Abstract
Xylose reductase (XR) is a key enzyme in d-xylose metabolism, catalyzing the reduction of d-xylose to xylitol. An NADH-preferring XR was purified to homogeneity from Candida parapsilosis KFCC-10875, and the xyl1 gene encoding a 324-amino-acid polypeptide with a molecular mass of 36,629 Da was subsequently isolated using internal amino acid sequences and 5′ and 3′ rapid amplification of cDNA ends. The C. parapsilosis XR showed high catalytic efficiency (kcat/Km = 1.46 s−1 mM−1) for d-xylose and showed unusual coenzyme specificity, with greater catalytic efficiency with NADH (kcat/Km = 1.39 × 104 s−1 mM−1) than with NADPH (kcat/Km = 1.27 × 102 s−1 mM−1), unlike all other aldose reductases characterized. Studies of initial velocity and product inhibition suggest that the reaction proceeds via a sequentially ordered Bi Bi mechanism, which is typical of XRs. Candida tropicalis KFCC-10960 has been reported to have the highest xylitol production yield and rate. It has been suggested, however, that NADPH-dependent XRs, including the XR of C. tropicalis, are limited by the coenzyme availability and thus limit the production of xylitol. The C. parapsilosis xyl1 gene was placed under the control of an alcohol dehydrogenase promoter and integrated into the genome of C. tropicalis. The resulting recombinant yeast, C. tropicalis BN-1, showed higher yield and productivity (by 5 and 25%, respectively) than the wild strain and lower production of by-products, thus facilitating the purification process. The XRs partially purified from C. tropicalis BN-1 exhibited dual coenzyme specificity for both NADH and NADPH, indicating the functional expression of the C. parapsilosis xyl1 gene in C. tropicalis BN-1. This is the first report of the cloning of an xyl1 gene encoding an NADH-preferring XR and its functional expression in C. tropicalis, a yeast currently used for industrial production of xylitol.
Xylitol, a five-carbon sugar alcohol, is used as an alternative natural sweetener in food products. It is an anticariogenic sweetener with the same order of sweetness as sucrose (29) and is an insulin-independent sugar substitute for diabetics, as it does not require insulin for its metabolism (21). Though xylitol is currently produced by chemical hydrogenation of xylose using Ni/Al2O3 as a catalyst, the chemical production of xylitol is uneconomical because of the requirement for high temperature and pressure as well as the formation of by-products that require expensive separation and purification steps (18). Xylitol can also be produced microbiologically using xylose-utilizing yeast (28, 31, 44-46, 58). In xylose-fermenting yeast, d-xylose is metabolized through an inducible pathway in which it is first reduced by an NAD(P)H-dependent xylose reductase (XR) to xylitol. Xylitol is then oxidized by an NAD-dependent xylitol dehydrogenase to xylulose, which is phosphorylated to xylulose 5-phosphate and then channeled through the pentose phosphate pathway and can be converted into ethanol via glycolysis or further metabolized via the tricarboxylic acid cycle and respiratory pathway (60). Under anaerobic conditions or at very low oxygen transfer rates, NAD-linked xylitol dehydrogenase is considerably inhibited, thus leading to xylitol accumulation rather than the conversion of d-xylose to xylulose (20, 22, 56).
Since xylose is among the most abundant sugars in terms of biomass (3), there is considerable interest in improving its metabolic utilization, for which XR activity is essential (6, 54). XR, a component of the aldose reductase (ALR) family, is a member of the aldo-keto reductase (AKR) superfamily of enzymes. The majority of known AKRs are monomeric, but most XRs function as noncooperative, tightly associated dimers with a subunit molecular mass of 33 to 40 kDa (49, 61). While the majority of AKRs have no well-defined role assigned to them, many are believed to act as general detoxification catalysts by reducing reactive carbonyl-containing compounds (24). It has also been proposed that they function in osmotic regulation by controlling levels of intracellular polyols (4).
Several XR genes have been cloned and expressed in various hosts (1, 2, 16, 55, 59). Up to now, the primary structures of yeast XRs, whose physiological role can clearly be assigned to the conversion of d-xylose, are known (37). Yeast ALRs can be classified into two groups according to whether they are strictly specific for NADPH or show dual coenzyme specificity (42). d-Xylose-fermenting Pichia stipitis (58), Candida shehatae (28), and Candida tenuis (44) XRs have dual coenzyme specificity, which distinguishes these enzymes from most ALRs exhibiting a strict requirement for NADPH. The dual-coenzyme-specific XRs reported, however, are still primarily NADPH dependent, especially in a coenzyme mixture simulating physiological conditions.
Recently, a panel of mutant strains of Candida was screened for high xylitol production, and two new high-producing isolates were deposited as Candida parapsilosis KFCC-10875 and Candida tropicalis KFCC-10960 (33, 46, 47). The C. tropicalis strain exhibited the highest xylitol production yield and rate (46); however, it has been suggested that XRs that are mostly or largely NADPH dependent, including the XR of C. tropicalis, are limited by cofactor availability for xylose conversion and consequently result in low xylitol production. Since C. tropicalis XR is known to be NADPH dependent, regeneration of NADPH via glucose metabolism through the oxidative pentose phosphate pathway has been the main concern in the production of xylitol in C. tropicalis (46). However, supplementation of cosubstrates including glucose for regenerating the reduced cofactor may also result in the accumulation of by-products, such as ethanol and glycerol, which complicate purification (26, 46).
While several XRs, including C. tropicalis XR, have been cloned and characterized (1, 2, 16, 55, 59), C. parapsilosis XR has never been cloned, purified, or characterized. The isolation of an XR with a strong preference for NADH over NADPH has also never been reported. Since NADH is more prevalent in the cell, XR enzymes with dual coenzyme specificity or with a preference for NADH may improve the xylose conversion process (30). In this study, we describe the cloning of the xyl1 gene from C. parapsilosis mutant strain KFCC-10875 encoding an XR with unusual dual coenzyme specificity and its functional expression in C. tropicalis, an organism currently used for industrial xylitol production.
MATERIALS AND METHODS
Materials.
d-Xylose, d-glucose, xylitol, Coomassie brilliant blue R-250, and the column chromatographic support media including Cibacron Blue 3GA affinity resin were obtained from Sigma Chemical Co. (St. Louis, Mo.). Enzyme cofactors (NAD, NADH, NADP, and NADPH) were purchased from Boehringer Mannheim (Indianapolis, Ind.), and various restriction enzymes and modified enzymes were obtained from Bioneer (Deajeon, Korea). All other chemicals were of analytical grade or higher.
Microorganisms and media.
C. parapsilosis (KFCC-10875) was used as a source for the xyl1 gene, and C. tropicalis (KFCC-10960) was used as a host strain for xyl1 gene expression and xylitol production. The properties and the optimal culture media for C. tropicalis KFCC-10960 (46) and C. parapsilosis KFCC-10875 (47) were previously reported. Escherichia coli MC1061 was used as the host for gene cloning. The wild-type and recombinant E. coli cells were subcultured regularly in Luria-Bertani (LB) medium at 37°C, supplemented with ampicillin (50 μg/ml) as appropriate.
Purification and gene cloning of C. parapsilosis XR.
XR was purified to homogeneity from cells of C. parapsilosis as described elsewhere (38). The purified protein was separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and transferred from the gel onto a polyvinylidene difluoride (PVDF) membrane filter (Millipore, Bedford, Mass.) in transfer buffer (10 mM 3-[cyclohexylamino]-1-propanesulfonic acid [CAPS] [pH 11], 10% methanol). The membrane was fully washed with deionized distilled water and then stained with Coomassie brilliant blue R-250. PVDF-blotted protein bands corresponding to the XR were sequenced with a protein sequencer (model 610A; Applied Biosystems). To determine internal amino acid sequences, the purified protein was digested with CNBr (51) and the resulting peptide fragments were fractionated by reverse-phase high-performance liquid chromatography (HPLC) on a Capcell Pak C18 MG column (4.6 by 250 mm; Shiseido), and their N-terminal amino acid sequences were analyzed. The partial XR gene was amplified by PCR using Taq DNA polymerase and two primers designed from the internal peptide 1 and peptide 3 amino acid sequences, namely, F1 [5′-AA(AG)ACTTTGTC(AT)GATTTGAA(TC)TTGG-3] and R1 [5′-ATC(AG)ATAAC(TA)GACAA(AG)TTTTG(AT)GC-3′]. The complete nucleotide sequence was determined by 5′ and 3′ rapid amplification of cDNA ends (RACE) using a 5′/3′ RACE kit (Roche Diagnostics GmbH, Manheim, Germany) and subcloned into the T-overhang vector, pGEM-T Easy (Promega). For 5′ RACE, the following gene-specific primers were designed: SP1 (5′-ATTCTGTCTGGGGAGTTGG-3′) and SP2 (nested) (5′-TCTTGGAATGACAGCGATGCC-3′) and SP3 (further nested) (5′-CTCTGGGTCTGATATCTC-3′). For 3′ RACE, SP4 (5′-CGTCGACTTGTTCTACATCC-3′) was used as a gene-specific primer. Standard techniques for nucleic acid manipulation were used (52). Restriction enzymes and other modifying enzymes were purchased from Bioneer.
Construction of the integrative plasmid and yeast transformation.
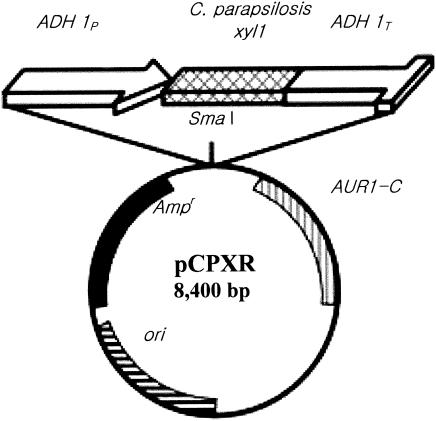
The C. parapsilosis xylI coding region was amplified by PCR using two synthetic primers (GGTACCATGTCTACTGCTACTGC and GTCGACTTAAACAAAAAATGGAATG) with engineered KpnI and SacI sites of primers. The amplified xylI gene was inserted into the KpnI and SacI sites of pAUR123 vector (Takara, Tokyo, Japan) containing an ampicillin and aureobasidin A resistance gene, the AOX1 promoter, and transcription termination fragment. For preparation of the integrative plasmid for expression in C. tropicalis, two primers were designed to amplify from the promoter region to the terminator region (CTGGATCCTCTAGATCCC and GGATCCGTGTGGAAGAACG). The amplified xylI gene, promoter, and termination fragment were inserted into the SmaI site of pAUR101 vector (Fig. 1). Transformation of C. tropicalis was performed according to the lithium acetate method (53). The transformants were selected on a yeast-peptone-dextrose plate containing aureobasidin A (0.1 μg/ml).
FIG. 1.
Physical map of yeast expression vector pCPXR. ADH 1P, alcohol dehydrogenase (ADH) promoter; ADH 1T, ADH terminator; AUR1-C, yeast selection marker; Ampr, bacterial selection marker; ori, bacterial origin of replication.
Culture conditions for fermentor experiments.
A suspension of frozen cells was inoculated into a 500-ml flask containing 75 ml of growth medium and incubated at 30°C and 250 rpm for 18 h. The culture broth (5%, vol/vol) was transferred to a 5-liter jar fermentor (KoBiotech Co., Incheon, Korea) containing an initial working volume of 2.5 liters of fermentation medium. Fermentor experiments were performed at 30°C; the pH of the culture broth was controlled at 5.0 during the fermentation. As soon as the cell mass reached about 15 g/liter, the carbon source, a solution of xylose (300 g/liter), or the mixed solution of xylose and glucose (300 and 45 g/liter, respectively) was fed continuously by adjusting the peristaltic pump (101U/R; Watson-Marlow) speed in the range of 5 to 40 ml/h. The aeration rate was fixed at 0.5 vol/vol/min (vvm). The agitation speed was gradually increased from 300 to 800 rpm to maintain the level of dissolved oxygen (DO) above 20% until the cell mass reached 15 g/liter and then reduced to 350 rpm to limit the concentration of DO.
Analytical methods.
Optical density was measured at 660 nm (Beckman DU 530), and dry cell weight was determined gravimetrically after the culture broth was centrifuged at 6,000 × g, washed with buffer, and dried overnight at 105°C. One optical density unit was found to be equivalent to 0.227 g (dry cell weight) liter−1. Concentrations of glucose, xylose, xylitol, and glycerol were measured by HPLC equipped with the shodex Sugar Pak I column (Waters, Milford, Mass.) with a refractive-index detector (Waters). The column was eluted with distilled water at a temperature of 90°C and a flow rate of 0.5 ml/min.
Nucleotides were separated and determined on a Capcell Pak C18 MG column (4.6 by 250 mm; Shiseido) with a UV detector (Waters model 486). The eluent consisted of two elements: 0.1 M KH2PO4 (pH 6.0) (buffer A) and 0.1 M KH2PO4 containing 2.5 M CH3OH (pH 6.0) (buffer B). The most suitable gradient was 11 min at 100% buffer A to 25% buffer B, 8 min from 25 to 90% buffer B, 6 min from 90 to 100% buffer B, and finally 6 min at 100% buffer B. The flow rate was 1 ml/min at room temperature, and detection was at 254 nm.
XR activity assay.
The activity of XR was determined spectrophotometrically by monitoring the change in A340 upon oxidation or reduction of NAD(P)(H) at 37°C. Unless indicated otherwise, the XR assay mixture (1.0 ml) for reduction contained 50 mM phosphate (pH 7.0), 0.05 mM NAD(P)H, 20 mM xylose, and enzyme solution (0.1 ml). This reaction mixture was allowed to stand for 1 min to eliminate the endogenous oxidation of NAD(P)H, and the reaction was started by the addition of 0.1 ml of substrate. One unit of enzyme activity is defined as the amount of enzyme required to consume 1 μmol of NAD(P)H per min under the specified conditions. Activities are expressed as units per milligram of protein, and the results presented show the means of triplicate assays.
Protein database search and sequence alignment.
The amino acid sequences deduced from the XR gene sequences of C. parapsilosis were compared with related enzymes from other sources using the BLAST Network at the National Center for Biotechnology Information. The multiple sequence alignment between the C. parapsilosis XR and related enzymes was performed with the program CLUSTAL W.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been submitted to GenBank and assigned the accession number AY193716.
RESULTS
Purification and internal amino acid sequences of C. parapsilosis XR.
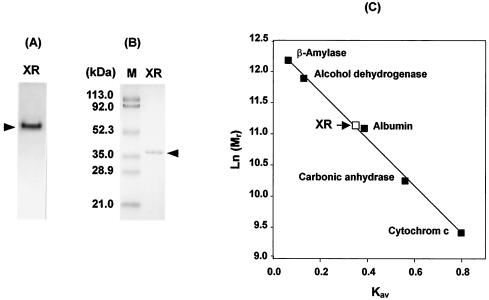
The purification procedure resulted in a 47.9-fold purification of XR with a recovery of 8.9% and specific activity of 41.7 U/mg of protein (Table 1). The yield of the purified enzyme was 1.25 mg, starting from 215 g (wet weight) of C. parapsilosis cells. At this purification step, C. parapsilosis XR was found to be homogeneous by native and SDS-PAGE (Fig. 2A and B). The molecular weights of the XR as determined by SDS-PAGE (Fig. 2B) and gel filtration chromatography (Fig. 2C) on a Superose-12 HR were 36.4 and 69.0 kDa, respectively, indicating that the enzyme is homodimeric.
TABLE 1.
Purification of xylose reductase from the cell extract of C. parapsilosis
| Procedure | Total protein (mg) | Total activity (U) | Specific activity (U/mg of protein) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extraction | 425 | 369 | 0.87 | 100 | 1.00 |
| Ammonium sulfate fractionation | 121 | 266 | 2.20 | 72.1 | 2.53 |
| DEAE-cellulose ion exchange chromatography | 28.2 | 90.2 | 3.20 | 24.4 | 3.68 |
| Sephadex G-100 gel filtration chromatography | 3.36 | 51.7 | 15.4 | 14.0 | 17.7 |
| Cibacron Blue 3GA affinity chromatography | 1.25 | 40.1 | 32.1 | 10.9 | 36.9 |
| Preparative electrophoresis | 0.79 | 32.9 | 41.7 | 8.9 | 47.9 |
FIG. 2.
PAGE and determination of the molecular mass of XR purified from C. parapsilosis. Shown are results from native PAGE (A) and SDS-PAGE (B). The enzyme solution was run on a 10% (wt/vol) polyacrylamide slab gel as described in Materials and Methods. The arrow indicates the protein band containing XR. (C) Determination of the molecular mass of native C. parapsilosis XR by gel filtration chromatography. The chromatography runs were performed as described in Materials and Methods. The marker proteins used and their Mrs were as follows: β-amylase, 200 kDa; alcohol dehydrogenase, 150 kDa; bovine serum albumin, 66 kDa; carbonic anhydrase, 29 kDa; cytochrome c, 12.4 kDa. The arrow indicates the position for the XR molecular mass from C. parapsilosis. Kav = (Ve − Vo)/(Vt − Vo) where Ve is the elution volume of protein, Vo is the elution volume of Blue Dextran 2000, and Vt is total bed volume.
The pure enzyme (2.0 μg) was separated by SDS-10% PAGE and blotted onto a PVDF membrane. Automated Edman degradation of the enzyme protein was unsuccessful, implying that the N terminus of the enzyme was blocked. Then, to analyze the internal amino acid sequence, C. parapsilosis XR was digested with CNBr, and the resulting peptide fragments were separated by reverse-phase HPLC as described in Material and Methods. The N-terminal amino acid sequences of three peptide fragments were as follows: peptide 1, KTLSDLNLDYVD; peptide 2, DLKRALDTPVLL; and peptide 3, AQNLSVIDFEL.
Substrate and coenzyme specificity.
d-Erythrose, d-ribose, d-arabinose, d-glucose, and d-fructose (all at 50 mM) were examined as alternative substrates for C. parapsilosis XR (Table 2). While d-arabinose, d-erythrose, and d-ribose showed slight activity (20% or less, compared with d-xylose-NADH activity), the other sugar and sugar phosphate did not serve as substrates for XR in the presence of either NADPH or NADH. Other XRs, such as C. tropicalis XR (61) and Pachysolen tannophilus XR (19), reduce other sugar substrates with 30∼120% of their d-xylose reduction activity. The C. parapsilosis XR did not show any activity towards d-glucuronate and benzoquinone, typical substrates used to characterize aldehyde reductases and carbonyl reductases of the AKR superfamily, respectively (23). These results confirm that C. parapsilosis XR is an ALR of the AKR superfamily.
TABLE 2.
Substrate and coenzyme specificity of xylose reductase purified from C. parapsilosisa
| Substrate | Specific activity (U/mg of protein) | Km (mM) | kcat/Km (s−1 mM−1) |
|---|---|---|---|
| d-Xylose | 32.1 | 31.5 | 1.5 × 100 |
| d-Xyloseb | 2.62 | 244.3 | 1.9 × 10−2 |
| d-Erythrose | 6.54 | 151.7 | 8.9 × 10−2 |
| d-Ribose | 2.67 | 302.0 | 4.3 × 10−2 |
| d-Arabinose | 1.79 | 285.4 | 2.8 × 10−2 |
| d-Glucose | 0.34 | —e | — |
| d-Fructose | 0.28 | — | — |
| Glucuronate | NDd | — | — |
| Benzoquinone | ND | — | — |
| Xylitolc | 1.29 | 326.3 | 3.1 × 10−2 |
The purified enzyme was assayed in the standard assay condition for reduction with various substrates. Each value represents the mean of triplicate measurements and varied from the mean by not more than 10%.
NADPH was used instead of NADH.
Assay was performed in the standard assay condition for oxidation.
ND, not detectable.
—, not determined.
HPLC analysis of the reaction products formed after the enzyme (10 U) was incubated at 37°C in a mixture of buffer and d-xylose (1 mM) confirmed the identity of the enzyme under examination, with and without NADH. Each mixture was then immediately chromatographed isocratically on CarboPacTM MA1 column with 500 mM NaOH at a flow rate of 0.4 ml/min and detected with a Bio-LC ED50A electrochemical detector (Dionex, Sunnyvale, Calif.). The HPLC chromatogram showed a significant decrease in d-xylose content for the sample containing NADH, accompanied by formation of a new peak that chromatographed with authentic xylitol; this substance was absent when the enzyme extract and NADH were incubated without d-xylose and showed the same mass profile as authentic xylitol (data not shown). The peaks of the residual substrate xylose (27.5 min) and the product xylitol (12.4 min) were assigned based on the retention times of the standard samples.
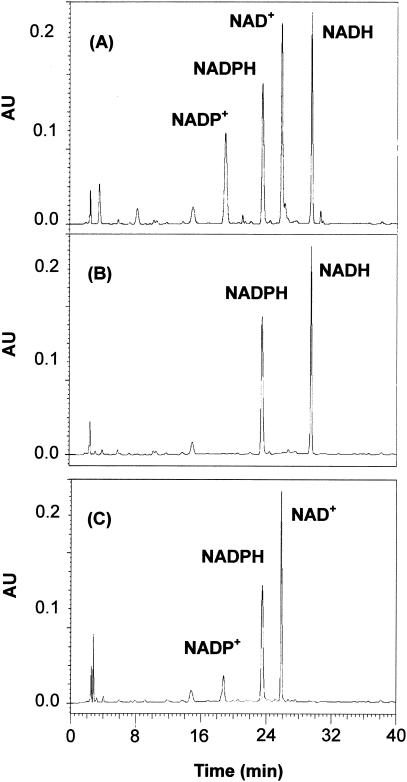
Most of the polyol oxidizing and reducing enzymes described to date are pyridine nucleotide linked, requiring either NADH or NADPH as a coenzyme. Although the ALRs, including yeast XRs, generally have a greater affinity for NADPH than for NADH (58), C. parapsilosis XR strongly preferred NADH (Km = 3.3 μM; kcat/Km = 1.39 × 104 s−1 mM−1) to NADPH (Km = 36.5 μM; kcat/Km = 1.27 × 102 s−1 mM−1). HPLC analysis of the residual coenzymes in a reaction mixture containing NADPH and NADH confirmed the preferential utilization of NADH by C. parapsilosis XR (Fig. 3).
FIG. 3.
HPLC analysis of the reaction products of C. parapsilosis XR. The enzyme (10 U) was incubated in a mixture of buffer and d-xylose (30 mM) with 0.5 mM NADH and 0.5 mM NADPH at 37°C for 1 min. The sample was eluted on a Capcell Pak C18 MG column (4.6 by 250 mm; Shiseido) with a UV detector (Waters model 486). The eluent consisted of two elements: 0.1 M KH2PO4 (pH 6.0) (buffer A) and 0.1 M KH2PO4 containing 2.5 M CH3OH (pH 6.0) (buffer B). The most suitable gradient was 11 min at 100% buffer A to 25% buffer B, 8 min from 25 to 90% buffer B, 6 min from 90 to 100% buffer B, and finally 6 min at 100% buffer B. The flow rate was 1 ml/min at room temperature, and detection was at 254 nm. Shown are the HPLC profiles of authentic standards NAD+, NADP+, NADH, and NADPH (A) and HPLC profiles of the initial reaction mixture (B) and the final reaction products (C). The residual coenzyme NADPH (23.6 min) and the products NAD+ (25.9 min) and NADP+ (18.7 min) are illustrated.
Optimum pH and thermal stability.
The optimum pH for reduction of d-xylose by purified C. parapsilosis XR was 6.0, with 81 and 65% of maximum activity at pH 5.0 and 7.0, respectively. The optimum pH for oxidation was 8.0, with 85 and 68% of the maximum activity at pH 7.0 and 9.0, respectively. Maximal reductase activity at pH 6.0 and an alkaline pH optimum for xylitol oxidation are common features of similar ALRs isolated from diverse microorganisms (39). In addition, the rate of the reverse reaction with 100 mM xylitol was insignificant (<5% of the forward reaction rate with 50 mM d-xylose), as is typical of an AKR. The stability of XR was tested in standard buffer containing 1 mM dithiothreitol (DTT). Preparations were incubated at 4, 20, 30, 45, and 50°C and retained 50% of their initial activities after 60 days, 8 days, 3 days, 4.5 h, and 2 min, respectively.
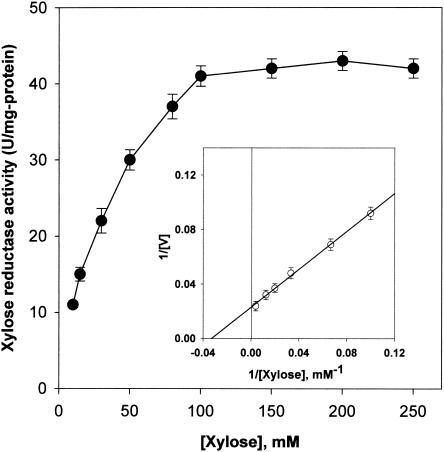
Kinetics.
Initial velocities were determined in the standard assay mixture at pH 6.0. The concentration of d-xylose varied from 1 to 300 mM. Figure 4 shows typical Michaelis-Menten-type kinetics for XR activity increasing with d-xylose concentration. The maximum enzyme activity was obtained at a d-xylose concentration of about 200 mM under the experimental conditions. The Lineweaver-Burk plot (inset in Fig. 4) obtained for the conversion of d-xylose under standard assay conditions shows that the Km for d-xylose is 31.5 mM, which is comparable to the values for XRs from various sources. The catalytic efficiency (kcat/Km = 1.40 s−1 mM−1) of C. parapsilosis XR was greater than those of C. tenuis (0.30 s−1 mM−1), Saccharomyces cerevisiae (0.12 s−1 mM−1), or human AR (0.017 s−1 mM−1) (43). These properties may be important for supporting the large accumulation of xylitol observed in this yeast strain.
FIG. 4.
Effects of substrate concentration on the activities of XR. XR activity of the enzyme (1 U) was measured in the presence of the indicated concentrations of d-xylose and 0.25 mM NADH at pH 6.0. The inset shows Lineweaver-Burk plot of initial velocity versus various fixed d-xylose concentrations. Each value represents the mean of triplicate measurements and varied from the mean by not more than 10%.
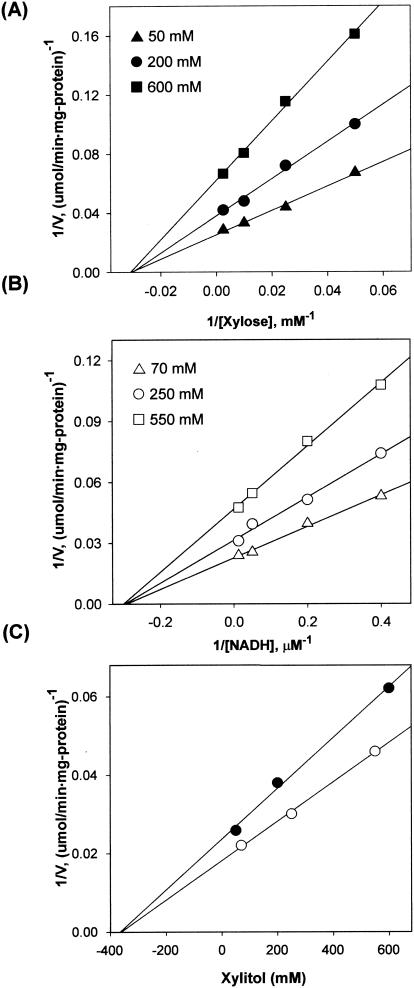
The product inhibition studies, under nonsaturating conditions, showed that the inhibition by xylitol was noncompetitive against NADH and d-xylose (Fig. 5). The secondary plots for noncompetitive inhibition with NADH and d-xylose are shown in Fig. 5C, indicating that the xylitol product binds to C. parapsilosis XR with a Ki of 368 mM. The inhibition by NAD+ was competitive with NADH (Ki = 180 μM) and was noncompetitive with d-xylose (Ki = 325 μM). Under nonsaturating conditions, only an ordered Bi Bi reaction mechanism exhibits a pattern of inhibition in which one product is a noncompetitive inhibitor for both substrates and the other product is competitive and noncompetitive for the respective substrates (50). The results of the initial-velocity and product inhibition studies suggest that the enzyme reaction proceeds via a sequentially ordered Bi Bi mechanism, as proposed for other ALRs (7), in which coenzyme NADH binds first to the enzyme, d-xylose binds to the binary enzyme-coenzyme complex, and xylitol leaves the enzyme before the release of NAD+ (14, 17).
FIG. 5.
Graphical analysis of the inhibition of C. parapsilosis XR by xylitol. The effects of increasing xylitol (product) concentration on the apparent Km and Vmax values for d-xylose and NADH were examined. Analysis of these data by double-reciprocal plots indicated that xylitol inhibited XR noncompetitively with respect to d-xylose (A) and NADH (B). Also shown are the secondary plots for noncompetitive inhibition with d-xylose and NADH (C). The xylitol product binds to XR with a Ki of 368 mM.
Stereospecificity of hydride transfer.
The stereospecificity of the hydride transfer step was examined using stereospecifically labeled NADH, and the oxidized NAD+ generated during the reaction was analyzed by 1H nuclear magnetic resonance. When (4R)-[4-d]NADD was used as a coenzyme for the reduction of d-xylose, the H-4 signal at δ 8.73 was retained in the NAD+ species, indicating the transfer of the pro-R deuterium at the C-4 position of the nicotinamide ring. By contrast, the δ 8.73 signal was absent after incubation with (4S)-[4-d]NADD due to the depletion of (4R)-hydrogen in the C. parapsilosis XR-catalyzed oxidation of NADD. Therefore, C. parapsilosis XR specifically transfers the 4-pro-R hydrogen from the C-4 of the nicotinamide ring to the re face of the carbonyl carbon of the substrate, which is typical of members of the AKR superfamily (5).
Effects of various chemicals.
The XR activity was measured in the presence of metal ions (1 mM) or various other compounds. XR activity was not stimulated by MgCl2, MnCl2, ZnCl2, CaCl2, NiCl2, CoCl2, or FeCl2 (each at 1 mM concentration), and it was neither inhibited nor activated by EDTA at concentrations ranging from 1 to 10 mM. By contrast, Cu2+ ion caused significant inhibition of C. parapsilosis XR (Ki = 34 μM). Pretreatment of C. parapsilosis XR with the same concentration of EDTA protected against the inhibitory effect of Cu2+, and the XR activity previously inactivated with Cu2+ was completely restored by addition of EDTA. The protection and reactivation of the enzyme with EDTA suggest that the effect of Cu2+ is reversible. Dependence of the enzyme activity on sulfhydryl compounds has been reported for several reductases purified from C. tenuis (44) and pig lens (11). We also examined the effects of sulfhydryl compounds on the C. parapsilosis XR. The addition of 1 mM 2-mercaptoethanol, glutathione, cysteine, or DTT to the reaction mixture increased the enzyme activity by 13, 21, 29, and 43%, respectively. These results suggest that sulfhydryl compounds, including DTT, the best reductant, keep the active enzyme in a reduced state.
Cloning and characterization of C. parapsilosis XR gene.
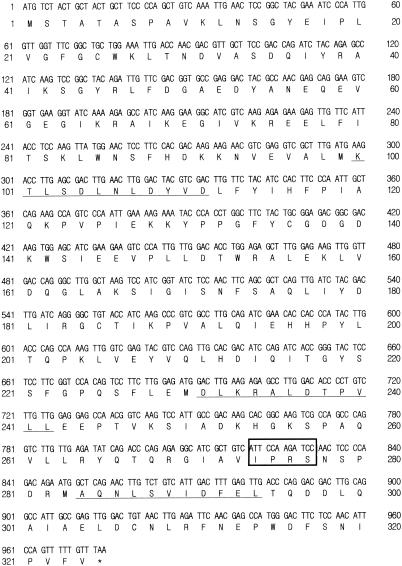
PCR was carried out using cDNA from C. parapsilosis as a template and degenerate primers F1 and R1 designed from the internal peptide 1 and peptide 3 amino acid sequences (KTLSDLNLDYVD and AQNLSVIDFEL), respectively. The resulting 0.6-kb DNA fragment was subcloned and sequenced. The complete cDNA nucleotide sequences were determined by 5′ and 3′ RACE, and the full-length cDNA encoding XR was successfully cloned. The cDNA sequence contained a 975-bp open reading frame encoding a putative 324-amino acid protein (Fig. 6) with a calculated isoelectric point of pH 5.19 and molecular mass of 36,629 Da which exhibited significant homology to the reported sequences of XRs from various sources (25, 27, 62). The calculated molecular mass was in good agreement with that determined by SDS-PAGE (Fig. 2B).
FIG. 6.
Nucleotide and deduced amino acid sequences of the C. parapsilosis XR gene (xyl1). The deduced amino acid sequence is given below the nucleotide sequence. The amino acid sequence of C. parapsilosis XR determined by Edman degradation is indicated by the underline. An asterisk marks the termination codon.
A BLAST search showed a close relationship between C. parapsilosis XR and a subset of XR family enzymes, and the entire open reading frames had very similar GC content and structural organization. The homology of C. parapsilosis XR and XRs from C. tropicalis (62), C. shehatae, Pichia guilliermondii, C. tenuis, P. stipitis, and Kluyveromyces lactis were 76.5, 66.4, 65.4, 63.9, 62.3, and 61.9, respectively. Multiple sequence alignment of the XRs, including C. parapsilosis XR, from various sources revealed several regions of sequence conservation, namely, an N-terminal region (LxxGxxxPxxGxG), an active site region (GxxxxDxAxxY, which contains the conserved Asp-49 and Tyr-54), a region including the active site (LxxxxxxxxDxxxxH, which contains the conserved His-116), and a 7-amino acid peptide (G133FYCGDG139) that is reminiscent of the Wierenga coenzyme-binding motif of other dehydrogenases (32). Each N-terminal sequence of XRs from various sources contains a highly conserved LNSG region (positions 12 to 15, according to the C. parapsilosis XR numbering). The serine residue at position 14 (C. parapsilosis XR numbering) in this motif is replaced by an asparagine in C. shehatae and K. lactis, and the asparagine residue at position 13 is replaced by a serine in C. tenuis (25). The catalytic tetrad of human AR (Tyr-54, Asp-49, Lys-83, and His-116) is completely conserved in C. parapsilosis XR, where Tyr-54 acts as the acid base catalyst, and Asp-49, Lys-83, and His-116 play an important role in facilitating hydride transfer (8). It appears that this mechanism is conserved, as most active ALRs identified to date possess these four amino acids.
Transformation and expression of C. parapsilosis xyl1 gene in C. tropicalis.
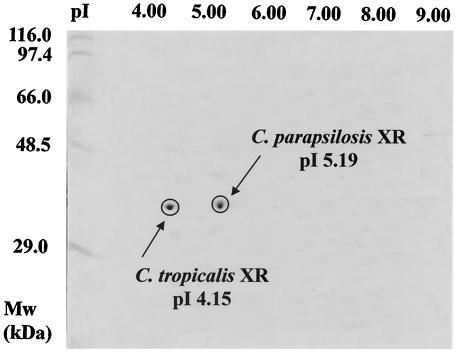
The C. parapsilosis xyl1 gene was expressed in C. tropicalis under the transcriptional control of the ADH1 promoter. While C. tropicalis XR is largely NADPH dependent, the C. tropicalis BN-1 harboring the C. parapsilosis xyl1 gene exhibited a dual coenzyme specificity for both NADPH (specific activity = 0.53 U/mg of protein) and NADH (specific activity = 0.58 U/mg of protein). HPLC analysis of the residual coenzymes in a reaction mixture containing NADPH and NADH confirmed the dual utilization of NADH and NADPH by C. tropicalis BN-1 XR (Fig. 3B), indicating that the C. parapsilosis xyl1 gene was functionally expressed in C. tropicalis. The functional expression of the C. parapsilosis xyl1 gene was further confirmed by two-dimensional electrophoresis of the partially purified XRs from C. tropicalis BN-1, which displayed two distinct spots corresponding to the pI values of each protein, 5.19 and 4.15 for C. parapsilosis XR and C. tropicalis XR, respectively (Fig. 7).
FIG. 7.
Two-dimensional electrophoresis of partially purified XR from C. tropicalis BN-1.
Xylose fermentation in C. tropicalis harboring the C. parapsilosis xyl1 gene.
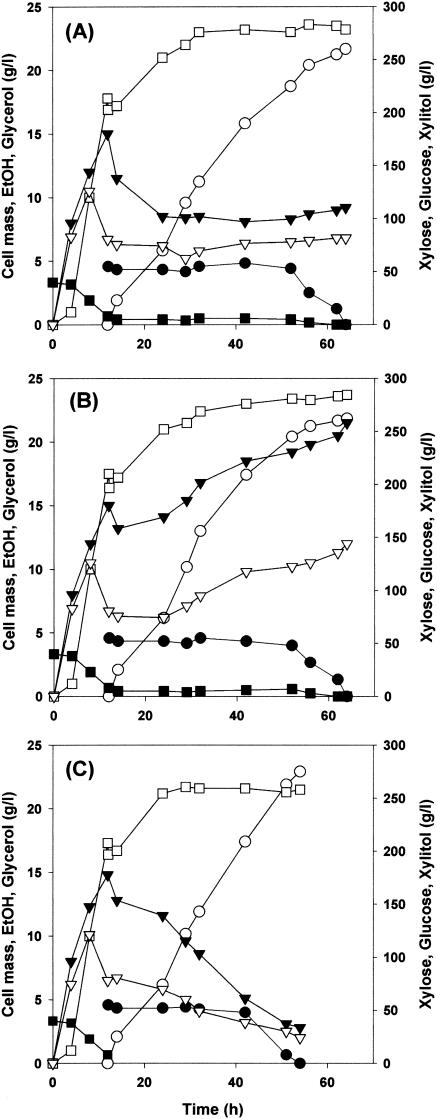
In a previous report (46), the use of a mixture of xylose and glucose as cosubstrates to increase the yield of xylitol from xylose in C. tropicalis cultures was discussed. In the present study, when C. tropicalis BN-1 and the wild-type cultures were fed with the mixture of xylose and glucose, there was no significant difference in the xylitol production profiles of the two strains (Fig. 8). Production of ethanol and glycerol, however, was significantly higher in the BN-1 recombinant than in the wild type, which suggests that surplus reduction equivalents derived from glucose metabolism were consumed for ethanol and glycerol synthesis. When a solution containing only xylose was fed, C. tropicalis BN-1 exhibited a significant increase in xylitol production yield (91.6%, wt/wt) and productivity (5.09 g/liter/h) and a significant decrease in ethanol and glycerol production (Fig. 8) relative to the wild type and the recombinant C. tropicalis BN-1 culture with xylitol-glucose mixture.
FIG. 8.
Fed batch fermentation profiles of C. tropicalis wild and mutant strain. Shown are result for C. tropicalis (A), C. tropicalis BN-1 with xylose and glucose (B), and C. tropicalis BN-1 with xylose (C). •, xylose; ○, xylitol; ▪, glucose; □, cell mass; ▾, ethanol; ▿, glycerol.
DISCUSSION
Table 3 compares the properties of NAD(P)H-linked XRs from various sources. The C. parapsilosis XR from the present study had a Km value (31.5 mM) for d-xylose comparable to those of XRs from various microorganisms, and this value was also similar to the values reported for ALRs from rabbit muscle (29 mM) (15) and human muscle (49 mM) (43). Although C. parapsilosis XR is similar to the homologous XRs described in other microorganisms, it has several novel properties. In general, ALRs, including XRs, are characterized by broad substrate specificities and low catalytic efficiency. The C. parapsilosis XR, however, shows high substrate specificity for d-xylose compared to XRs from other sources and differs from other XRs in that it has a high catalytic efficiency (kcat/Km = 1.40 s−1 mM−1). These properties of C. parapsilosis XR may partially explain the high production of xylitol observed in this strain (47).
TABLE 3.
Properties of aldose reductases from various organisms
| Organism (source or reference) | Mr, subunit (kDa) | Mr, native (kDa) | Coenzyme | pI | Km (Xylose) (mM) | kcat/Km (s−1 mM−1) | Km (NAPDH) (μM) | Km (NADH) (μm) | Optimal pH |
|---|---|---|---|---|---|---|---|---|---|
| C. parapsilosis (this work) | 36.6 | 69.0 | NADH >> NAPDH | 5.19 | 31.5 | 1.46 | 36.5 | 3.3 | 6.0 |
| C. tropicalis (61) | 36.5 | 58.0 | NADPH | 4.15 | 30 to 37 | —a | 9 to 14 | — | 6.0 |
| C. tenuis (44) | 43 | 48 | NADPH > NADH | 4.70 | 72 | 0.30 | 4.8 | 25.4 | 6.0 |
| S. cerevisiae (34) | 33 | 33 | NADPH | 6.60 | 27.9 | 0.12 | 0.013 | — | 5.0 |
| P. stipitis (58) | 34 | 65 | NADPH > NADH | 5.76 | 42 | 0.60 | 9 | 21 | 6.0 |
| P. tannophilus (19) | 38 | 38 | NADPH | 4.87 | 162 | 0.062 | 59 | — | 7.0 |
—, not reported.
Since most anabolic reductive reactions require NADPH rather than NADH and since transhydrogenase is absent in yeast (35), the overall process of assimilation leads to the production of a considerable surplus of NADH (12, 48). Most ALRs also have a strong preference for NADPH over NADH (10). As far as we are aware, no members of the ALR family are specific for NADH only or show a higher affinity for NADH than for NADPH. Only a few XR members can use both cofactors (13, 44, 58), with a very low activity ratio of NADH/NADPH (41). The C. parapsilosis XR, however, shows much higher catalytic efficiency with NADH (kcat/Km = 1.39 × 104 s−1 mM−1) than with NADPH (kcat/Km = 1.27 × 102 s−1 mM−1), unlike most XRs, including the XR of C. tropicalis (61, 62), making this enzyme even more interesting. The tetra-amino acid motif Ile-Pro-Lys-Ser is conserved among NADPH-dependent reductases, and the lysine residue in this motif is involved in NADPH binding (9). Although the motif is present in C. parapsilosis XR (boxed in Fig. 6), the lysine residue in this motif is replaced by an arginine. The Km values for NADPH were estimated to be 0.013 to 14 μM for XRs from other sources (34, 44) and 36.5 μM for C. parapsilosis XR. The difference in the Km values could be due to the amino acid sequence differences in the NADPH-binding region. Oligonucleotide-directed mutagenesis studies to address questions about the structure-function relationship regarding coenzyme specificity are presently under way.
Previously, high production of xylitol by the newly isolated mutants of C. parapsilosis and C. tropicalis was reported (33, 46, 47). C. tropicalis exhibited the highest xylitol production yield and rate (46). As noted above, the NADPH dependence of most XRs, including the XR from C. tropicalis, is likely to be a limiting factor for xylose conversion to xylitol (30). Thus, the cloning and characterization of a C. parapsilosis XR with NADH-preferring coenzyme specificity has an important bearing on our attempts to improve the kinetics and efficiency of xylose fermentation by C. tropicalis. A cosubstrate including glucose was supplemented to regenerate the reduced cofactor in C. tropicalis cultures. However, glucose supplementation in cultures of C. tropicalis BN-1 (harboring the NADH-preferring xyl1 gene) resulted in accumulation of by-products such as ethanol and glycerol, as has been suggested previously (26). When a solution containing only xylose was used as a feeding solution, C. tropicalis BN-1 exhibited a significant increase in xylitol production and a significant decrease in ethanol and glycerol production (Fig. 8). Ethanol, which can freely diffuse through the cell membrane, not only lowers the yield of xylitol but can also inhibit the hexose transport system noncompetitively, because it interferes with hydrophobic regions in the cell membrane (36). Glycerol is generally produced during anaerobic fermentation to oxidize excess NADH (48). The decrease in ethanol and glycerol formation is probably due to the consumption of reduction equivalents in the XR reaction rather than in the alcohol dehydrogenase or glycerophosphate dehydrogenase reaction and to the supply of reduction equivalents by anaerobic ethanol oxidation (40, 57). Glycerol and ethanol accumulation during the culture period represents a serious problem for the purification of xylitol from the fermentation broth. Glycerol, in particular, represents a serious hindrance to the industrial production of xylitol by C. tropicalis, since it lowers the yield of xylitol, necessitates additional purification steps, including chromatography, lowers the purification yield, and thus increases production costs.
In this study, we cloned, characterized, and transformed the C. parapsilosis NADH-preferring xyl1 gene into C. tropicalis. The resulting recombinant, C. tropicalis BN-1, produced less ethanol and glycerol than the wild strain, consequently facilitating xylitol purification while increasing overall xylitol production. Our results improve the understanding of xylitol biosynthesis in C. parapsilosis and C. tropicalis and should contribute to better industrial production of xylitol by biological processes. Since NADH is less costly, more prevalent in the cell, and more stable than NADPH, improvements in industrial processing could be obtained if the natural cofactor specificity of the enzyme could be broadened to enable the use of NADH. Therefore, the C. parapsilosis XR, an ALR with a strong preference for NADH and a high catalytic efficiency, is a good candidate for studying cofactor specificity as well as for industrial applications such as use as a novel biocatalyst for the enzymatic synthesis of sugar alcohols to reduce downstream purification. However, definitive proof of the characteristics of the NADH-preferring C. parapsilosis XR requires complete X-ray crystallographic analysis of the enzyme or enzyme-coenzyme complex and extensive site-directed mutagenesis study.
Acknowledgments
We thank Eun-Hye Kim (Korea Basic Science Institute, Seoul, Korea) for the help in 1H-nuclear magnetic resonance analysis and Ji-Hyun Yoo for thorough proofreading.
This work was supported by a grant (M1011100007-01A160000510) from the Ministry of Science and Technology, Korea.
REFERENCES
- 1.Amore, R., P. Kotter, C. Kuster, M. Ciriacy, and C. P. Hollenberg. 1991. Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase-encoding gene (XYL1) from the xylose-assimilating yeast Pichia stipitis. Gene 109:89-97. [DOI] [PubMed] [Google Scholar]
- 2.Anderlund, M., P. Radstrom, and B. Hahn-hagerdal. 2001. Expression of bifunctional enzymes with xylose reductase and xylitol dehydrogenase activity in Sacchromyces cerevisiae alters product formation during xylose fermentation. Metab. Eng. 3:226-235. [DOI] [PubMed] [Google Scholar]
- 3.Aspinall, G. O. 1980. Chemistry of cell-wall polysaccharides, p. 473-500, In J. Preiss (ed.), The biochemistry of plants, vol. 3. Academic Press, New York, N.Y.
- 4.Bedford, J. J., S. M. Bagnasco, P. F. Kador, H. W. Harris, and M. B. Burg. 1987. Characterization and purification of a mammalian osmoregulatory protein, aldose reductase, induced in renal medullary cells by high extracellular NaCl. J. Biol. Chem. 262:14255-14259. [PubMed] [Google Scholar]
- 5.Bernd, N., M. Peter, N. Wilfried, and P. Michael. 2001. Structural and functional properties of aldose xylose reductase from the d-xylose-metabolizing yeast Candida tenuis. Chem. Biol. Interact. 130-132:583-595. [DOI] [PubMed] [Google Scholar]
- 6.Billard, P., S. Me′nart, R. Fleer, and M. Bolotin-Fukuhara. 1995. Isolation and characterization of the gene encoding xylose reductase from Kluyveromyces lactis. Gene 162:93-97. [DOI] [PubMed] [Google Scholar]
- 7.Boghosian, R. A., and E. T. McGuinness. 1981. Quick brain aldose reductase: a kinetic study using the centrifugal fast analyzer. Int. J. Biochem. 13:909-914. [DOI] [PubMed] [Google Scholar]
- 8.Bohren, K. M., C. E. Grimshaw, C. J. Lai, D. H. Harrison, D. Ringe, G. A. Petsko, and K. H. Gabbay. 1994. Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme. Biochemistry 33:2021-2032. [DOI] [PubMed] [Google Scholar]
- 9.Bohren, K. M., J. L. Page, R. Shankar, S. P. Henry, and K. H. Gabbay. 1991. Expression of human aldose and aldehyde reductases. Site-directed mutagenesis of a critical lysine 262. J. Biol. Chem. 266:24031-24037. [PubMed] [Google Scholar]
- 10.Bolen, P. L., G. T. Hayman, and H. S. Shepherd. 1996. Sequence and analysis of an aldose (xylose) reductase gene from the xylose-fermenting yeast Pachysolen tannophilus. Yeast 12:1367-1375. [DOI] [PubMed] [Google Scholar]
- 11.Branlant, G. 1982. Properties of an aldose reductase from pig lens. Comparative studies of an aldehyde reductase from pig lens. Eur. J. Biochem. 129:99-104. [DOI] [PubMed] [Google Scholar]
- 12.Bruinenberg, P. M., P. H. M. deBo, J. P. van Dijken, and W. A. Scheffers. 1984. NADH-linked aldose reductase; the key to anaerobic alcoholic fermentation of xylose by yeast. Appl. Microbiol. Biotechnol. 19:256-260. [Google Scholar]
- 13.Chen, J., P. C. Turner, and H. H. Rees. 1999. Molecular cloning and characterization of hemolymph 3-dehydroecdysone 3β-reductase from the cotton leafworm, Spodoptera littoralis. A new member of the third superfamily of oxidoreductases. J. Biol. Chem. 274:10551-10556. [DOI] [PubMed] [Google Scholar]
- 14.Cleland, W. W. 1970. Steady state kinetics, p. 1-65. In P. D. Boyer (ed.), The enzymes, 3rd ed., vol. 2. Academic Press, New York, N.Y.
- 15.Cromlish, J. A., and T. G. Flynn. 1983. Purification and characterization of two aldose reductase isoenzymes from rabbit muscle. J. Biol. Chem. 258:3416-3424. [PubMed] [Google Scholar]
- 16.Dahn, K. M., B. P. Davis, P. E. Pittman, W. R. Kenealy, and T. W. Jeffries. 1996. Increased xylose reductase activity in the xylose-fermenting yeast Pichia stipitis by overexpression of XYL1. Appl. Biochem. Biotechnol. 57-58:267-276. [DOI] [PubMed] [Google Scholar]
- 17.Diamandis, E. P., C. L. Grass, R. Uldall, D. Mendelssohn, and D. Maini. 1992. An enzymatic method for measuring serum mannitol and its use in hemodialysis patients. Clin. Biochem. 25:457-462. [DOI] [PubMed] [Google Scholar]
- 18.Dieters, W. December1975. Xylitol production from d-xylose. Swiss patent 560 175.
- 19.Ditzelmuller, G., C. P. Kubicek, W. Wohrer, and M. Rohr. 1984. Xylose metabolism in Pachysolen tannophilus: purification and properties of xylose reductase. Can. J. Microbiol. 30:1330-1336. [Google Scholar]
- 20.Du Preez, J. C., B. van Driessel, and B. A. Prior. 1989. Effect of aerobiosis on fermentation and key enzyme levels during growth of Pichia stipitis, Candida shehatae, and Candida tenuis on d-xylose. Arch. Microbiol. 152:143-147. [Google Scholar]
- 21.Emodi, A. 1978. Xylitol: its properties and food application. Food Technol. 32:20-32. [Google Scholar]
- 22.Furlan, S. A., P. Bouilloud, and H. F. De Castro. 1994. Influence of oxygen on ethanol and xylitol production by xylose fermenting yeasts. Process Biochem. 29:657-662. [Google Scholar]
- 23.Griffin, B. W. 1992. Functional and structural relationships among aldose reductase, L-hexonate dehydrogenase (aldehyde reductase), and recently identified homologous proteins. Enzyme Microbiol. Technol. 14:690-695. [DOI] [PubMed] [Google Scholar]
- 24.Grimshaw, C. E. 1992. Aldose reductase: model for a new paradigm of enzymic perfection in detoxification catalysts. Biochemistry 31:10139-10145. [DOI] [PubMed] [Google Scholar]
- 25.Häcker, B., A. Habenicht, M. Kiess, and R. Mattes. 1999. Xylose utilization: cloning and characterization of the xylose reductase from Candida tenuis. Biol. Chem. 380:1395-1403. [DOI] [PubMed] [Google Scholar]
- 26.Hallborn, J., M. Walfridsson, U. Airaksinen, H. Ojamo, B. Hahn-Hagerdahl, M. Pentilla, and S. Keranen. 1991. Xylitol production by recombinant Saccharomyces cerevisiae. Bio/Technology 9:1090-1095. [DOI] [PubMed] [Google Scholar]
- 27.Handumrongkul, C., D. P. Ma, and J. L. Silva. 1998. Cloning and expression of Candida guilliermondii xylose reductase gene (xyl1) in Pichia pastoris. Appl. Microbiol. Biotechnol. 49:399-404. [DOI] [PubMed] [Google Scholar]
- 28.Ho, N. W. Y., F. P. Lin, S. Huang, P. C. Andrews, and G. T. Tsao. 1990. Purification, characterization, and amino terminal sequence of xylose reductase from Candida shehatae. Enzyme Microbiol. Technol. 12:33-39. [DOI] [PubMed] [Google Scholar]
- 29.Hyvonen, L., P. Koivistoinen, and F. Voirol. 1982. Food technological evaluation of xylitol. Adv. Food Res. 28:373-403. [Google Scholar]
- 30.Jeffries, T. W., and N. Q. Shi. 1999. Genetic engineering for improved xylose fermentation by yeast. Adv. Biochem. Eng. Biotechnol. 65:117-161. [DOI] [PubMed] [Google Scholar]
- 31.Jeppson, H., K. Holmgren, and B. Hahn-Hagerdal. 1999. Oxygen-dependant xylitol metabolism in Pichia stipitis. Appl. Microbiol. Biotechnol. 53:92-97. [Google Scholar]
- 32.Jornvall, H., B. Persson, M. Krook, S. Atrian, R. Gonzlez-Duarte, J. Jeffery, and D. Ghosh. 1995. Short-chain dehydrogenases/reductases (SDR). Biochemistry 34:6003-6013. [DOI] [PubMed] [Google Scholar]
- 33.Kim, S. Y., and D. K. Oh. 1999. Evaluation of xylitol production from corn cob hemicellulose hydrolysate by Candida parapsilosis. Biotechnol. Lett. 21:891-895. [Google Scholar]
- 34.Kuhn, A., C. van Zyl, A. van Tonder, and B. A. Prior. 1995. Purification and partial characterization of an aldo-keto reductase from Saccharomyces cerevisiae. Appl. Environ. Microbiol. 61:1580-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagunas, R., and J. M. Gancedo. 1973. Reduced pyridine-nucleotides balance in glucose-growing Saccharomyces cerevisiae. Eur. J. Biochem. 37:90-94. [DOI] [PubMed] [Google Scholar]
- 36.Leäo, C., and N. van Uden. 1982. Effects of ethanol and other alkanols on the glucose transport system of Saccaromyces cerevisiae. Biotechnol. Bioeng. 24:2601-2604. [DOI] [PubMed] [Google Scholar]
- 37.Lee, H. 1998. The structure and function of yeast xylose (aldose) reductase. Yeast 14:77-984. [DOI] [PubMed] [Google Scholar]
- 38.Lee, J. K., K. W. Hong, and S. Y. Kim. 2003. Purification and properties of a NADPH-dependent erythrose reductase from the newly isolated Torula corallina. Biotechnol. Progress 19:495-500. [DOI] [PubMed] [Google Scholar]
- 39.Lewis, D. H., and D. C. Smith. 1967. Sugar alcohols (polyols) in fungi and green plants. I. Distribution, physiology and metabolism. New Phytol. 66:143-184. [Google Scholar]
- 40.Meinander, N., B. Hahn-Hagerdal, M. Linko, P. Linko, and H. Ojamo. 1994. Fed-batch xylitol production with recombinantXYL-1-expressing Saccharomyces cerevisiae using ethanol as a co-substrate. Appl. Microbiol. Biotechnol. 42:334-339. [Google Scholar]
- 41.Metzger, M. H., and C. P. Hollenberg. 1995. Amino acid substitutions in the yeast Pichia stipitis xylitol dehydrogenase coenzyme-binding domain affect the coenzyme specificity. Eur. J. Biochem. 228:50-54. [DOI] [PubMed] [Google Scholar]
- 42.Mishra, P., and A. Singh. 1993. Microbial pentose utilization. Adv. Appl. Microbiol. 39:91-152. [DOI] [PubMed] [Google Scholar]
- 43.Morjana, N. A., C. Lyons, and T. G. Flynn. 1989. Aldose reductase from human psoas muscle. Affinity labeling of an active site lysine by pyridoxal 5′-diphospho-5′-adenosine. J. Biol. Chem. 264:2912-2919. [PubMed] [Google Scholar]
- 44.Neuhauser, W., D. Haltrich, K. D. Kulbe, and B. Nidetzky. 1997. NAD(P)H-dependent aldose reductase from the xylose-assimilating yeast Candida tenuis. Biochem. J. 326:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh, D. K., S. Y. Kim, and J. H. Kim. 1998. Increase of xylitol production rate by controlling redox potential in Candida parapsilosis. Biotechnol. Bioeng. 58:440-444. [DOI] [PubMed] [Google Scholar]
- 46.Oh, D. K., and S. Y. Kim. 1998. Increase of xylitol yield by feeding xylose and glucose in Candida tropicalis. Appl. Microbiol. Biotechnol. 50:419-425. [DOI] [PubMed] [Google Scholar]
- 47.Oh, D. K., and S. Y. Kim. 1997. Xylitol production from xylose by Candida tropicalis DS-72. Korean J. Appl. Microbiol. Biotechnol. 25:311-316. [Google Scholar]
- 48.Oura, E. 1977. Reaction products of yeast fermentations. Process Biochem. 12:19-21. [Google Scholar]
- 49.Rizzi, M., P. Erlemann, N. A. Bui-Thanh, and H. Dellweg. 1988. Xylose fermentation by yeasts. 4. Purification and kinetic studies of xylose reductase from Pichia stipitis. Appl. Microbiol. Biotechnol. 29:148-154. [Google Scholar]
- 50.Rudolph, F. B. 1979. Product inhibition and abortive complex formation. Methods Enzymol. 63:411-436. [DOI] [PubMed] [Google Scholar]
- 51.Ruzicka, F. J., K. W. Lieder, and P. A. Frey. 2000. Lysine 2,3-aminomutase from Clostridium subterminale SB4: mass spectral characterization of cyanogen bromide-treated peptides and cloning, sequencing, and expression of the gene kamA in Escherichia coli. J. Bacteriol. 182:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 53.Schiestl, R. H., and D. Gietz. 1989. High efficiency transformation of intact yeast cells by single stranded nucleic acids as carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 54.Schneider, H., H. Lee, M. Barbosa, C. Kubicek, and A. James. 1989. Physiological properties of a mutant of Pachysolen tannophilus deficient in NADPH-dependent d-xylose reductase. Appl. Environ. Microbiol. 55:2877-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takuma, S., N. Nakashima, M. Tantirungkij, S. Kinoshita, H. Okada, T. Seki, and T. Yoshida. 1991. Isolation of xylose reductase gene of Pichia stipitis and its expression in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 28:327-340. [DOI] [PubMed] [Google Scholar]
- 56.Vandeska, E., S. Kuzmanov, and T. W. Jeffries. 1995. Xylitol formation and key enzyme activities in Candida boidinii under different oxygen transfer rates. J. Ferment. Bioeng. 80:513-516. [Google Scholar]
- 57.Van Zyl, C., B. A. Prior, S. G. Kilian, and J. L. F. Kock. 1989. d-Xylose utilization by Saccharomyces cerevisiae. J. Gen. Microbiol. 135:2791-2798. [DOI] [PubMed] [Google Scholar]
- 58.Verduyn, C., R. Van Kleef, J. Frank, H. Schreuder, J. P. van Dijken, and W. A. Scheffers. 1985. Properties of the NAD(P)H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem. J. 226:669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walfridsson, M., M. Anderlund, X. Bao, and B. Hahn-hagerdal. 1997. Expression of different levels of enzymes from Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilization. Appl. Microbiol. Biotechnol. 48:218-224. [DOI] [PubMed] [Google Scholar]
- 60.Webb, S. R., and H. Lee. 1990. Regulation of d-xylose utilization by hexoses in pentose-fermenting yeasts. Biotechnol. Adv. 8:685-697. [DOI] [PubMed] [Google Scholar]
- 61.Yokoyama, S., T. Suzuki, K. Kawai, H. Horitsu, and K. Takamizawa. 1995. Purification, characterization and structure analysis of NADPH dependent d-Xylose reductases from Candida tropicalis. J. Ferment. Bioeng. 79:217-223. [Google Scholar]
- 62.Yokoyama, S. I., Y. Kinoshita, T. Suzuki, K. Kawai, H. Horitsu, and K. Takamizawa. 1995. Cloning and sequencing of two d-xylose reductase genes (xyrA and xyrB) from Candida tropicalis. J. Ferment. Bioeng. 80:603-605. [Google Scholar]