuclease, which can resolve joint molecule structures in Escherichia coli (17). This type of endonucleolytic activity would be predicted to be important in the resolution of any putative TNE reaction intermediate. To examine this possibility, we utilized two MRE11 mutant genes, mre11-D56N and mre11-H125N, both of which result in mutations in the nuclease domains of the protein. These mutations are located in either motif II (D56N) or motif III (H125N) (see reference 17 for details). The two genes were cloned into an expression vector under control of the endogenous MRE11 promoter, creating plasmids pSM312:mre11-D56N and pSM304:mre11-H125N, respectively. These MRE11 variants were introduced into LSY678(Int)Hyg(rep) or LSY678(Int)Hyg(ins), both of which contained an integrated hygromycin gene with either a replacement or insertion mutation. This complementation experiment was similar to the one described above, except that the expression of each MRE11 gene was under control of its endogenous promoter rather than a constitutive promoter. To initialize the reaction, Hyg3S/74NT was electroporated into each strain, and correction events were scored by determining the number of hygromycin-resistant colonies. The results indicate that addition of pSM258:mre11(wt) increased the TNE activity on a chromosomal target in a wild-type background (Table 5). In contrast, pSM304:mre11-H125N and pSM312:mre11-D56N failed to elevate the TNE activity to an appreciable extent. These results indicate that increasing the amount of wild-type MRE11 gene expression, even in a wild-type cell, can enhance TNE activity but that overexpression of an Mre11 mutant devoid of nuclease activity does not enhance this activity. The data also suggest that neither mre11-D56N nor mre11-H125N can act as a dominant negative protein in the presence of wild-type MRE11.
TABLE 5.
TNE activitya
| Strain | Expression construct | No. of Hygr colonies | Correction efficiency (105) |
|---|---|---|---|
| LSY678(Int)Hyg(rep) | 1,290 ± 119 | 3.21 ± 0.29 | |
| LSY678(Int)Hyg(rep) | pSM258:mre11(wt) | 3,796 ± 318 | 9.58 ± 0.48 |
| LSY678(Int)Hyg(rep) | pSM304:mre11-H125N | 2,072 ± 219 | 3.52 ± 0.53 |
| LSY678(Int)Hyg(rep) | pSM312:mre11-D56N | 1,668 ± 117 | 4.40 ± 0.97 |
| LSY678(Int)Hyg(ins) | 716 ± 64 | 2.56 ± 0.28 | |
| LSY678(Int)Hyg(ins) | pSM258:mre11(wt) | 992 ± 109 | 6.16 ± 0.68 |
| LSY678(Int)Hyg(ins) | pSM304:mre11-H125N | 556.17 ± 72.28 | 3.52 ± 0.49 |
| LSY678(Int)Hyg(ins) | pSM312:mre11-D56N | 492.28 ± 59.04 | 3.13 ± 0.45 |
LSY678(Int)Hyg(rep) and LSY678(Int)Hyg(ins) containing episomal plasmid pSM258:mre11(wt), pSM304:mre11-H125N, or pSM312:mre11-D56N were targeted with 5 μg of Hyg3S/74NT, and then the correction efficiency was assessed by determining colony growth on YPD plates containing hygromycin, as described in Materials and Methods. The values are averages ± standard deviations for three experiments.
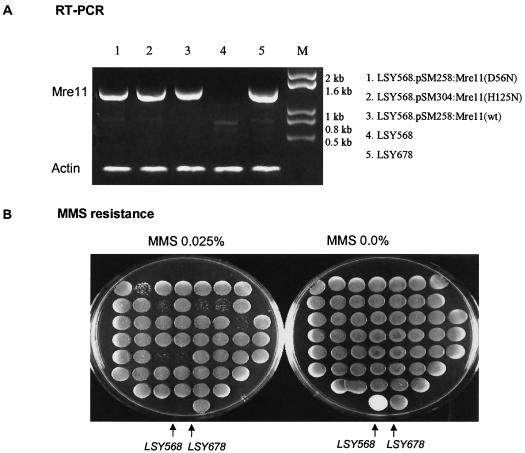
The same experiment was carried out with strain LSY678(Int)Hyg(ins), which, as indicated above, requires repair of a frameshift mutation to generate Hygr colonies. Hygromycin resistance was enabled, as shown by the removal of the inserted (A) residue directed by Hyg3S/74NT, and overexpression of wild-type MRE11 elevated the repair of the frameshift mutation, whereas both mutant MRE11 genes failed to do this (Table 5). Functional expression of the wild-type gene was confirmed by measuring MMS resistance in LSY568(Δmre11) and performing RT-PCR analyses of each gene. The wild-type gene was able to confer normal levels of resistance, but neither mutant construct was able to protect the strain and each expression construct was active, producing the correct transcript at approximately the same level (Fig. 3A).
FIG. 3.
Nuclease-deficient MRE11 expression and activity confirmed in a targeted nucleotide alteration reaction. (A) RT-PCR amplification of MRE11 gene expressed from different constructs, as shown on the right. Note that LSY568 is Δmre11, while LSY678 is wild type. (B) MMS-sensitive LSY678, LSY568, and 5′-FOA-resistant colonies were spotted on duplicate YPD plates and YPD plates containing 0.025% MMS and cultured for 48 h at 30°C (see Materials and Methods). The cells having a wild-type MRE11 function conferred resistance to MMS, while the cells carrying integrated nuclease-deficient MRE11 were sensitive.
While the data support the notion that the nuclease function of Mre11 is important in the TNE reaction, true measurement of the effect of MRE11 was complicated by the presence of the wild-type MRE11 gene (Table 5). It is difficult to assess the impact of overexpression of a mutant protein in the context of a wild-type background. To eliminate the background effect of endogenous MRE11, we replaced the endogenous MRE11 gene in wild-type strains LSY678(Int)Hyg(rep) and LSY678(Int)Hyg(ins) with mre11-H125N and mre11-D56N, respectively. This was accomplished with two integrative plasmids, pSM438:mre11-H125N and pSM444:mre11-D56N, which carry a URA3 marker and which were linearized with BstEII and electroporated into both LSY678(Int)Hyg(rep) and LSY678(Int)Hyg(ins). By a single crossover event, the Mre11 mutant plasmids became integrated into the chromosome at the MRE11 locus, creating a direct repeat in the chromosome. Homologous recombination between the two direct repeats was induced by selection for 5′-FOA resistance in order to excise one copy of the MRE11 gene and the integrated plasmid, including the uracil marker. As determined by this method, a single copy of the MRE11 gene, either wild type or mutant, remained in the chromosome, as shown by the response to 5′-FOA. Yeast cells bearing a mutant mre11 gene exhibited severe sensitivity to MMS, while the cells bearing a wild-type MRE11 gene exhibited resistance to MMS. As shown in Fig. 3B, LSY678(matα), LSY568(Δmre11), and the integrated 5′-FOA-resistant mutant strains were spread onto plates containing MMS. LSY678 showed resistance, while LSY568(Δmre11) was sensitive, as predicted. The colonies of the LSY678 strain showing sensitivity to MMS were isolated and confirmed by DNA sequence analyses to contain the nuclease-defective MRE11 (data not shown).
Each of these strains was then electroporated with Hyg3S/74NT, and the cells were spread on hygromycin and aureobasidin plates by using the standard protocol. Strains bearing either MRE11::mre11-H125N or MRE11::mre11-D56N supported a low level of TNE, as shown by a significant reduction in the correction efficiency compared to that of the strain bearing the wild-type MRE11 gene (Table 6). Again, the reduction was more pronounced when the frameshift mutation in LSY678(Int)Hyg(ins) was used as the target of Hyg3S/74NT. This pattern is consistent with other data presented in this paper and previously. The data are also consistent with the results described previously, except that in this study the requirement for the nuclease function of Mre11 was more stringent since no wild-type Mre11 was present to mask the effect (Table 5). Taken together, the data suggest that Mre11 is an important part of the TNE reaction and that its inherent nuclease function may be a critical biochemical function.
TABLE 6.
Targeted nucleotide alteration activitya
| Strain | No. of Hygr colonies | Correction efficiency (105) | Difference (fold) |
|---|---|---|---|
| LSY678(Int)Hyg(rep) | 660.45 ± 59.43 | 3.09 ± 0.34 | 1 |
| LSY678(Int)Hyg(rep) MRE11::mre11-H125N | 218.22 ± 33.96 | 0.57 ± 0.05 | 0.18 |
| LSY678(Int)Hyg(rep) MRE11::mre11-D56N | 190.62 ± 22.86 | 0.38 ± 0.04 | 0.12 |
| LSY678(Int)Hyg(ins) | 350.57 ± 19.53 | 1.41 ± 0.11 | 1 |
| LSY678(Int)Hyg(ins) MRE11::mre11-H125N | 32.65 ± 3.15 | 0.17 ± 0.02 | 0.12 |
| LSY678(Int)Hyg(ins) MRE11::mre11-D56N | 17.08 ± 1.51 | 0.06 ± 0.00 | 0.04 |
The number of hygromycin-resistant colonies and correction efficiency were calculated for each integrated MRE11-bearing strain (LSY678). Three independent experiments were carried out in duplicate.
DISCUSSION
Understanding the mechanism of TNE directed by MSO vectors in S. cerevisiae is a long-term goal of our laboratory. To this end, we have undertaken a reductionist approach and have begun to analyze the reaction in yeast strains deficient in genes known to be involved in recombination or repair pathways. Previously, we used a genetically based assay system to identify RAD51 and RAD54 as key players in the first phase of the TNE reaction, while RAD52 was found to act in a more suppressive fashion during this period. Here we provide evidence that MRE11 is a critical component of this developing pathway.
Several lines of evidence support the hypothesis that Mre11 is involved in this reaction. First, LSY568, which is deficient in MRE11, exhibits an 80 to 90% reduction in TNE activity on episomal targets, independent of whether the mutation is a point or frameshift mutation. Second, complementation of the Δmre11 strain with the wild-type MRE11 gene rescues the mutant phenotype and produces high levels of activity; in some cases the levels of activity exceed those observed in the wild-type strain. MRE11 was also found to be critical for repair of chromosomal mutations, and upon overexpression of MRE11 the activity was rescued and even elevated four- to sixfold. Third, rescue of the MRE11 mutation occurs when expression is regulated by either a constitutive promoter or the endogenous promoter. Fourth, TNE is enhanced by overexpression of wild-type MRE11 but not by MRE11 genes bearing mutations in the nuclease domain. Finally, when the endogenous MRE11 gene is replaced with either of the nuclease-deficient MRE11 genes, the repair of point and frameshift mutations, directed by Hyg3S/74NT, is reduced by almost 90%.
Recent studies on the biochemical activity of Mre11 indicated that it can cleave both DNA bubble structures and DNA hairpins and may play a role in genome stability. Thus, a role for Mre11 in TNE should center around its activity on the D-loop reaction intermediate, a configuration resembling both a DNA bubble and a hairpin. In fact, cleavage, perhaps at the interface of single-stranded and double-stranded DNA regions, may lead to productive resolution of the D-loop structure. Trujillo and Sung (25) also provided evidence that Mre11, in conjunction with Rad50, may catalyze nucleolytic scission at secondary structures formed as by-products of strand separation. Furthermore, the nuclease cleavage reaction appears to be specific, with the cut site appearing at the 3′ end of the single-stranded duplex DNA junction. Rad50 can stimulate the cleavage by Mre11, but the influence of Xrs2, the third member of the complex (26), is not clear at this time (7).
The normal cellular requirement for Mre11 activity could arise during DNA replication when secondary structures appear as a result of strand separation and fork movement. Mre11 may help resolve any blocks, such as structural aberrations encountered by a stalled replication fork, a function performed by the SbcCD complex in E. coli (23). More recently, Lobachev et al. (15) found that yeast cells deficient in MRE11, RAD50, XRS2, or SAE2 can have inserted duplications resulting from a reaction in which extended hairpins are cleaved and processed. It is plausible, therefore, that the D loop created by assimilation of the oligonucleotide into the target helix resembles the structural configuration that is acted upon naturally by the endonucleolytic activity of Mre11. These results are consistent with an alternative mechanism for TNE described by Storici and colleagues (24). In this process, designated delitto perfecto, site-specific mutagenesis directs precise DNA alterations by using short oligonucleotides. Since this reaction is coupled to homologous recombination, it is likely that such a scenario would also require an Mre11-like nuclease function to complete the process. Thus, our data obtained with Mre11 are consistent with the delitto perfecto pathway of recombination.
Our data also provide a necessary explanation of a critical step in one model of TNE, which was recently described by Igoucheva et al. (10). These workers used single-stranded oligonucleotides to correct a mutation in the LacZ gene inserted as a target in a mammalian cell line. At the heart of this model is an integration step in which the oligonucleotide assimilates into the target helix. The most critical enzymatic activity is the type of nucleolytic cleavage that Mre11 can easily provide. In fact, this integration scenario clearly explains the need for Mre11 found in this study. This result places Mre11 among the most important molecules in the process of gene manipulation and TNE. It is likely that overexpressing Mre11 in other eukaryotic cells would readily increase the TNE efficiency.
The Mre11-Rad50 protein complex could stabilize the D-loop structure by binding to each end of the assimilated oligonucleotide. It has been shown previously that a complex of Mre11 and Rad50 binds to ends of linear DNA molecules, tethering them together (5). Such activity would essentially increase the stability of the conjoined molecules configured into a D loop during the TNE process. Support for this notion comes from amino acid sequence analyses and alignments, which classify Rad50 as a member of the family of proteins involved in the structural maintenance of chromosomes. Mre11 and Rad50 also participate in the nonhomologous recombination pathway, a pathway that would require DNA end binding and DNA condensation (see reference 19 for a review). DNA condensation could enhance targeted nucleotide exchange by bringing the oligonucleotide vector into close proximity with the DNA target. However, the data presented here are not consistent with this explanation, as the DNA binding domains of Mre11 appear not to be essential for the reaction. While the exact site of Mre11 activity has not been fully elucidated, we provide evidence in this paper that Mre11 is needed for significant levels of TNE and that the endonucleolytic activity of Mre11 is likely to be the keystone function provided by this enzyme, perhaps for vector integration. As the frequency of TNE increases, the potential applications for this technique also rise. Our efforts in piecing together the mechanism of this reaction in a methodical fashion should help us reach this goal.
Acknowledgments
This work was supported by National Institutes of Health grant 1R01 DK56134.
We are grateful to Anja van Brabant, Peter Cheng, Lorraine Symington, Michael A. Resnick, and Kirill S. Lobachev for advice, suggestions, and/or reagents.
REFERENCES
- 1.Biet, E., J. S. Sun, and M. Dutreix. 2003. Stimulation of D-loop formation by polypurine/polypyrimidine sequences. Nucleic Acids Res. 31:1006-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brachman, E. E., and E. B. Kmiec. 2002. The 'biased' evolution of targeted gene repair. Curr. Opin. Mol. Ther. 4:171-176. [PubMed] [Google Scholar]
- 3.Brachman, E. E., and E. B. Kmiec. 2003. Targeted gene repair of cyc1 mutations in Saccharomyces cerevisiae directed by modified single-stranded DNA oligonucleotides. Genetics 163:527-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole-Strauss, A., H. Gamper, W. K. Holloman, M. Munoz, N. Cheng, and E. B. Kmiec. 1999. Targeted gene repair directed by the chimeric RNA/DNA oligonucleotide in a mammalian cell-free extract. Nucleic Acids Res. 27:1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Jager, M., J. van Noort, D. C. van Gent, C. Dekker, R. Kanaar, and C. Wyman. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8:1129-1135. [DOI] [PubMed] [Google Scholar]
- 6.Drury, M., and E. B. Kmiec. 2003. DNA pairing is an important step in the process of targeted nucleotide exchange. Nucleic Acids Res. 31:899-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis, H. M., D. Yu, T. DiTizio, and D. L. Court. 2001. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 98:6742-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuse, M., Y. Nagase, H. Tsubouchi, K. Murakami-Murofushi, T. Shibata, and K. Ohta. 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17:6412-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamper, H. B., A, Cole-Strauss, R. Metz, H. Parekh, R. Kumar, and E. B. Kmiec. 2000. A plausible mechanism for gene correction by chimeric oligonucleotides. Biochemistry 39:5808-5816. [DOI] [PubMed] [Google Scholar]
- 10.Igoucheva, O., V. Alexeev, M. Pryce, and K. Yoon. 2003. Transcription affects formation and processing of intermediates in oligonucleotide-mediated gene alteration. Nucleic Acids Res. 31:2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johzuka, K., and H. Ogawa. 1995. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139:1521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, L., S. Cheng, A. J. van Brabant, and E. B. Kmiec. 2002. Rad51p and Rad54p, but not Rad52p, elevate gene repair in Saccharomyces cerevisiae directed by modified single-stranded oligonucleotide vectors. Nucleic Acids Res. 31:2742-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, L., M. C. Rice, M. Drury, S. Cheng, H. Gamper, and E. B. Kmiec. 2002. Strand bias in targeted gene repair is influenced by transcriptional activity. Mol. Cell. Biol. 22:3852-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, L., M. C. Rice, and E. B. Kmiec. 2001. In vivo gene repair of point and frameshift mutations directed by chimeric RNA/DNA oligonucleotides and modified single-stranded oligonucleotides. Nucleic Acids Res. 29:4238-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 16.Moerschell, R. P., S. Tsunasawa, and F. Sherman. 1988. Transformation of yeast with synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA 85:524-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogawa, H., K. Johzuka, T. Nakagawa, S. H. Leem, and A. H. Hagihara. 1995. Functions of the yeast meiotic recombination genes, MRE11 and MRE2. Adv. Biophys. 31:67-76. [DOI] [PubMed] [Google Scholar]
- 19.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parekh-Olmedo, H., M. Drury, and E. B. Kmiec. 2002. Targeted nucleotide exchange in Saccharomyces cerevisiae directed by short oligonucleotides containing locked nucleic acids. Chem. Biol. 9:1073-1084. [DOI] [PubMed] [Google Scholar]
- 21.Rice, M. C., M. Bruner, K. Czymmek, and E. B. Kmiec. 2001. In vitro and in vivo nucleotide exchange directed by chimeric RNA/DNA oligonucleotides in Saccharomyces cerevisiae. Mol. Microbiol. 40:857-868. [DOI] [PubMed] [Google Scholar]
- 22.Santana, E., A. E. Peritz, S. Iyer, J. Uitto, and K. Yoon. 1998. Different frequency of gene targeting events by the RNA-DNA oligonucleotide among epithelial cells. J. Invest. Dermatol. 111:1172-1177. [DOI] [PubMed] [Google Scholar]
- 23.Sharples, G. J., and D. R. Leach. 1995. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 17:1215-1217. [DOI] [PubMed] [Google Scholar]
- 24.Storici, F., L. K. Lewis, and M. A. Resnick. 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19:773-776. [DOI] [PubMed] [Google Scholar]
- 25.Trujillo, K. M., and P. Sung. 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276:35458-35464. [DOI] [PubMed] [Google Scholar]
- 26.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 27.Wahlestedt, C., P. Salmi, L. Good, J. Kela, T. Johnsson, T. Hokfelt, C. Broberger, F. Porreca, J. Lai, K. Ren, M. Ossipov, A. Koshkin, N. Jakobsen, J. Skouv, H. Oerum, M. H. Jacobsen, and J. Wengel. 2000. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. USA 97:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto, T., R. P. Moerschell, L. P. Wakem, S. Komar-Panicucci, and F. Sherman. 1992. Strand-specificity in the transformation of yeast with synthetic oligonucleotides. Genetics 131:811-819. [DOI] [PMC free article] [PubMed] [Google Scholar]