Abstract
Escherichia coli is being developed as a biocatalyst for bulk chemical production from inexpensive carbohydrates derived from lignocellulose. Potential substrates include the soluble xylodextrins (xyloside, xylooligosaccharide) and xylobiose that are produced by treatments designed to expose cellulose for subsequent enzymatic hydrolysis. Adjacent genes encoding xylobiose uptake and hydrolysis were cloned from Klebsiella oxytoca M5A1 and are functionally expressed in ethanologenic E. coli. The xylosidase encoded by xynB contains the COG3507 domain characteristic of glycosyl hydrolase family 43. The xynT gene encodes a membrane protein containing the MelB domain (COG2211) found in Na+/melibiose symporters and related proteins. These two genes form a bicistronic operon that appears to be regulated by xylose (XylR) and by catabolite repression in both K. oxytoca and recombinant E. coli. Homologs of this operon were found in Klebsiella pneumoniae, Lactobacillus lactis, E. coli, Clostridium acetobutylicum, and Bacillus subtilis based on sequence comparisons. Based on similarities in protein sequence, the xynTB genes in K. oxytoca appear to have originated from a gram-positive ancestor related to L. lactis. Functional expression of xynB allowed ethanologenic E. coli to metabolize xylodextrins (xylosides) containing up to six xylose residues without the addition of enzyme supplements. 4-O-methylglucuronic acid substitutions at the nonreducing termini of soluble xylodextrins blocked further degradation by the XynB xylosidase. The rate of xylodextrin utilization by recombinant E. coli was increased when a full-length xynT gene was included with xynB, consistent with xynT functioning as a symport. Hydrolysis rates were inversely related to xylodextrin chain length, with xylobiose as the preferred substrate. Xylodextrins were utilized more rapidly by recombinant E. coli than K. oxytoca M5A1 (the source of xynT and xynB). XynB exhibited weak arabinosidase activity, 3% that of xylosidase.
Cellulose and hemicellulose (primarily methylglucuronoxylan) are the most abundant carbohydrate constituents of woody biomass and agricultural residues (2, 9). High cost associated with the depolymerization of these polymers into monomeric sugars is a primary obstacle preventing their use as a feedstock for chemicals and automotive fuels (31, 46). All native lignocellulosic materials must be pretreated to solubilize hemicellulose constituents and expose cellulose surfaces prior to enzymatic degradation. Although hemicellulose can be depolymerized by mineral acids, conditions required for complete hydrolysis generate toxins that complicate biological utilization (20, 29, 33, 49). Conditions which are less severe generate soluble xylodextrins (xylooligosaccharides) that must be further degraded prior to entering pentose metabolism.
Xylodextrin utilization has been demonstrated in a variety of bacteria (10, 15, 32, 40, 43, 44, 47). Bacillus stearothermophilus contains a gene cluster involved in the transport and metabolism of large soluble products from methylglucuronoxylan (40). To facilitate the bioconversion of xylodextrins into useful chemicals, such as ethanol, genes encoding xylosidase and xylanase have been expressed in Saccharomyces cerevisiae (23, 25, 26) but with limited success in xylan fermentation. Ethanologenic strains of Escherichia coli KO11 and Klebsiella oxytoca M5A1(pLOI555) expressing bacterial xylosidase and xylanase genes have been shown to metabolize xylan and soluble xylodextrins by using a complicated two-step process (6). None of these studies, however, have included heterologous genes encoding xylobiose uptake systems to facilitate metabolism.
Isoprimeverose, a xyloside dimer composed of xylose linked α1,6 to glucose, is transported by a proton symport in Lactobacillus pentosus (8, 16, 17). This gene has been cloned and expressed at high levels in L. lactis for detailed investigations of transport. Xylobiose uptake in Streptomyces lividans appears to utilize a different mechanism involving an ATP-dependent transport system (19). Fungi such as Aureobasidium pullulans transport xylobiose by an uncharacterized, energy-dependent permease (30).
Previous investigations in this and other laboratories developed ethanologenic strains of E. coli and K. oxytoca M5A1 that metabolize all of the monomeric sugar constituents in lignocellulose (37, 38, 45). Subsequent studies characterized genes from M5A1 encoding a cellobiose phosphotransferase system and phosphocellobiase (27) and added these genes to ethanologenic derivatives of E. coli (34). M5A1 was also found to hydrolyze chromogenic xylosides and to metabolize xylobiose (6), consistent with the presence of an efficient xylosidase activity and uptake system.
In this paper we report the cloning and characterization of the xynTB operon encoding a xylobiose/cation symport and a new xylosidase (glycosyl hydrolase family 43). Functional expression of these K. oxytoca M5A1 genes in ethanologenic E. coli KO11 enabled the metabolism of soluble β-1,4-linked xylodextrins containing up to six xylosyl residues.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Cultures of K. oxytoca M5A1 and E. coli were grown at 37°C in Luria-Bertani (LB) medium supplemented with sugar as indicated. Ampicillin (100 μg/ml) was used for plasmid selection. Bacterial cell mass was estimated by measuring optical density at 550 nm (OD550) by using a Bausch & Lomb Spectronic 70 spectrophotometer (330 mg of dry cell weight per liter at an OD550 of 1.0).
TABLE 1.
Description of strains and plasmids used in this study
| Strains and plasmids | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli KO11 | Δfrd, CmR, carrying the Zymomonas mobilis pdc adhB cassette | 38 |
| E. coli TOP10F′ | lacIqlacZΔM15 | Invitrogen |
| K. oxytoca M5A1 | Wild type | 37 |
| Plasmids | ||
| pCR2.1-TOPO | 3.9 Kbp, KmR AmpR, pUC origin, TA cloning vector | Invitrogen |
| pNEB193 | AmpR cloning vector similar to pUC19 | New England Biolabs |
| pLOI3701 | pUC18 derivative with ∼7-kbp DNA fragment, xynT′ xynB | This study |
| pLOI3702 | pUC18 derivative with ∼6.3-kbp DNA fragment, xynT′ xynB | This study |
| pLOI3703 | pUC18 derivative with ∼5.6-kbp DNA fragment, xynT′ xynB | This study |
| pLOI3704 | pCR2.1-TOPO derivative with ∼6.0-kbp DNA fragment, xynT′ xynB | This study |
| pLOI3705 | pCR2.1-TOPO derivative with ∼6.0-kbp DNA fragment, xynTB operon | This study |
| pLOI3706 | derivative of pLOI3705 with all Klebsiella DNA removed between the two vector EcoRI sites | This study |
| pLOI3707 | pNEB193 derivative carrying the 3.6-kbp AseI-PstI fragment with the complete xynTB coding regions (lacking regulatory sites); transcribed opposite to the lac promoter | This study |
| pLOI3708 | pNEB193 derivative containing the 3.6-kbp AseI-PstI fragment with the complete xynTB coding regions (lacking regulatory sites); transcribed in the same direction as the lac promoter | This study |
| pLOI3709 | Derivative of pLOI3708, ΔxynT, xynB (918-bp ClaI internal deletion of xynT) | This study |
Isolation of clones containing K. oxytoca xylosidase gene.
A pUC18 library containing 4- to 6-kbp Sau3AI fragments of K. oxytoca chromosomal DNA (27) was transformed into E. coli DH5α. A second K. oxytoca library was prepared from 6- to 9-kbp Sau3AI fragments by using vector pCR2.1-TOPO (Invitrogen, Carlsbad, Calif.). Both libraries were screened for xylosidase activity (fluorescent colonies) by using LB plates containing ampicillin and 10 μg of 4-methylumbelliferyl 7-β-d-xylopyranoside (MUX)/ml.
Preparation of GAX1 and GAX2 standards.
Aldouronic acid standards, 2′-O-(4-O-methyl-α-d-glucopyranosyluronic acid)-d-xylose (methylglucuronoxylose [GAX1]) and 2′-O-(4-O-methyl-α-d-glucopyranosyluronic acid)-d-xylobiose (methylglucuronoxylobiose [GAX2]), were prepared from 4-O-methylglucuronoxylan using a modification of methods described by Jones et al. (21). 4-O-methylglucuronoxylan was isolated from sweet gum (Liquidambar styraciflua) by alkaline extraction and was structurally defined by 13C nuclear magnetic resonance (NMR) spectroscopy (22). After partial acid hydrolysis in 0.1 N H2SO4 at 122°C (30 min), GAX1 and GAX2 were purified by using gel filtration (BioGel P2 [Bio-Rad, Hercules, Calif.] in 0.05 M formic acid). Products were identified by 1H and 13C NMR (K. Zuobi-Hasona, F. M. St. John, J. D. Rice, and J. F. Preston, unpublished data). Uncoupled NMR spectra were obtained by using a Nicollet NT-300 spectrometer at 25°C in the Fourier transform mode at 300 MHz for 1H and 75.45 MHz for 13C. Structural assignments for peaks were made on the basis of those reported for aldouronic acids (1H/13C 2D-NMR) from sunflower (3) and birchwood (7) hemicellulose.
Preparation of soluble xylodextrin.
A mixture of xylodextrins was prepared by partial acid hydrolysis of birchwood methylglucuronoxylan (Sigma, St. Louis, Mo.) with trifluoroacetic acid (13). Approximately 38 mg of methylglucuronoxylan was mixed with 7.5 ml of 1.6 N trifluoroacetic acid in a 15-ml screw-cap tube. Xylan was dispersed by using an ultrasonic water bath prior to hydrolysis at 100°C for 90 min (manual mixing). After cooling to room temperature, hydrolysates were neutralized over a 15-min period by adding anion exchange resin (∼1.5 g of Amberlite IRA 400 in the -OH form). Neutralized hydrolysates were filtered, lyophilized, and dissolved in 150 μl of distilled water (∼125 mg of total carbohydrate per ml). Based on thin-layer chromatography and densitometry, approximately half of the soluble xylodextrins contained six or fewer sugar residues: 7.4% xylose, 11.5% xylobiose (X2), 9.6% xylotriose (X3), 5.7% xylotetrose (X4), 7.4% xylopentose (X5), and 3.9% xylohexose (X6). Additional compounds were identified as methylglucuronoxylosides but were not quantified.
Fermentation of xylodextrins.
Seed cultures of K. oxytoca M5A1- and E. coli KO11-harboring plasmids were grown in 250-ml flasks containing 50 ml of LB broth (5% xylose) for 12 h (37°C, 125 rpm). Sufficient culture was harvested by centrifugation to provide 0.17 mg of dry cell weight (approximately 1 ml of cells at an OD550 of 0.5), washed twice with 1 ml of LB lacking sugar, and resuspended in 50 μl of filter-sterilized LB containing 50% (vol/vol) soluble xylodextrins (hydrolysate). Small samples (10 μl) were removed during incubation at 37°C and were stored frozen. Xylodextrins were separated by thin-layer chromatography as described previously (50). After visualizing with N-(1-naphthyl)ethylenediamine reagent (4), relative amounts were estimated by densitometry using Quantity One Software and a VersaDoc Imaging System Model 1000 (Bio-Rad).
Measurement of xylosidase and arabinosidase activities.
Cultures of K. oxytoca M5A1 and E. coli KO11 were grown to half maximal density (OD550 of ∼2.0) in LB containing 5% total sugar (glucose, xylose, or a combination of both). Sufficient culture was harvested by centrifugation to provide approximately 0.33 mg of dry cell weight, washed with 50 mM sodium phosphate buffer (pH 6.8), and resuspended in 1 ml of the same buffer. Cells were permeablized by mixing with 1 drop of 0.1% sodium dodecyl sulfate (SDS) and 2 drops of chloroform for 10 s using a Vortex mixer. Dilutions of permeabilized cells were assayed at 37°C using either 1 mM p-nitrophenyl β-d-xylopyranoside (pNP-XP) or 1 mM p-nitrophenyl-α-l-arabinofuranoside (pNP-AF) as substrate (1 ml total volume). Reactions were terminated by adding 2 ml of 0.5 M sodium carbonate. Protein concentrations were measured by using the Bradford reagent (Bio-Rad). Activity is expressed as nanomoles of p-nitrophenol released per minute per milligram of protein.
Construction of a xynTB expression plasmid.
The 3.6-kbp AseI-PstI fragment from pLOI3705 containing the ribosomal binding site and full coding region for xynT and xynB (lacking the CRP and XylR regulatory regions) was gel purified. This DNA fragment was blunt ended, ligated behind the lac promoter (PmeI site) in pNEB193 (New England Biolabs, Beverly, Mass.), and transformed into E. coli TOP10F′ (Invitrogen). Both orientations were recovered and were designated pLOI3708 (forward with respect to lac promoter) and pLOI3707 (reverse). For measurement of xylosidase activity, cultures were grown in LB broth lacking sugar. At an OD550 of 0.7, each culture was divided into two flasks. Isopropyl β-d-thiogalactopyranoside (IPTG; 1 mM) was added to one while the other served as a control. After 2 h of further incubation, cells were harvested by centrifugation and were assayed for β-xylosidase and α-arabinosidase activities. A portion of each culture was also harvested and washed in TE buffer (10 mM Tris-Cl, 1 mM EDTA [pH 8.0]) for protein analysis by SDS-polyacrylamide gel electrophoresis (PAGE) (10 to 15% acrylamide gradient) using a PhastGel system (Amersham, Piscataway, N.J.).
Phylogenetic analyses.
Homologs of XynB and XynT were identified by BLASTP search (1). Protein sequences were aligned using ClustalX version 1.81 (42). Phylogenetic trees were constructed using MEGA (v.2.1) (http://www.megasoftware.net) (24). Phylogenetic relationships were inferred by using the neighbor-joining algorithm and were tested by bootstrap analysis with 1,000 repetitions.
Nucleotide sequence accession number.
The sequence for the K. oxytoca xynTB operon and upstream region was submitted to GenBank and was assigned accession number AY297960.
RESULTS AND DISCUSSION
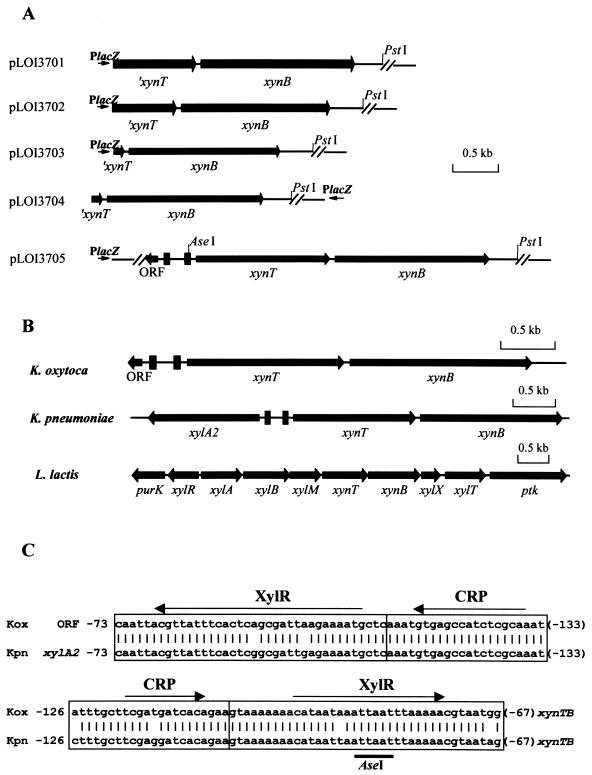
Isolation of the K. oxytoca xynTB operon. Five unique clones exhibiting β-xylosidase activity were initially isolated as fluorescent colonies (MUX positive) from the 4- to 6-kbp K. oxytoca library. Two were weakly fluorescent and contained the previously characterized K. oxytoca casB gene encoding phosphocellobiase (27). The three remaining clones (Fig. 1A) contained an identical open reading frame (ORF) encoding a 559-amino-acid product (64 kDa) that shared homology with a putative xylosidase (XynB) from Lactococcus lactis (accession number NP_267661). This new K. oxytoca gene was also designated xynB. Based on a SignalP analysis (http://www.cbs.dtu.dk/services/SignalP/) (35), K. oxytoca xynB was predicted to encode a cytoplasmic protein that lacks a signal peptide. All three MUX-positive clones (pLOI3701, pLOI3702, and pLOI3703) also included an upstream incomplete ORF that resembled the carboxy terminus of a transport protein in L. lactis.
FIG. 1.
Genes encoding β-xylosidase and xylobiose uptake. (A) K. oxytoca library clones. AseI and PstI restriction sites were used to subclone the full-length xynT and xynB genes lacking CRP and XylR regulatory sequences (solid bars). The directions of the vector lacZ promoters are also shown here. A single scale bar denotes size for all plasmids. (B) Comparison of gene organization in the xynTB region. Each solid rectangle represents a contiguous set of CRP and XylR regulatory sequences. Individual scale bars are included for each plasmid. (C) Comparison of CRP and XylR regulatory sequences associated with the xynTB operon and respective upstream genes in K. oxytoca M5A1 and K. pneumoniae. Sequences are numbered relative to start codons for the unidentified ORF (K. oxytoca M5A1), xylA2 (K. pneumoniae), and xynT (both organisms). Thin arrows indicate direction of transcription for the respective genes.
A second K. oxytoca gene library was constructed to facilitate isolation of the complete sequence for the putative transport protein. One MUX-positive clone was recovered that contained both genes (pLOI3705). An additional clone was recovered that contained a truncated transport gene (pLOI3704). Using the TMpred program (http://www.ch.embnet.org/software/TMPRED_form.html), the putative transporter gene was predicted to encode a membrane protein (484 amino acids) containing at least 10 transmembrane helices. This translated sequence was very similar to XynT in L. lactis (accession number NP_267662; 83% identity). Based on the proximity of the two K. oxytoca genes and their concordant direction of transcription, both genes are presumed to form a single transcriptional unit, the xynTB operon (Fig. 1B).
A divergently transcribed, incomplete ORF was also identified that began 425 bp upstream from the start codon for K. oxytoca xynT (Fig. 1B). This ORF exhibited no significant homology to other sequences in the database. Comparison of K. oxytoca xynT and xynB to the unannotated sequence for K. pneumoniae (http://genome.wustl.edu) readily identified corresponding genes. In K. pneumoniae, however, a xylA homolog denoted xylA2 resides upstream from the xynTB region (Fig. 1B) (28). In L. lactis, this upstream region contains a xylose mutarotase (xylM) required for the efficient metabolism of xylan (14). Based on an in silico analysis of the K. pneumoniae genome (28), potential regulatory sites for catabolite repression (CRP) and for xylose induction by XylR were predicted to be within the ORF-xynTB intergenic region. These sequences were very similar in K. oxytoca M5A1 and K. pneumoniae (Fig. 1C) despite the difference in upstream genes (unidentified ORF and xylA2, respectively). In contrast, the gram-positive regulatory sites for xylose (XylR) and CRP (CcpA) were absent in the intergenic region of xylR-xylABM xynTBX in L. lactis (39).
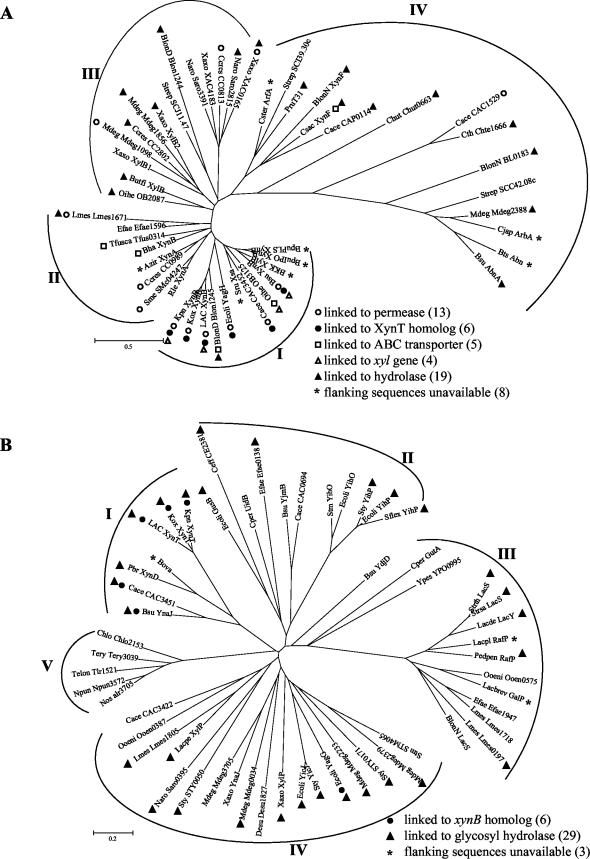
Sequence comparisons and phylogenetic analyses of XynB homologs.
A BLASTP search of protein sequences identified more than 70 homologs of XynB, 48 of which were selected for phylogenetic analyses (Fig. 2A). Experimental verification of hydrolase activity is limited in this group and includes only xynB from Bacillus pumilus (26, 47), xylB from Bacillus sp. strain KK-1 (10), xsa from Selenomonas ruminantium (44), xylB from Butyrivibrio fibrisolvens (43), arbA from Cellvibrio japonicus (36), and Abn from Bacillus thermodenitrificans TS-3 (41). Xylosidase activity was reported in GenBank entries for xynF from Caldicellulosiruptor saccharolyticum and xynA from Azospirillum irakense without supporting data. Most XynB homologs are from gram-positive organisms consistent with an early gram-positive ancestor.
FIG.2.
Unrooted phylogenetic trees of XynB (A) and XynT (B) homologs. Trees have been provisionally assigned into different groups based on similarities in primary structure of XynB (groups I to IV) and XynT (groups I to V), respectively, and the functions of neighboring genes. Abbreviations for the organisms and their XynB and XynT homologs are listed in alphabetical order with accession numbers in parentheses: Azir, Azospirillum irakense, XynA (AAF66622); Bha, Bacillus halodurans, XynB (BAB07402); BKK, Bacillus sp. strain KK-1, XylB (AAC27699); BlonD, B. longum DJO10A, Blon1245 (ZP_00121429), Blon1244 (ZP_00121428); BlonN, B. longum NCC2705, XynF (AAN25335), BL0183 (AAN24037), LacS (NP_696148); BpuPLS, B. pumilus strain PLS, XynB (AAC97375); BpuIPO, B. pumilus strain IPO, XynB (S19729); Bsu, B. subtilis, XynB (AAB41091), AbnA (CAA99586), YnaJ (NP_389639), YjmB (NP_389113), YdjD (NP_388497); Bts, Bacillus thermodenitrificans TS-3, Abn (BAB64339); Butfi, B. fibrisolvens, XylB (A49776); Cace, C. acetobutylicum, CAC3452 (AAK81382), CAC1529 (NP_348156), CAP0115 (NP_149278), CAC3451 (NP_350041), CAC0694 (NP_347331), CAC3422 (NP_350012); Ccres, C. crescentus CB15, CC0989 (AAK22973), CC2802 (AAK24766), CC0813 (AAK22798); Ceff, Corynebacterium efficiens YS-314, CE2381 (NP_738991); Chlo, Chloroflexus aurantiacus, Chlo2153 (ZP_00019154); Chut, Cytophaga hutchinsonii Chut0663 (ZP_00117295); Cjap, C. japonicus, ArbA (CAA71485); Cper, C. perfringens, UidB (NP_561069), GutA (NP_561685); Csac, Caldicellulosiruptor saccharolyticus, XynF (AAB87371); Cster, Caldicellulosiruptor stercorarium, ArfA (CAD48310); Cth, Caldicellulosiruptor thermocellum ATCC 27405, Chte1666 (ZP_00061257); Desu, Desulfitobacterium hafniense, Desu1827 (ZP_00098712); Ecoli, E. coli K12, YagH (P77713), YagG (NP_414804), GusB (AAA68924), YihO (NP_418312), YihP (AAC76874), YicJ (AAC76681); Efae, Enterococcus faecium, Efae1596 (ZP_00036717), Efae0138 (ZP_00035300), Efae1947 (ZP_00037059); Kpn, K. pneumoniae unfinished genome sequence (NC_002941; Washington University, http://genome.wustl.edu); LAC, L. lactis subsp. lactis IL-1403, XynB (AAK05603), XynT (NP_267662); Lacpe, L. pentosus, XylP (P96792); Lacbrev, L. brevis, GalP (AAK54067); Lacpl, L. plantarum, RafP (AAL09166); Lacde, L delbrueckii subsp. bulgaricus, LacY (P22733); Lmes, L. mesenteroides subsp. mesenteroides ATCC 8293, Lmes1671 (ZP_00064177), Lmes1718 (ZP_00064224), Lmes0197 (ZP_00062720), Lmes1805 (ZP_00064310); Mdeg, M. degradans 2-40, Mdeg1098 (ZP_00065724), Mdeg2388 (ZP_00066989), Mdeg1856 (ZP_00066469), Mdeg2233 (ZP_00066837), Mdeg0034 (ZP_00064673), Mdeg3705 (ZP_00068288), Mdeg2379 (ZP_00066980); Naro, Novosphingobium aromaticivorans, Saro3391 (ZP_00096353), Saro2815 (ZP_00095782), Saro0395 (ZP_00093393); Nos, Nostoc sp. strain PCC 7120, alr3705 (NP_487745); Npun, N. punctiforme, Npun3572 (ZP_00109128); Oihe, Oceanobacillus iheyensis, OB3125 (BAC15081), OB2087 (BAC14043); Ooeni, Oenococcus oeni MCW, Ooen0387 (ZP_00069379), Ooen0575 (ZP_00069564); Pbr, P. bryantii, XynD (CAD21012); Pedpen: Pediococcus pentosaceus RafP (P43466); Pru T31, Prevotella ruminicola strain T31 (BAA78558); Rle, Rhizobium leguminosarum bv. trifolii, XynA (AAL14914); Sflex, Shigella flexneri 2a strain 301, YihP (NP_709677); Sme, Sinorhizobium meliloti, SMc04247 (CAC46496); Sru, Selenomonas ruminantium, Xsa (AAB97967); Stm, Salmonella enterica serotype Typhimurium LT2, STM4065 (NP_462946), and YihO (NP_462897); Strep, Streptomyces coelicolor A3 (2), SCJ11.47 (CAB52932), SCC42.08c (CAB92901), and SCI39.30c (CAD55182); Strsa, Streptococcus salivarius, LacS (AAL67293); Strth, S. thermophilus A147, LacS (P23936); Sty, S. enterica subsp. enterica serovar Typhi, STY0171 (NP_454763), YicJ (NP_458178), YihP (NP_458032), and STY0050 (NP_454653); Telon, Thermosynechococcus elongatus BP-1, Tlr1521 (NP_682311); Tery, Trichodesmium erythraeum IMS101, Tery3039 (ZP_00073765); Tfusca, Thermobifida fusca, Tfus0314 (ZP_00056973); Xaxo, Xanthomonas axonopodis pv. citri strain 306, XylB1 (AAM36146), XylB2 (AAM39065), XAC0165 (AAM35057), XAC4183 (AAM39018), XylP (NP_644556), YnaJ (NP_644495); Ypes, Yersinia pestis, YPO0995 (NP_404610).
Operons that include xylobiose transporters adjacent to hydrolases appear common in nature and may provide an evolutionary advantage (Fig. 2A). Among the 39 XynB homologs for which flanking sequences were available, 12 xylosidase genes were adjacent to a putative permease and 5 were adjacent to a putative ATP-dependent ABC transporter. Of the 12 with adjacent permeases, 5 encoded homologs of K. oxytoca XynT. All appear to contain operons resembling K. oxytoca xynTB: B. subtilis (ynaJ-xynB), E. coli (yagG-yagH), C. acetobutylicum (CAC3451-CAC3452, xynD), L. lactis (xynT-xynB), and K. pneumoniae (xynT-xynB). Nineteen of the XynB homologs were found adjacent to other glycosyl hydrolase genes, a particularly common feature in organisms that degrade xylan and cellulose. Four were adjacent to genes directly involved in xylose catabolism.
XynB homologs can be readily organized into four groups based on the phylogenetic tree structure (Fig. 2A) and their genomic neighboring genes. Group I is the most cohesive group and includes K. oxytoca XynB and 11 other members. This group contains all organisms with putative xynTB operons and all organisms with xynB adjacent to xylose catabolism genes. Two members of this group are adjacent to putative ABC transporters. Within Group I, K. oxytoca XynB shared the highest homologies (Fig. 3) with XynB from K. pneumoniae (identifier RKP04766, 89% identity; Integrated Genomics, Inc.) (28), XynB from L. lactis (81% identity), and Blon1245 (68% identity) from Bifidobacterium longum. These four genes form a closely related subgroup with lower homology to other xylosidases (<50% identity). The E. coli homolog YagH also showed significant homology (46% identity). The similarities of XynB and XynT to respective homologs in L. lactis and K. pneumoniae are consistent with a gram-positive ancestor of L. lactis. It is interesting that the cryptic homologs in E. coli are located in a cluster of genes surrounded by insertion sequences which may indicate a transposon-mediated acquisition. Limited sequence is available in the region encoding XynB homologs for four members of this group. B. subtilis and C. acetobutylicum also contained adjacent homologs of XynT and XynB that are more distantly related.
FIG. 3.
Comparison of translated sequences for K. oxytoca XynB (KOX) and homologs from K. pneumoniae (KPN), L. lactis (LAC), B. longum (BLON; Blon1245), and E. coli K12 YagH. Residues which are identical in three of the five sequences are shaded in black. Solid circles above residues D15, F85, D128, and E188 in KOX correspond to residues D18, F114, D158, and E221 from C. japonicus ArbA that are involved in catalytic activity and substrate binding.
XynB homologs in groups II and III are more diverse than those in Group I. Group II primarily contains XynB homologs that are adjacent to transport genes unrelated to K. oxytoca XynT. In groups III and IV, most homologs are located adjacent to hydrolases for carbohydrate polymers. Group IV also includes XynB homologs that exhibit arabinase/arabinosidase activities (ArbA from C. japonicus [36], Abn from B. thermodenitrificans TS-3 [41], and XynF from C. saccharolyticus).
All XynB homologs contain the COG3507 domain characteristic of glycosyl hydrolase family 43 (5). The ArbA protein from C. japonicus is the only member in family 43 with a known three-dimensional structure (36). Although overall sequence identity to K. oxytoca XynB was less than 23%, residues (D38, F114, D158, and E221) that are involved in catalytic activity and ligand binding in C. japonicus ArbA were found to align with residues D15, F85, D128, and E188 in K. oxytoca XynB (Fig. 3). Accepting substitutions of F to Y or W, further comparisons revealed that the identities and spacings of these four residues were conserved in all but 2 of the 48 homologs examined.
Sequence comparisons and phylogenetic analyses of XynT homologs.
A BLASTP search using K. oxytoca XynT identified 100 homologous transporters (Fig. 2B), all of which contain the MelB domain (COG2211) characteristic of Na+/melibiose symporters. For most members, additional sequence data was available for adjacent genes. Multiple XynT homologs were found in some organisms, including three in C. acetobutylicum, B. subtilis, and Leuconostoc mesenteroides, four in Microbulbifer degradans, and two in both Xanthomonas axonopodis and X. campestris. None of the XynT homologs have previously been shown to function in xylobiose transport. XylP from L. pentosus has been shown to transport an α-xyloside, isoprimeverose[α-d-xylopyranosyl-(1,6)-d-glucopyranose] (8).
Based on primary structure and adjacent genes, XynT homologs can be organized into at least five groups (Fig. 2B). Group I contains seven members, including XynT from K. oxytoca. As with XynB, K. oxytoca XynT was most similar to K. pneumoniae and L. lactis homologs (94 and 83% identity, respectively), providing further support for the cotransfer of xynT and xynB. Two other transporters, XynD from Prevotella bryantii strain B14 (accession number CAD21012) and a partial ORF from Bacteroides ovatus (S55894), also shared significant homology with K. oxytoca XynT (39 and 41% identity, respectively). The other two members of this group (B. subtilis YnaJ and C. acetobutylicum CAC3451) were more distantly related to XynT (both 34% identity) but contained adjacent genes encoding homologs of K. oxytoca XynB. Remaining members of the XynT tree exhibited lower homologies to K. oxytoca XynT (less than 30% identity). All but Group V (cyanobacteria) included transporters that are adjacent to hydrolases. Lactose, raffinose, and galactoside symports were found in Group III.
Regulation of K. oxytoca xynB expression.
The presence of putative XylR and CRP sites in the promoter region of the K. oxytoca xynTB operon (Fig. 1C) indicates possible regulation by xylose (XylR) and by glucose (CRP). Since E. coli and K. oxytoca are closely related with extensive similarity in regulatory systems and protein sequences, the effect of these sugars was investigated in both organisms (Table 2). Expression of xynB was controlled more tightly in M5A1 than in KO11(pLOI3705). However, activity levels were consistently higher in KO11. Expression of xynB was induced 56-fold in K. oxytoca by xylose in the absence of glucose and was repressed to basal levels in the presence of both sugars. In KO11(pLOI3705), xylose induced xynB expression by 19-fold in the absence of glucose. In the presence of both sugars, expression was partially repressed (sixfold induction) in KO11(pLOI3705). These results demonstrate that the native xynTB operon may be subject to CRP, inducer exclusion, and induction by xylose in both K. oxytoca and KO11. Differences in xylosidase activities and the extent of regulation can be attributed largely to the higher gene dosage of xynTB in KO11 harboring pLOI3705, a high-copy plasmid.
TABLE 2.
β-Xylosidase activities in permeabilized cell preparations of K. oxytoca M5A1 and recombinant E. colia
| Strains and properties | Growth media | β-Xylosidase activity (nmol min−1 mg−1) |
|---|---|---|
| K. oxytoca M5A1 | LB + 5% glucose | 0.5 |
| LB + 2.5% glucose + 2.5% xylose | 0.5 | |
| LB + 5% xylose | 28 | |
| E. coli KO11(pLOI3705) | LB + 5% glucose | 7.5 |
| LB + 2.5% glucose + 2.5% xylose | 43 | |
| LB + 5% xylose | 140 (5.0) | |
| E. coli TOP10F′ (pLOI3708) | LB, no sugar, without IPTG | 520 |
| LB, no sugar, with IPTG | 3,100 (105) | |
| E. coli KO11(pLOI3708) | LB + 5% xylose | 1,020 |
| E. coli KO11(pLOI3709) | LB + 5% xylose | 1,100 |
| E. coli KO11 (pNEB193) | LB + 5% xylose | 0.4 |
| E. coli TOP10F′ | LB, no sugar | <0.4 |
Values represent an average of two or more experiments which differed by less than 25%. Arabinosidase activities are shown in parentheses.
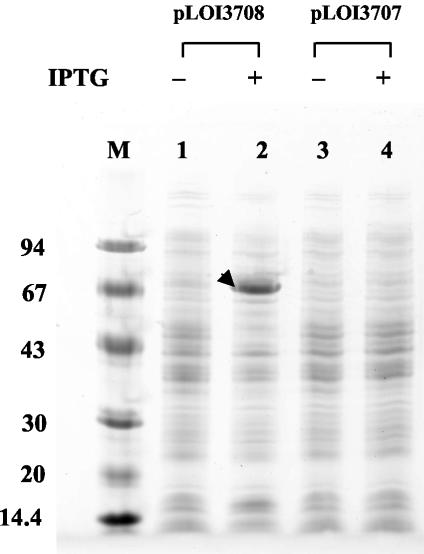
Additional plasmids were constructed to remove the native regulatory elements and place the xynTB operon directly under the control of the lac promoter in pNEB193. Despite the presence of lacIq in the host, E. coli TOP10F′(pLOI3708) expressed much higher levels of xylosidase in the presence and absence of IPTG than recombinant KO11(pLOI3705) containing the native regulatory elements. Little or no xylosidase activity was observed in TOP10F′ or in KO11(pLOI3706), which lacks K. oxytoca DNA. The high level of xynB induction in TOP10F′(pLOI3708) was apparent in protein gels (Fig. 4). A prominent 65-kDa protein band corresponding to XynB was clearly visible in IPTG-induced cells carrying the xynTB genes. This band was absent in an analogous construct in which the direction of xynTB transcription was reversed (pLOI3707). Although no new band corresponding to XynT (54 kDa) was detected, this membrane protein may be expressed at lower levels or may be poorly solubilized.
FIG. 4.
SDS-PAGE comparison of proteins in recombinant strains of E. coli TOP10F′ containing xynTB coding regions in the forward (pLOI3708) and reverse (pLOI3707) orientations with respect to the lac promoter. Cells were grown and harvested as described for xylosidase assays. Lanes: M, protein standards with molecular mass in kilodaltons indicated on the left; 1, TOP10F′(pLOI3708) without IPTG; 2, TOP10F′(pLOI3708) with 1 mM IPTG; 3, TOP10F′(pLOI3707) without IPTG; 4, TOP10F′(pLOI3707) with 1 mM IPTG. The arrow denotes a 65-kDa band in lane 2 corresponding to XynB.
Several xylosidases have been reported to also serve as efficient arabinosidases (43, 44). However, this does not appear to be the case for XynB. Recombinant XynB was 30-fold less active with p-nitrophenyl-α-l-arabinofuranoside as a substrate than the xylopyranoside derivative (Table 2) and was inactive with p-nitrophenyl-α-l-arabinopyranoside (data not shown).
Xylodextrin utilization by ethanologenic derivatives of E. coli strain KO11.
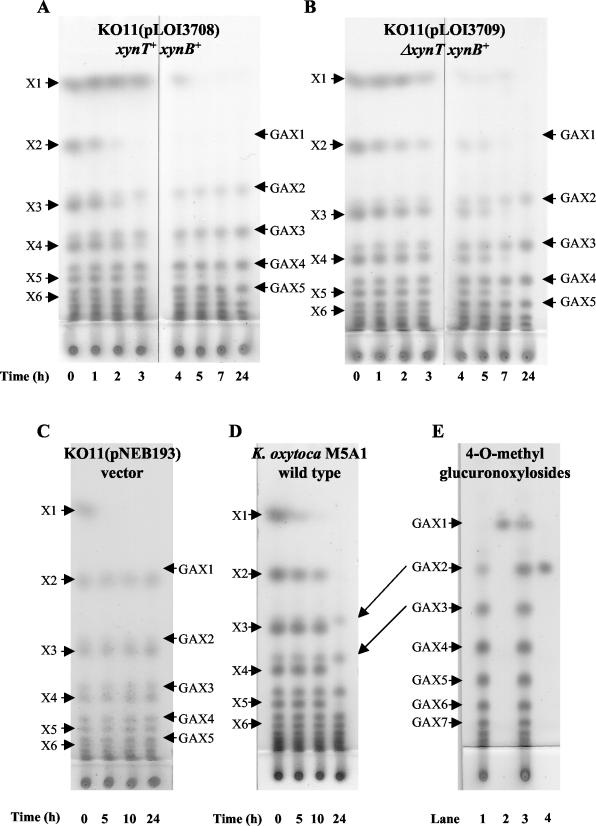
The biological activities of XynT and XynB were confirmed in small-scale fermentations using a homologous series of soluble xylodextrins (partial acid hydrolysate of birchwood methylglucuronoxylan) as natural substrates (Fig. 5A and B). KO11(pLOI3708) harboring the full-length xynT and xynB genes metabolized xylodextrins more rapidly than KO11(pLOI3709) containing an internal deletion in xynT. Differences between these strains were particularly evident between 2 to 7 h of incubation. Since both strains expressed equivalent levels of xylosidase activity (Table 2), the increased rate of metabolism by KO11(pLOI3708) can be attributed to XynT-mediated transport of xylobiose or xylodextrins. After 24 h of incubation, xylodextrins X2 through at least X6 had been fully metabolized by both strains. Similar results were also obtained for a comparison between the library clones pLOI3704 with a truncated xynT gene and pLOI3705 containing the full xynTB operon (data not shown). Only xylose was metabolized by control strains of E. coli containing the vector alone (Fig. 5C). Native K. oxytoca M5A1 metabolized xylodextrins (Fig. 5D) more slowly than recombinant E. coli strains expressing the xynB or the xynTB operon, consistent with observed lower levels of xylosidase activity (Table 2).
FIG.5.
Thin-layer chromatograms of xylodextrins during fermentation. β-Xylosidase activities for all strains are provided in Table 2. (A) KO11(pLOI3708) containing the full coding regions for xynT and xynB. (B) KO11(pLOI3709) containing the coding region for xynB and a nonfunctional xynT (internal deletion). Note that xylosidase activities in permeabilized cell preparations of KO11(pLOI3708) and KO11(pLOI3709) were equivalent. (C) KO11(pNEB193) containing the vector alone. (D) K. oxytoca M5A1. (E) Cochromatography of GAX1 and GAX2 standards with spent medium from a 24-h xylodextrin fermentation with KO11(pLOI3708). Lanes: 1, spent medium; 2, GAX1 alone; 3, spent medium plus GAX1 and GAX2; 4, GAX2 alone. The labels X1 to X6 refer to the number of xylosyl residues, xylose to xylohexose. Methylglucuronoxylosides containing 1 to 7 xylosyl residues are labeled GAX1 to GAX7, respectively. Arrows connecting panels D and E are included to illustrate positions corresponding to GAX2 and GAX3 in both chromatograms. Xylodextrins were identified by comparison to xylobiose, xylotriose, and cellobiosides as standards (6). Longer methylglucuronoxylosides (GAX3 to GAX7) were inferred by migration rates relative to xylodextrins with equivalent numbers of carbohydrate residues.
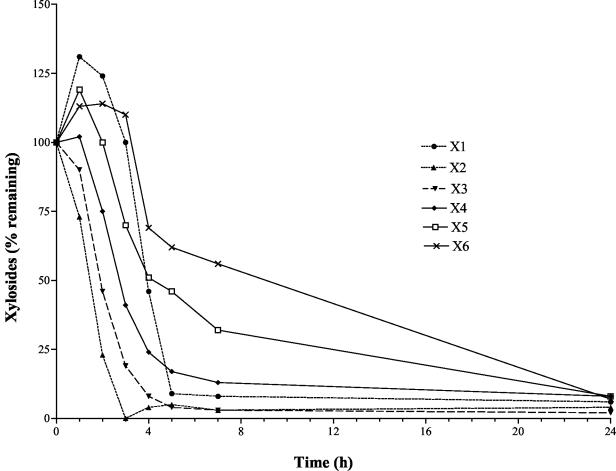
Thin-layer chromatography and densitometry were used to monitor xylodextrin utilization by KO11(pLOI3708) (Fig. 6). The rate of utilization was inversely related to chain length, with xylobiose being metabolized most rapidly. Xylose accumulated initially together with smaller amounts of xylopentose and xylohexose. Despite the production of xylobiose and xylotriose as intermediates during the degradation of longer xylodextrins, both were near the lower limit of detection at incubation times longer than 3 h (Fig. 5A). After 24 h, xylodextrins up to six residues in length had been metabolized.
FIG. 6.
Relative utilization of mixed xylodextrins by KO11(pLOI3708). Xylodextrins were separated by thin-layer chromatography and quantified by densitometry. The labels X1 to X6 refer to the number of xylosyl residues, xylose to xylohexose. Data are expressed as percentages of initial values at time zero.
At least seven soluble components of birchwood methylglucuronoxylan hydrolysate were not metabolized by both K. oxytoca and the recombinant E. coli expressing XynB activities. Most were partially masked by more abundant xylodextrins but were clearly evident after fermentation (Fig. 5A, B, and D). Since the 4-O-methylglucuronic acid substitution of xylan is known to increase the resistance of substituted xylodextrins to chemical hydrolysis (21) and to block degradation by xylanases (11, 12, 48), GAX1 and GAX2 standards were prepared for comparison to the unknown components. Cochromatography of these standards with 24-h broth from xylodextrin fermentations with KO11(pLOI3708) identified the two most mobile spots as GAX1 and GAX2 (Fig. 5E). GAX1 migrated just above xylobiose. Although only a small amount of GAX1 was present before or after fermentation, GAX2 was a prominent component after fermentation, positioned immediately above the xylotriose region. GAX1 and GAX2 appear to represent part of a homologous series of compounds that migrate immediately above each xylodextrin containing an equal number of glycosyl residues (GAX1-GAX7). Resistance to hydrolysis by XynB xylosidase is consistent with a methylglucuronic acid substitution at the nonreducing terminus (18).
Acknowledgments
This research was supported by grants from the U.S. Department of Agriculture (01-35504-10669 and 00-52104-9704), the U.S. Department of Energy (FG02-96ER20222), and the Consortium for Plant Biotechnology Research (GO12026-161). DNA sequences were kindly provided by Jack Shelton (Microbiology Sequencing Facility) and the ICBR DNA Sequencing Core at the University of Florida.
Footnotes
University of Florida Agricultural Experiment Station publication no. R-09658.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aristidou, A., and M. Penttilä. 2000. Metabolic engineering applications to renewable resource utilization. Curr. Opin. Biotechnol. 11:187-198. [DOI] [PubMed] [Google Scholar]
- 3.Bazus, A., L. Rigal, A. Gaset, T. Fontaine, J.-M. Wieruszeski, and B. Fournet. 1993. Isolation and characterization of hemicelluloses from sunflower hulls. Carbohydr. Res. 243:323-332. [DOI] [PubMed] [Google Scholar]
- 4.Bounias, M. 1980. N-(1-napththyl)ethylenediamine dihydrochloride as a new reagent for nanomole quantification of sugars on thin-layer plates by a mathematical calibration process. Anal. Biochem. 106:291-295. [DOI] [PubMed] [Google Scholar]
- 5.Bourne, Y., and B. Henrissat. 2001. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr. Opin. Struct. Biol. 11:593-600. [DOI] [PubMed] [Google Scholar]
- 6.Burchhardt, G., and L. O. Ingram. 1992. Conversion of xylan to ethanol by ethanologenic strains of Escherichia coli and Klebsiella oxytoca. Appl. Environ. Microbiol. 58:1128-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavagna, F., H. Deger, and J. Puls. 1984. 2D-NMR analysis of the structure of an aldotriuronic acid obtained from birch wood. Carbohydr. Res. 129:1-8. [Google Scholar]
- 8.Chaillou, S., P. W. Postma, and P. H. Pouwels. 1998. Functional expression in Lactobacillus plantarum of xylP encoding the isoprimeverose transporter of Lactobacillus pentosus. J. Bacteriol. 180:4011-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandrakant, P., and V. S. Bisaria. 1998. Simultaneous bioconversion of cellulose and hemicellulose to ethanol. Crit. Rev. Biotechnol. 18:295-331. [DOI] [PubMed] [Google Scholar]
- 10.Chun, Y. C., K. W. Jung, J.-C. Lee, S.-H. Park, H. K. Chung, and K.-H. Yoon. 1998. Molecular cloning and the nucleotide sequence of a Bacillus sp. KK-1 β-xylosidase gene. J. Microbiol. Biotechnol. 8:28-33. [Google Scholar]
- 11.Dahlman, O., A. Jacobs, A. Liljenberg, and A. I. Olsson. 2000. Analysis of carbohydrates in wood and pulps employing enzymatic hydrolysis and subsequent capillary zone electrophoresis. J. Chromatogr. A 891:157-174. [DOI] [PubMed] [Google Scholar]
- 12.Davis, M., B. Rosin, L. L. Landucci, and T. W. Jeffries. 1997. Characterization of UV absorbing products released from Kraft pulps by xylanases, p. 435-442. In 1997 Biological sciences symposium. TAPPI Press, Atlanta, Ga.
- 13.Doner, L. W. 1988. High-performance thin-layer chromatography of starch, cellulose, xylan, and chitin hydrolysates. Methods Enzymol. 160:176-180. [Google Scholar]
- 14.Erlandson, K. A., S. C. Delamarre, and C. A. Batt. 2001. Genetic evidence for a defective xylan degradation pathway in Lactococcus lactis. Appl. Environ. Microbiol. 67:1445-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasparic, A., J. Martin, A. S. Daniel, and H. J. Flint. 1995. A xylan hydrolase gene cluster in Prevotella ruminicola B14: sequence relationships, synergistic interactions, and oxygen sensitivity of a novel enzyme with exoxylanase and β-(1,4)-xylosidase activities. Appl. Environ. Microbiol. 61:2958-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heuberger, E. H., L. M. Veenhoff, R. H. Duurkens, R. H. Friesen, and B. Poolman. 2002. Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J. Mol. Biol. 317:591-600. [DOI] [PubMed] [Google Scholar]
- 17.Heuberger, E. H., E. Smits, and B. Poolman. 2001. Xyloside transport by XylP, a member of the galactoside-pentoside-hexuronide family. J. Biol. Chem. 276:34465-34472. [DOI] [PubMed] [Google Scholar]
- 18.Hurlbert, J. C., and J. F. Preston. 2001. Functional characterization of a novel xylanase from corn strains of Erwinia chrysanthemi. J. Bacteriol. 183:2093-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurtubise, Y., F. Shareck, D. Kluepfel, and R. Morosoli. 1995. A cellulase/xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17:367-377. [DOI] [PubMed] [Google Scholar]
- 20.Ingram, L. O., H. C. Aldrich, A. C. C. Borges, T. B. Causey, A. Martinez, F. Morales, A. Saleh, S. A. Underwood, L. P. Yomano, S. W. York, J. Zaldivar, and S. Zhou. 1999. Enteric bacterial catalysts for fuel ethanol production. Biotechnol. Prog. 15:855-866. [DOI] [PubMed] [Google Scholar]
- 21.Jones, J. K. N., C. B. Purves, and T. E. Timmel. 1961. Constitution of a 4-O-methylglucuronoxylan from the wood of trembling aspen (Populus tremuloides Michx). Can. J. Chem. 39:1059-1066. [Google Scholar]
- 22.Kardosova, A., M. Matulova, and A. Malovikova. 1998. (4-O-methyl-α-d-glucurono)-d-xylan from Rudbeckia fulgida var. sullivanti (Boynton and Beadle). Carbohydr. Res. 308:99-105. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J.-H., B.-W. Kim, K.-H. Yoon, and S.-W. Nam. 2000. Expression of Bacillus sp. β-xylosidase gene (xylB) in Saccharomyces cerevisiae. Biotechnol. Lett. 22:1025-1029. [Google Scholar]
- 24.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 25.La Grange, D. C., M. Claeyssens, I. S. Pretorius, and W. H. Van Zyl. 2000. Coexpression of the Bacillus pumilus beta-xylosidase (xynB) gene with the Trichoderma reesei β-xylanase 2 (xyn2) gene in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 54:195-200. [DOI] [PubMed] [Google Scholar]
- 26.La Grange, D. C., I. S. Pretorius, and W. H. Van Zyl. 1997. Cloning of the Bacillus pumilus β-xylosidase gene (xynB) and its expression in Saccharomyces cerevisiae. Appl. Environ. Biotechnol. 47:262-266. [DOI] [PubMed] [Google Scholar]
- 27.Lai, X., F. C. Davis, R. B. Hespell, and L. O. Ingram. 1997. Cloning of cellobiose phosphoenolpyruvate-dependent phosphotransferase genes: functional expression in recombinant Escherichia coli and identification of a putative binding region for disaccharides. Appl. Environ. Microbiol. 63:355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laikova, O. N., A. A. Mironov, and M. S. Gelfand. 2001. Computational analysis of the transcriptional regulation of pentose utilization systems in the gamma subdivision of Proteobacteria. FEMS Microbiol. Lett. 205:315-322. [DOI] [PubMed] [Google Scholar]
- 29.Larsson, S., E. Palmqvist, B. Hahn-Hagerdal, C. Tengborg, K. Stenberg, G. Zacchi, and N. O. Nilvebrant. 1999. The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb. Technol. 24:151-159. [Google Scholar]
- 30.Lubomír, K., and B. Peter. 1998. Disaccharides permeases: constituents of xylanolytic and mannanolytic systems of Aureobasidium pullulans. Biochim. Biophys. Acta 1425:560-566. [DOI] [PubMed] [Google Scholar]
- 31.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilzation: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mai, V., J. Wiegel, and W. W. Lorenz. 2000. Cloning, sequencing, and characterization of the bifunctional xylosidase-arabinosidase from the anaerobic thermophile Thermoanaerobacter ethanolicus. Gene 247:137-143. [DOI] [PubMed] [Google Scholar]
- 33.Martinez, A., M. E. Rodriguez, M. L. Wells, S. W. York, J. F. Preston, and L. O. Ingram. 2001. Detoxification of dilute acid hydrolysates of lignocellulose with lime. Biotechnol. Prog. 17:287-293. [DOI] [PubMed] [Google Scholar]
- 34.Moniruzzaman, M., X. Lai, S. W. York, and L. O. Ingram. 1997. Isolation and molecular characterization of high-performance cellobiose-fermenting spontaneous mutants of ethanologenic Escherichia coli KO11 containing the Klebsiella oxytoca casAB operon. Appl. Environ. Microbiol. 63:4633-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 36.Nurizzo, D., J. P. Turkenburg, S. J. Charnock, S. M. Roberts, E. J. Dodson, V. A. Mackie, E. J. Taylor, H. J. Gilbert, and G. J. Davies. 2002. Cellvibrio japonicus α-l-arabinanase 43A has a novel five-blade β-propeller fold. Nat. Struct. Biol. 9:665-668. [DOI] [PubMed] [Google Scholar]
- 37.Ohta, K., D. S. Beall, J. P. Mejia, K. T. Shanmugam, and L. O. Ingram. 1991. Metabolic engineering of Klebsiella oxytoca M5A1 for ethanol production from xylose and glucose. Appl. Environ. Microbiol. 57:2810-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta, K., D. S. Beall, J. P. Mejia, K. T. Shanmugam, and L. O. Ingram. 1991. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl. Environ. Microbiol. 57:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodionov, D. A., A. A. Mironov, and M. S. Gelfand. 2001. Transcriptional regulation of pentose utilisation systems in the Bacillus/Clostridium group of bacteria. FEMS Microbiol. Lett. 205:305-314. [DOI] [PubMed] [Google Scholar]
- 40.Shulami, S., O. Gat, A. L. Sonenshein, and Y. Shoham. 1999. The glucuronic acid utilization gene cluster from Bacillus stearothermophilus T-6. J. Bacteriol. 181:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takao, M., A. Yamaguchi, K. Yoshikawa, T. Terashita, and T. Sakai. 2002. Molecular cloning of the gene encoding thermostable endo-1, 5-α-l-arabinase of Bacillus thermodenitrificans TS-3 and its expression in Bacillus subtilis. Biosci. Biotechnol. Biochem. 66:430-433. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Utt, E. A., C. K. Eddy, K. F. Keshav, and L. O. Ingram. 1991. Sequencing and expression of the Butyrivibrio fibrisolvens xylB gene encoding a novel bifunctional protein with β-d-xylosidase and α-l-arabinofuranosidase activities. Appl. Environ. Microbiol. 57:1227-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitehead, T. R., and M. A. Cotta. 2001. Identification of a broad-specificity xylosidase/arabinosidase important for xylooligosaccharide fermentation by the ruminal anaerobe Selenomonas ruminantium GA192. Curr. Microbiol. 43:293-298. [DOI] [PubMed] [Google Scholar]
- 45.Wood, B. E., and L. O. Ingram. 1992. Ethanol production from cellobiose, amorphous cellulose, and crystalline cellulose by recombinant Klebsiella oxytoca containing chromosomally integrated Zymomonas mobilis genes for ethanol production and plasmids expressing thermostable cellulase genes from Clostridium thermocellum. Appl. Environ. Microbiol. 58:2103-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyman, C. E. 2003. Potential synergies and challenges in refining cellulosic biomass to fuels, chemicals, and power. Biotechnol. Prog. 19:254-262. [DOI] [PubMed] [Google Scholar]
- 47.Xu, W.-Z., Y. Shima, S. Negoro, and I. Urabe. 1991. Sequence and properties of β-xylosidase from Bacillus pumilus IPO: contradiction of the previous nucleotide sequence. Eur. J. Biochem. 202:1197-1203. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida, S., A. Kuno, N. Saito, M. Aoyama, and I. Kusakabe. 1998. Structure of xylan from culms of bamboo grass (Sasa senanensis Rehd.). J. Wood Sci. 44:457-462. [Google Scholar]
- 49.Zaldivar, J., A. Martinez, and L. O. Ingram. 2000. Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 68:524-530. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, S., and L. O. Ingram. 2001. Simultaneous saccharification and fermentation of amorphous cellulose to ethanol by recombinant Klebsiella oxytoca SZ21 without supplemental cellulase. Biotechnol. Lett. 23:1455-1462. [Google Scholar]