Abstract
Arbuscular mycorrhizae are beneficial for crops grown under low-till management systems. Increasingly, it is becoming apparent that bacteria associated with mycorrhizae can enhance the beneficial relationship between mycorrhizae and plants. However, it has been difficult to study these relationships by conventional techniques. In this study actively growing bacteria were identified in soil from an undisturbed fallow field known to contain arbuscular mycorrhizae by using molecular tools to eliminate the need for cultivation. A thymidine analog, bromodeoxyuridine (BrdU), was added to the soil and incubated for 2 days. DNA was extracted, and the newly synthesized DNA was isolated by immunocapture of the BrdU-containing DNA. The active bacteria in the community were identified by 16S rRNA gene PCR amplification and DNA sequence analysis. Based on 16S rRNA gene sequence information, a selective medium was chosen to isolate the corresponding active bacteria. Bacillus cereus strain VA1, one of the bacteria identified by the BrdU method, was isolated from the soil and tagged with green fluorescent protein. By using confocal microscopy, this bacterium was shown to clearly attach to arbuscular mycorrhizal hyphae. This study was the first to use this combination of molecular and traditional approaches to isolate, identify, and visualize a specific bacterium that is active in fallow soil and associates with arbuscular mycorrhizal hyphae.
The increasing demand for low-input agriculture has resulted in greater interest in soil microorganisms able to increase soil fertility and/or to stimulate plant nutrition and health. For example, mycorrhizal fungi are known to be beneficial for the growth of many plant species (34). Mycorrhizal associations vary widely in structure and function, but the most common interaction is the arbuscular mycorrhizal (AM) association formed between the roots of most higher plants and zygomycete fungi belonging to the order Glomales. It is estimated that more than 80% of all terrestrial plants form this type of association, including many plants that are important in agriculture and horticulture (27, 52). AM fungi are obligate biotrophs, and their life cycle depends on their ability to colonize a host plant. Fungal growth ceases after approximately 25 to 30 days of culture in the absence of the host plant (27). AM fungi have not been cultured in the absence of the host plants, and this has hampered mass production of these organisms and their utilization in crop systems (33).
In recent years, several bacteria have been reported to be associated with the mycorrhizospheres of different host plants (3, 4, 14, 16, 17). The mycorrhizosphere, or more specifically the mycorrhizal hyphosphere, is the volume of soil influenced by the mycorrhizal hyphae, in contrast to the rhizosphere, which is the volume of soil influenced by plant roots (14). The importance of bacterial associations with mycorrhizae has been highlighted recently by the identification of mycorrhization helper bacteria (MHB), which improve both ectomycorrhiza formation and plant growth (18). Garbaye (18) proposed five different underlying mechanisms to explain the phenomenon of MHB: (i) enhancement of the susceptibility of the plant to mycorrhizal colonization, (ii) enhancement of the root-fungus recognition process, (iii) nutritional enhancement of fungal growth, (iv) a beneficial change in the content of the rhizosphere soil, and (v) stimulation of germination of fungal propagules.
It has been proposed that active bacterial attachment and colonization of the rhizoplane or mycorrhizal surfaces are required for MHB to influence ectomycorrhizal plant growth. For example, the MHB strain BBc6 was shown to clearly attach to hyphae of Laccaria laccata, Suillus bovinus, and Paxillus involutus, in contrast to nonhelper bacteria, which did not attach to hyphae (50). It has been suggested that attachment of some plant growth-promoting rhizobacteria to hyphal walls requires polar flagella (50).
There have been reports of bacteria that specifically associate with ectomycorrhizae (7, 12, 19, 20), as well as reports of bacteria that have positive effects on AM (8, 12, 39, 43, 44, 46). For example, Duponnois and Plenchette (12) concluded that the MHB Pseudomonas monteilii strain HR13 significantly promoted both ectomycorrhizal and endomycorrhizal colonization of different Acacia species. Mamatha et al. (39) identified a Bacillus coagulans strain that was able to increase mycorrhiza levels in AM fungus-inoculated plants and proposed that this organism should be included in the MHB group. In another study, Budi et al. (8) reported the discovery of a new Paenibacillus strain, isolated from the mycorrhizosphere of sorghum, that improves AM formation while acting as a biological control agent against soilborne fungal diseases. Although culturing was successful in this case, it is known that most microorganisms in soil cannot be grown on the culture media available at this time. There are potentially many unknown bacteria that form beneficial associations with AM that cannot be cultured.
One way to overcome this obstacle is to use molecular tools to identify active bacteria associated with AM. A random analysis of rRNA or ribosomal DNA sequences from an environmental sample could lead to identification of the dominant organisms in the community but not necessarily the organisms involved in a particular physiological response (6). One promising tool that has been used recently for detection of active microbial populations in soil communities (6) relies on incorporation of the thymidine analogue bromodeoxyuridine (BrdU) into growing cells during DNA synthesis. The newly synthesized DNA can then be isolated by BrdU immunocapture (6). This approach permits identification of populations that grow in response to specified stimuli.
The aim of this study was to identify actively growing bacteria in fallow soil known to contain AM by using the BrdU approach. The information obtained was used to isolate active bacteria and to determine their specific associations with AM hyphae in the mycorrhizosphere.
MATERIALS AND METHODS
Field sampling.
Soil was collected on two sampling days, 1 July 2002 and 30 August 2002, from an undisturbed fallow field outside Uppsala, Sweden. The soil was a sandy loam previously planted with clover. This area represents a typical low-input ecological region, where abundant arbuscular mycorrhizae are expected. Soil samples were obtained by removing the top 20 cm of soil containing most plant material and collecting the underlying layer with deeper roots and associated mycorrhizal hyphae. The samples were sieved (2 mm) and then immediately stored on ice. After arrival at the laboratory, the soil samples were stored at −20°C.
AM staining.
AM staining was performed for each soil sample in order to confirm the presence of AM hyphae in the soil. Three different clover roots were washed, heated to 90°C in 10% (wt/vol) KOH for 1 h, rinsed with water, and soaked in 1% HCl at room temperature for 30 min. The HCl was replaced by a trypan blue solution (0.05% [wt/vol] trypan blue, 50% [vol/vol] glycerol, 0.5% [vol/vol] HCl; Sigma), and the preparations were heated at 60°C for 1 h; the roots were rinsed with double-distilled water and destained in acidified glycerol (50% [vol/vol] glycerol, 0.5% [vol/vol] HCl). The roots were applied to glass slides and analyzed by using an Axiophot epifluorescence microscope (Zeiss, Oberkochen, Germany). Quantification of AM colonization was performed as described by McGonigle et al. (41) by examining 100 intersections per slide for triplicate root samples.
DNA extraction.
Except for control soil, which received no supplements, 1-g soil samples were mixed with 200 μl of 100 mM BrdU (Sigma) and incubated in petri dishes at room temperature. After 2 days of incubation, DNA was extracted from the samples with a Fast DNA kit for soil (Bio 101, Vista, Calif.), as described by the manufacturer.
Immunochemical purification of BrdU-containing DNA.
Immunochemical purification of BrdU-labeled DNA was performed by a modification of the method described by Urbach et al. (57). Monoclonal anti-BrdU antibodies (diluted 1:10 in phosphate-buffered saline [PBS]; Boehringer Mannheim, Indianapolis, Ind.) were mixed at a 1:9 ratio with herring sperm DNA (1.25 mg/ml in PBS; Promega, Madison, Wis.), which had been heated to 100°C for 1 min, quickly frozen in dry ice-ethanol, and thawed. The mixture was incubated for 40 min at room temperature. Dynabeads coated with goat anti-mouse immunoglobulin G (DYNAL, Lake Success, N.Y.) were washed three times with PBS-bovine serum albumin (BSA) (PBS containing 1 mg of acetylated BSA per ml) by using a magnetic particle concentrator (DYNAL) and were resuspended in PBS-BSA at the initial concentration. Denatured soil DNA (1 μg [total weight] in 10 μl of PBS that was heated to 100°C for 1 min, frozen on ice, and thawed) was mixed with 10 μl of a herring sperm DNA-antibody mixture and incubated for 30 min at room temperature. Samples were subsequently mixed with 25-μl portions of paramagnetic beads and incubated for 30 min with constant agitation at room temperature. Then they were washed seven times with 0.5 ml of PBS-BSA. To elute the BrdU-containing DNA fraction, 100 μl of 1.7 mM BrdU (in PBS-BSA) was added, and the samples were incubated for 30 min at room temperature with constant agitation. Two microliters of glycogen (20 mg/ml; U.S. Biochemicals) was added to each bead supernatant, and the DNA was isolated by ethanol precipitation.
PCR.
To identify the BrdU-labeled DNA, PCR amplification of 16S rRNA gene fragments was performed by using bacterial primer 27f (5′-AGA GTT TGA TCM TGG CTC AG-3′) as the forward primer and bacterial primer 1541r (5′-AAG GAG GTG ATC CAG CC-3′) (31) as the reverse primer. Amplification was carried out by using a 50-μl reaction mixture containing 0.5 μl of Taq DNA polymerase (5 U/μl; Amersham/USB, Cleveland, Ohio), 5 μl of 10× buffer (Amersham/USB), 0.5 μl of a solution containing each deoxynucleoside triphosphate at a concentration of 20 mM, 41.6 μl of sterile distilled water, 0.7 μl of each primer (50 μM), and 1 μl of template. The thermocycling, which was conducted with a model 2400 thermal cycler (Perkin-Elmer, Norwalk, Conn.), started with an initial denaturation step of 95°C for 5 min. A total of 33 cycles consisting of 40 s at 94°C, 40 s at 55°C, and 60 s at 72°C was followed by a final primer extension step at 72°C for 7 min. The PCR was further optimized based on the program described above with annealing temperatures of 50, 55, and 56°C. The amplified DNA was verified by electrophoresis of aliquots (10 μl) of PCR mixtures in 1% agarose (containing 0.02% ethidium bromide) in 1× TBE buffer (0.1 M Tris, 0.09 M boric acid, 1 mM EDTA).
Cloning.
The amplified, approximately 1,500-bp PCR fragments were cloned by using a TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.) and following the manufacturer's instructions. Approximately 20 clones from each original sample were randomly chosen for screening.
RFLP analysis.
The inserts from the selected clones were amplified by using the M13f and M13r primers provided by the manufacturer (Invitrogen). These primers amplified the entire cloned PCR product plus a ∼250-bp region on either side of the insert. The template was added by touching each clone with a sterile loop and then touching the loop to the reaction mixture. The same amplification conditions were used that were used for full-length 16S rRNA amplification, except that the annealing temperature was changed to 50°C. The PCR products were digested with HaeIII and RsaI (Promega) in individual reactions. Ten-microliter portions of PCR products were incubated for 4 h at 37°C with the restriction enzyme mixtures. The resulting restriction fragment length polymorphism (RFLP) products were separated by gel electrophoresis in 2.2% agarose gels. After 2 h of electrophoresis at 90 V, each gel was stained with ethidium bromide (1 mg liter−1) and photographed under UV light. Clones that produced identical patterns were considered discrete operational taxonomic units.
Sequencing.
One clone of each RFLP group in each sample was chosen for sequence analysis. These clones were isolated from 3-ml overnight cultures in Luria broth supplemented with 50 μg of kanamycin ml−1 by using the Wizard Minipreps DNA purification system (Promega). Partial sequencing was performed with an Applied Biosystems International PRISM 377 DNA sequencer by using the M13f primer. Sequences were compared to sequences in the GenBank database by using BLAST software (2) to find the closest relatives.
Isolation.
Selective agar medium commercially available for isolation of Bacillus cereus was used to isolate B. cereus from the original soil collected (field soil collected on 30 August 2002). Soil samples (1 g) were shaken in 50 ml of sterile water for 5 min. The soil slurries were then serially diluted in sterile water, and 0.1-ml suspensions were inoculated in duplicate onto B.cereus selective agar (Oxoid). The cultures were incubated at 37°C for 2 days, and colonies that were morphologically distinct on the basis of pigment, shape, size, and surface texture were isolated by transfer of single colonies to B. cereus selective agar. Isolates were examined for purity by microscopy, and strains of interest were Gram stained. To determine whether any of these strains could be identical or similar to the active BrdU-labeled strain, the 16S rRNA gene region was PCR amplified (under the same conditions that were used for full-length 16S rRNA gene amplification) by using universal bacterial primers 27f and 1541r, and the PCR products were purified with a QIAquick PCR purification kit (Qiagen, Valencia, Calif.) and sequenced (ABI PRISM 377 DNA sequencer).
gfp tagging.
The multicopy plasmid pnf8 (15) was used to tag bacteria with the gfp gene encoding green fluorescent protein (GFP) (55). The pnf8 plasmid is a shuttle vector carrying the gfp-mut1 gene, driven by the strong, constitutive Listeria monocytogenes promoter Pdlt. Escherichia coli strain JM109 harboring the vector was grown at 37°C in Luria-Bertani (LB) medium supplemented with erythromycin (150 μg ml−1). B. cereus cells from the mid-log phase (optical density at 600 nm, 0.4) were washed four times with sterile water, and plasmid DNA was added to 100 μl of competent cells in a 2-mm electroporation cuvette (Techtum LAB AB, Umeå, Sweden). The DNA was electroporated into the cells by using a model ECM-600 electroporation system (BTX, San Diego, Calif.) set at 2.5 kV, 200 Ω, and 25 μF. The cells were then immediately transferred to 2 ml of LB medium and incubated for 3 h at 37°C before they were plated on selective medium (LB medium containing 10 μg of erythromycin ml−1). Tagged cells were grown in the presence of erythromycin to ensure stability of the plasmid and the fluorescent phenotype and then used in short-term incubation experiments (1 h) with AM fungal hyphae.
Flow cytometry.
Flow cytometry was used to monitor the eventual loss of the pnf8 plasmid from cell cultures. The intensity of GFP fluorescence in the cells was quantified over time by flow cytometry by using a Becton Dickinson FACScalibur system equipped with a 15-mV, air-cooled argon ion laser excitation light source (488 nm). GFP fluorescence was detected by using an HQ500LP filter (Chroma Technology Corp., Brattleboro, Vt.). B. cereus cells harboring the pnf8 plasmid were grown at 37°C in 10% LB medium both containing and not containing selective antibiotics. For determination of the number of cells which lost the GFP plasmid without selective pressure, the numbers of gfp-tagged and non-gfp-tagged cells in cultures with and without selective antibiotics, respectively, were compared. An internal standard (buffer containing a known concentration of polystyrene green fluorescent microspheres [diameter, 2.2 μm] with a coefficient of variation of < 5% [Duke Scientific, Palo Alto, Calif.]) was used to determine the total number of cells. Measurements were obtained once a day for 4 days for triplicate samples from each culture.
Microscopic studies of bacterial attachment to hyphae.
An AM fungal inoculant (Glomus dussii; BIORIZE France) was used for microscopic studies. In addition, hyphae from the original Uppsala soils were examined by microscopy. Some of the samples were stained with trypan blue in order to better visualize mycorrhizal hyphae by microscopy. Hyphae were observed in soil by stereomicroscopy, picked by using fine forceps, washed in distilled water, and applied to slides. A gfp-tagged bacterial colony was taken from a fresh LB agar plate supplemented with erythromycin (10 μg ml−1) and mixed with 500 μl of distilled water in a microcentrifuge tube. Two drops (approximately 100 μl) of this suspension was applied to the slide containing the hyphae, and the bacterial suspension was allowed to settle and dry. A drop of mounting medium (Vectashield; Vector Laboratories Inc., Burlingame, Calif.) was added to the slide before a coverslip was applied. Approximately 20 different hyphae from each of the two sources were examined. Microscopy was performed with a laser scanning confocal microscope (Leica TCS SP) by using excitation wavelengths of 488 nm (Ar) and 543 nm (HeNe). Two emission filters, LP 650 (red) and BP 505-550 (green), were used. The images were the result of pseudocolor merging of the outputs of the two channels.
Quantification of gfp-tagged cells attached to AM hyphae.
The following five gfp-tagged bacterial strains were applied to AM hyphae: B. cereus VA1(pnf8) (this study), B. cereus ATCC 14579(pnf8) (this study), Paenibacillus brasilensis strain PB177(pnf8) (I. von der Weid and J. K. Jansson, unpublished data), Arthrobacter chlorophenolicus A6G (13), and Pseudomonas fluorescens SBW25::gfp/lux (56). Cells were grown in nutrient broth until the early stationary phase, and then they were collected by centrifugation and quantified by microscopy. All bacterial suspensions were diluted in LB medium to obtain a concentration of approximately 107 cells per ml. G. dussii hyphae were picked from the commercial inoculum with fine forceps, and one hyphal fragment was added to individual microscope slides. A 10-μl aliquot of each bacterial suspension was added to slides containing the AM hyphae. The hyphae and bacteria were incubated for 2 min at room temperature, and then the hyphae were transferred with fine forceps to microscope slides containing approximately 10 μl of 1× PBS buffer to be washed. The slides containing hyphae and bacteria were gently shaken manually for approximately 10 s, and the hyphae were kept on the slides for 1 min. The hyphae were then transferred to fresh slides containing 5 μl of sterile water and immediately examined by epifluorescence microscopy. The number of gfp-tagged bacteria attached to 200 μm of hyphae was determined. To avoid bias, the hyphal fragment first seen in the field of vision in the microscope was assessed. Three individual hyphal segments were counted for each sample, and the average and standard deviation of these values were determined.
Nucleotide sequence accession numbers.
The nucleotide sequences of clones and the B. cereus VA1 isolate have been deposited in the EMBL nucleotide sequence database under accession numbers AJ539165 to AJ539174, AJ539226, and AJ563290.
RESULTS
AM staining.
The fungus-specific staining results confirmed the presence of abundant AM hyphae and vesicles inside clover roots in the fallow soil collected in Uppsala. The percentage of the clover root length colonized by mycorrhizal endophytes was approximately 60% for each soil sample (mean for three roots randomly examined) (results not shown).
PCR and RFLP analysis.
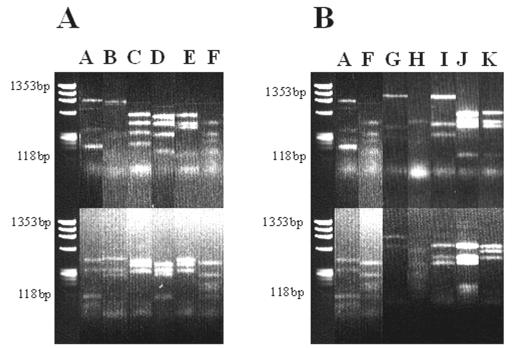
PCR products that were approximately 1,500 bp long were obtained from the DNA extracted from soil treated with BrdU (data not shown). The lack of PCR products in the preparations that were not treated with BrdU shows that the immunocapture process was specific (data not shown). The PCR products were cloned and analyzed, and approximately 50 clones were obtained from each soil sample. Twenty clones from sampling round 1 (1 July 2002) and 21 clones from round 2 (30 August 2002) were randomly collected. Prescreening of these clones by digestion with HaeIII and RsaI generated 11 distinct RFLP patterns, patterns A to K (Fig. 1). RFLP patterns A and F were found in soil obtained on both sampling dates (Fig. 1 and Table 1).
FIG. 1.
RFLP profiles of 16S rRNA gene fragments amplified from BrdU-labeled DNA extracted from field soil. The letters at the top correspond to distinct RFLP patterns. The preparations were digested with HaeIII (top) and RsaI (bottom). (A) Clones obtained from the sample collected on 1 July 2002. (B) Clones obtained from the sample collected on 30 August 2002.
TABLE 1.
RFLP patterns (operational taxonomic units) of screened clones and their closest matches to GenBank sequences
| RFLP pattern | No. of clones screened
|
Accession no. | Closest match in GenBank database (accession no.) | % Similarity for ∼800-bp sequence | Reference | |
|---|---|---|---|---|---|---|
| 1 July 2002 | 30 August 2002 | |||||
| A | 6 | 3 | AJ539165 | Paenibacillus amylolyticus (D85396) | 98 | 51 |
| B | 5 | 0 | AJ539166 | Paenibacillus sp. (AF324200) | 98 | Christner et al.a |
| C | 6 | 0 | AJ539167 | Bacillus psychrotolerans (AJ277983) | 99 | 1 |
| D | 1 | 0 | AJ539168 | Sporosarcina sp. (AF506059) | 98 | Nazaret et al.b |
| E | 1 | 0 | AJ539169 | Bacillus sp. (AF346496) | 99 | 5 |
| F | 1 | 1 | AJ539170 | Arthrobacter sp. (AB070602) | 98 | Tanaka et al.c |
| G | 0 | 2 | AJ539171 | Paenibacillus sp. (AB043868) | 99 | Nogi et al.d |
| H | 0 | 4 | AJ539172 | Arthrobacter aurescens (AF501335) | 100 | Jussila et al.e |
| I | 0 | 6 | AJ539173 | Paenibacillus sp. (AJ495806) | 99 | Wery et al.f |
| J | 0 | 4 | AJ539226 | Bacillus cereus (AY138278) | 100 | 49 |
| K | 0 | 1 | AJ539174 | Bacillus sp. (AJ438301) | 99 | DeClerck and De Vasg |
B. C. Christner, J. N. Reeve, E. Mosley-Thompson, and L. G. Thompson, unpublished data (http://www.ncbi.nlm.nih.gov/BLAST/).
S. Y. Nazaret, E. Brothier, and L. Y. Ranjard, unpublished data (http://www.ncbi.nlm.nih.gov/BLAST/).
N. Tanaka, T. Uchimura, and K. Komagata, unpublished data (http://www.ncbi.nlm.nih.gov/BLAST/).
Y. Nogi, H. Takami, K. Nakasone, and K. Horikoshi, unpublished data (http://www.ncbi.nlm.nih.gov/BLAST/).
M. M. Jussila, L. Suominen, and K. Lindstrom, unpublished data (http://www.ncbi.nlm.nih.gov/BLAST/).
N. Wery, U. Gerike, A. Sharman, J. B. Chaudhuri, D. W. Hough, and M. J. Danson, unpublished data (http://www.ncbi.nlm.nih.gov/BLAST/).
E. De Clerck and P. De Vos, unpublished data (http://www.ncbi.nlm.nih.gov/BLAST/).
Sequence identification.
Sequencing of the 5′ ends (total length, 750 to 800 bp) of the cloned PCR products of the RFLP-defined operational taxonomic units resulted in 11 distinct sequences (Table 1). All sequences were found to correspond closely to 16S rRNA gene sequences listed in the GenBank database. The most frequently obtained sequence type (sequences similar to the sequences of clones with RFLP pattern A) accounted for 22% of the screened clones obtained from the two rounds of soil sampling. One of the clones with RFLP pattern J was sequenced further (1,403 bp), and the sequence still exhibited 100% similarity to the B. cereus sequence in the GenBank database (accession number AY138278).
Isolation of B. cereus strain VA1.
We chose to isolate RFLP pattern J, which exhibited a high level of similarity to B. cereus based on 16S rRNA sequence information, from the original soil because of the similarity to B. cereus sequences in the GenBank database. Dilutions of the fallow soil were inoculated onto B. cereus selective medium, and nine colonies were selected and isolated. The cells were Gram stained and examined by microscopy. All isolates were gram positive and contained endospores typical of B. cereus. Six potential B. cereus isolates were selected for 16S rRNA gene sequence analysis. One isolate with 99.99% similarity to clones that produced RFLP pattern J was identified by aligning 1,361 bp, indicating that the clone and the isolate could be the same bacterium or at least closely related. Each cell of the isolate contained a central spore that did not distend the cell, and the organism was hemolytic. Based on this information, the bacterium was designated B. cereus strain VA1 (accession number AJ563290). The sequences of other isolates that were sequenced exhibited between 95 and 98% similarity to the sequence of the clone that produced RFLP pattern J.
GFP tagging and fluorescence stability.
B. cereus strain VA1 was tagged with the gfp gene (11) by using a shuttle vector optimized for expression of GFP in gram-positive bacteria (15) so that the cells could be visualized in association with mycorrhizal hyphae. The tagged strain was designated B. cereus VA1(pnf8). The GFP fluorescence intensity of the cells was monitored by flow cytometry during growth in nonselective medium to determine the stability of the plasmid vector. The cell population lost about 30% of the plasmids after 1 day, 40% after 2 days, 70% after 3 days, and 80% after 4 days (results not shown). In the control culture, which contained erythromycin for selection of the plasmid, no decrease in fluorescence intensity and hence no plasmid loss were observed.
In situ visualization of B. cereus strain VA1(pnf8) on mycorrhizal hyphae.
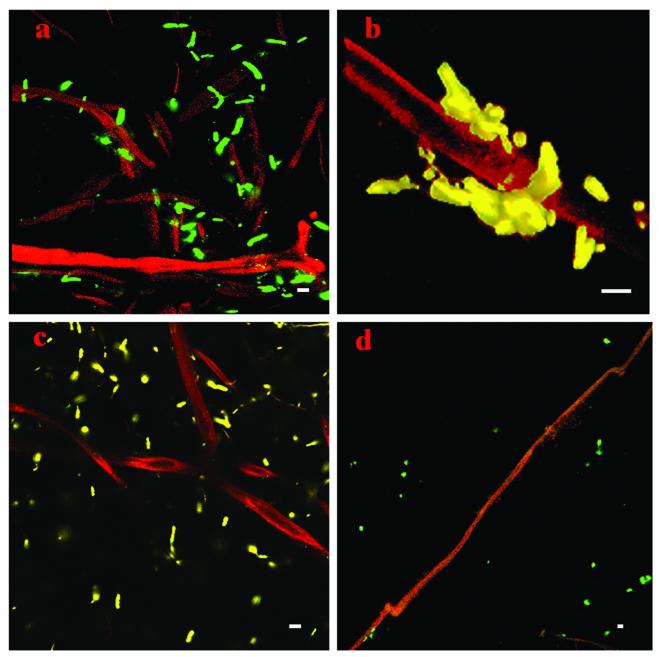
Colonization of mycorrhizal hyphae by B. cereus strain VA1(pnf8) was studied in situ with hyphae having two different origins: an AM inoculum (G. dussii) with distinct mycorrhizal hyphae and the original field soil, which was known to contain mycorrhizal hyphae. The colonization patterns obtained by using hyphae from the AM inoculum and the colonization patterns obtained by using the field soil were always very similar. Approximately 20 different hyphae from each of the two sources were examined. High concentrations of B. cereus strain VA1 cells were detected associated with fungal hyphae (Fig. 2). All hyphae examined had numerous B. cereus strain VA1 cell aggregates or microcolonies adhering to the outer cell walls. To determine whether attachment was a general phenomenon for B. cereus species, a control strain of B. cereus, ATCC 14579 (38), was tagged with the pnf8 plasmid, and this strain and a strain of gfp-tagged E. coli (DH5α::gfp/lux) (56) were applied separately to hyphae. Clear differences were observed in the patterns of association of B. cereus strain VA1(pnf8), B. cereus ATCC 14579(pnf8), and E. coli DH5α::gfp/lux cells (Fig. 2). The B. cereus VA1(pnf8) isolate showed a strong preference for attachment to the hyphae from the mycorrhizosphere in both the AM inoculum and the field soil, whereas B. cereus ATCC 14579(pnf8) and E. coli DH5α::gfp/lux cells were more randomly distributed throughout the sample. When hyphae were broken or had partially deteriorated, aggregates of B. cereus strain VA1(pnf8) were often found near cracks or degraded areas of the hyphae (data not shown).
FIG. 2.
Association of gfp-tagged B. cereus strain VA1 and controls with AM hyphae. (a) B. cereus strain VA1(pnf8) on mycorrhizal hyphae from an AM inoculum (G. dussii). (b) B. cereus strain VA1(pnf8) on a hyphal fragment from field soil. (c) B. cereus ATCC 14579(pnf8) on mycorrhizal hyphae (G. dussii). (d) E. coli DH5α::gfp/lux applied to mycorrhizal hyphae (G. dussii). Bars = 4 μm. Green cells are the result of pseudocolor rendering in the Adobe Photoshop 7.0 software.
Quantification of gfp-tagged cells attached to AM hyphae.
The number of B. cereus VA1 cells that attached to AM hyphae was determined by microscopy (Table 2). Other preexisting gfp-tagged bacteria of interest, including P. brasilensis strain PB177(pnf8), A. chlorophenolicus strain A6G, and P. fluorescens SBW25::gfp/lux, were applied to mycorrhizal hyphae as additional controls to determine whether attachment of B. cereus VA1 was a trait specific to this isolate. The Paenibacillus and Arthrobacter control strains were chosen since they are also gram-positive bacteria commonly found in soil and both Paenibacillus and Arthrobacter sequences were identified in the active fraction of the fallow soil in this study (Table 1). P. fluorescens strain SBW25 is a well-characterized, gram-negative, root-colonizing bacterium (5). It was found that the B. cereus VA1 isolate obtained in this study, as well as P. brasilensis PB177, attached to the hyphae at significantly higher levels than the other three bacteria [B. cereus ATCC 14579(pnf8), P. fluorescens SBW25::gfp/lux, and A. chlorophenolicus A6G], which were much less abundant on the hyphal surfaces (Table 2).
TABLE 2.
Numbers of gfp-tagged cells attached to 200 μm of AM hyphae
| Strain | No. of bacteria attached to 200 μm of hyphaea | SD | Reference |
|---|---|---|---|
| Bacillus cereus VA1(pnf8) | 109 | 6.66 | This study |
| B. cereus ATCC 14579(pnf8) | 32 | 5.51 | This study |
| P. brasilensis PB177(pnf8) | 97 | 5.69 | von der Weid and Janssonb |
| P. fluorescens SBW25::gfp/lux | 37 | 5.86 | 57 |
| A. chlorophenolicus A6G | 37 | 4.51 | 14 |
The values are means for triplicate samples from three different hyphae.
I. von der Weid and J. K. Jansson, unpublished data.
DISCUSSION
Understanding the mechanisms through which soil bacteria, mycorrhizae, and plants interact is crucial for the management of sustainable agricultural systems (25). The importance of bacterial associations with mycorrhizae has been highlighted recently since identification of MHB (18). Since MHB improve both mycorrhization and plant growth, they could be of practical interest for improving mycorrhizal inoculation techniques in agricultural systems. However, few MHB have been isolated for AM due to the difficulties in culturing AM fungi. Also, it is known that only a fraction of the bacteria in soil are culturable, although they may be viable and active. In this study these limitations were overcome by using a combination of traditional and molecular approaches.
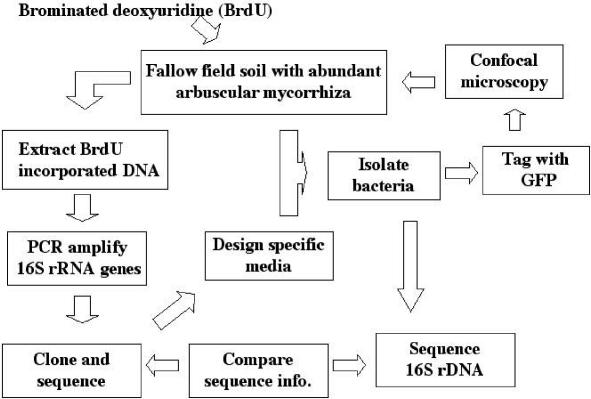
This is the first report to describe the BrdU immunocapture approach used to identify active bacterial populations in fallow soil and their subsequent association with AM hyphae in the mycorrhizosphere. An overview of the approach used in this study is shown in Fig. 3. Previously, it was found that E. coli and Bacillus subtilis strains could not incorporate exogenously supplied BrdU unless they had mutations affecting thymine synthesis (28, 37, 42). Therefore, it was widely assumed that wild-type bacteria do not take up BrdU (9, 57). Thus, the BrdU approach includes only the organisms capable of BrdU uptake and incorporation (6). Negative and positive results have since been reported (48, 57). Recently, it has been suggested that the majority of bacteria take up and incorporate [3H]thymidine, and therefore, it is likely that BrdU, which has been used successfully as a thymidine analog, can be taken up and incorporated in most organisms (6).
FIG. 3.
Outline of the approach used in the present study. rDNA, ribosomal DNA.
In this study the BrdU method allowed identification of metabolically active bacteria, and in combination with phylogenetic analysis of the 16S rRNA, the specific bacteria could be putatively identified to the species level.
RFLP analyses of resultant clones revealed clear groups. Two of the digestion patterns (patterns A and F) were found to be similar in soil samples obtained on both of the independent sampling days. Approximately 800 bp for each RFLP group was sequenced to differentiate distinct groups from each other. Only gram-positive bacteria (mostly Paenibacillus spp. and Bacillus spp.) were found, which is consistent with previous studies in which primarily gram-positive, rod-shaped bacteria associated with AM (8, 43) and ectomycorrhizal (47) fungi were found. The absence of gram-negative bacterial species in this study indicates that they are less active in the AM-enriched soils examined, since representatives of both gram-positive and gram-negative bacteria have been shown to incorporate [3H]thymidine (22, 36, 53, 58, 59) or BrdU (57) into their genomes. It has previously been shown by using other approaches that gram-negative bacteria, such as pseudomonads, are predominantly active in rhizospheres or upon nutrient addition in soil (10, 26), and thus they often behave as opportunistic r strategists (35). By contrast, the gram-positive bacteria in this study were active in bulk soil fractions containing mycorrhizae, typically a low-nutrient ecosystem that commonly favors k strategists.
One of the active bacteria identified by the BrdU approach was presumably isolated from the fallow field soil. The sequences of the 16S rRNA region of the clone obtained by the BrdU approach and this isolate exhibited 99.99% similarity. Only 1 bp was different, which could have been due to sequencing error. This indicated that the clone and the isolate were most likely the same bacterium or at least were closely related. Further taxonomic characterization confirmed that the isolate was a B. cereus strain, and it was designated B. cereus VA1. Of 41 clones examined, 4 were B. cereus strain VA1, based on the RFLP patterns, which indicates that they were among the dominant active populations in the community. Eventually, it would be interesting to perform extended studies to isolate additional active bacterial populations and to determine whether they also show association with mycorrhizal hyphae.
Attempts to chromosomally tag B. cereus strain VA1 with the gfp gene were unsuccessful (results not shown). Therefore, plasmid pnf8 (15), optimized for expression of GFP in gram-positive bacteria, was employed. The major drawback of plasmid tagging is the instability of the plasmid (23, 24). However, this was not a problem in this study, since the gfp-labeled bacteria were used in short-term trials with fungal hyphae. Also, pnf8 directed sufficiently high levels of GFP expression to enable detection of B. cereus strain VA1 both in pure cultures and in environmental samples by confocal microscopy.
gfp-tagged B. cereus strain VA1 specifically interacted with mycorrhizal hyphae. Results obtained with the field soil were compared to the results obtained with a more defined system by using an AM inoculant with spores and AM hyphae. In all cases, B. cereus VA1 showed quite similar patterns of attachment to hyphae. Aggregates of B. cereus VA1 were often detected on degraded areas of the hyphae, suggesting that the hyphae may serve as a source of nutrients. Since a large proportion of carbon in plants is transported to the external mycelium (32), it has been suggested that the extraradical hyphae can promote bacterial growth (29, 54) but can affect bacteria through different competitive actions as well (45). B. cereus strain VA1 could be actively involved in the degradation of the fungal hyphae or could form a commensal relationship with the hyphae and benefit from released carbon. Reductions in spore germination and hyphal length at the extramatrical stage have also been observed as effects of soil microorganisms (40, 60), suggesting that bacterial associations can sometimes be antagonistic.
Eventually, mycorrhizal symbiosis with and without B. cereus strain VA1 inoculation should be studied, and the data should be compared to confirm whether this bacterium serves as an MHB (18) for the symbiosis. Positive effects on AM symbiosis with plants after addition of sporulating bacilli (30) and fluorescent pseudomonads have been reported previously (43). The majority of interaction studies have been performed with soil microorganisms, and generally, a single-species AM fungal inoculum has been added to microcosm units. Such studies provide a first approximation for understanding the interactions which can occur under controlled laboratory conditions. However, the ecology of AM interactions with bacteria in the field might be quite different, for example, when there is a much greater diversity of AM fungi (30). Some bacterial isolates have been shown to provide a 50% increase in ectomycorrhizal development in field studies (21). Conversely, the mycorrhizal status of soils may selectively influence the persistence of bacterial inoculants and may affect the numbers of other indigenous bacteria (14).
Future work should help clarify the function and importance of the active microorganisms identified in this study in the AM symbiosis.
Acknowledgments
We thank Irene von der Weid for use of the gfp-tagged Paenibacillus strain and J. D. van Elsas for critical reading of the manuscript. Collaborators in the project, Roger Finlay, Berndt Gerhardsson, and Jonas Johansson, are acknowledged for useful discussions.
This work was supported by grants from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Program on Ecological Production).
REFERENCES
- 1.Abd El-Rahman, H. A., D. Fritze, C. Sproeer, and D. Claus. 2002. Two novel psychrotolerant species, Bacillus psychrotolerans sp. nov. and Bacillus psychrodurans sp. nov., which contain ornithine in their cell walls. Int. J. Syst. E vol. Microbiol. 52:2127-2133. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrade, G., R. G. Linderman, and G. J. Bethlenfalvay. 1998. Bacterial associations with the mycorrhizosphere and hyphosphere of the arbuscular mycorrhizal fungus Glomus mosseae. Plant Soil 202:79-87. [Google Scholar]
- 4.Andrade, G., K. L. Mihara, R. G. Linderman, and G. J. Bethlenfalvay. 1998. Soil aggregation status and rhizobacteria in the mycorrhizosphere. Plant Soil. 202:89-96. [Google Scholar]
- 5.Bailey, M. J., A. K. Lilley, I. P. Thompson, P. B. Rainey, and R. J. Ellis. 1995. Site directed chromosomal marking of a fluorescent pseudomonad isolated from the phytosphere of sugar beet; stability and potential for marker gene transfer. Mol. Ecol. 4:755-763. [DOI] [PubMed] [Google Scholar]
- 6.Borneman, J. 1999. Culture-independent identification of microorganisms that respond to specified stimuli. Appl. Environ. Microbiol. 8:3398-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowen, G. D., and C. Theodorou. 1979. Interactions between bacteria and ectomycorrhizal fungi. Soil Biol. Biochem. 11:119-386. [Google Scholar]
- 8.Budi, S. W., D. van Tuinen, G. Martinotti, and S. Gianinazzi. 1999. Isolation from the Sorghum bicolor mycorrhizosphere of a bacterium compatible with arbuscular mycorrhiza development and antagonistic towards soilborne fungal pathogens. Appl. Environ. Microbiol. 65:5148-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coote, J. G., and C. Binnie. 1986. Tolerance to bromodeoxyuridine in a thymidine-requiring strain of Bacillus subtilis. J. Gen. Microbiol. 132:481-492. [DOI] [PubMed] [Google Scholar]
- 10.Duineveld, B. M., G. A. Kowalchuk, A. Keijzer, J. D. van Elsas, and J. A. van Veen. 2001. Analysis of bacterial communities in the rhizosphere of Chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microb. 67:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn, A. K., and J. Handelsman. 1999. A vector for promoter trapping in Bacillus cereus. Gene 226:297-305. [DOI] [PubMed] [Google Scholar]
- 12.Duponnois, R., and C. Plenchette. 2003. A mycorrhiza helper bacterium enhances ectomycorrhizal and endomycorrhizal symbiosis of Australian Acacia species. Mycorrhiza 13:85-99. [DOI] [PubMed] [Google Scholar]
- 13.Elvang, A. M., K. Westerberg, C. Jernberg, and J. K. Jansson. 2001. Use of green fluorescent protein and luciferase biomarkers to monitor survival and activity of Arthrobacter chlorophenolicus A6 cells during degradation of 4-chlorophenol in soil. Environ. Microbiol. 3:32-42. [DOI] [PubMed] [Google Scholar]
- 14.Finlay, R. D. Action and interaction in the mycorrhizal hyphosphere: a re-evaluation of the role of mycorrhizal symbiosis in nutrient acquisition and plant ecology. In H. BassiriRad (ed.), Nutrient uptale by plants—an ecological perspective, in press. Springer-Verlag, Heidelberg, Germany.
- 15.Fortineau, N., P. Trieu-Cuot, O. Gaillot, E. Pellegrini, P. Berche, and J.-L. Gaillard. 2000. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res. Microbiol. 151:353-360. [DOI] [PubMed] [Google Scholar]
- 16.Founoune, H., R. Duponnois, J. M. Meyer, J. Thioulouse, D. Masse, J. L. Chotte, and M. Neyra. 2002. Interactions between ectomycorrhizal symbiosis and fluorescent pseudomonads on Acacia holosericea: isolation of mycorrhiza helper bacteria (MHB) from a Soudano-Sahelian soil. FEMS Microbiol. Ecol. 41:37-46. [DOI] [PubMed] [Google Scholar]
- 17.Garbaye, J. 1991. Biological interactions in the mycorrhizosphere. Experientia 47:370-375. [Google Scholar]
- 18.Garbaye, J. 1994. Helper bacteria: a new dimension to the mycorrhizal symbiosis. New Phytol. 128:197-210. [DOI] [PubMed] [Google Scholar]
- 19.Garbaye, J., and G. D. Bowen. 1987. Effect of different microflora on ectomycorrhizal inoculation of Pinus radiata. Can. J. For. Res. 17:941-943. [Google Scholar]
- 20.Garbaye, J., and G. D. Bowen. 1989. Stimulation of ectomycorrhizal infection of Pinus radiata by some microorganisms associated with the mantle of ectomycorrhizas. New Phytol. 112:383-388. [Google Scholar]
- 21.Garbaye, J., R. Duponnois, and J. L. Wahl. 1990. The bacteria associated with Laccaria laccata ectomycorrhizas or sporocarps: effects on symbiosis establishment on Douglas fir. Symbiosis 9:267-273. [Google Scholar]
- 22.Graupner, S., and W. Wackernagel. 2001. Pseudomonas stutzeri has two closely related pilA genes (type IV pilus structural protein) with opposite influences on natural genetic transformation. J. Bacteriol. 183:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halverson, L. J., M. K. Clayton, and J. Handelsman. 1993. Population biology of Bacillus cereus UW85 in the rhizosphere of field-grown soybeans. Soil Biol. Biochem. 25:485-493. [Google Scholar]
- 24.Halverson, L. J., M. K. Clayton, and J. Handelsman. 1993. Variable stability of antibiotic-resistance markers in Bacillus cereus UW85 in the soybean rhizosphere in the field. Mol. Ecol. 2:65-78. [DOI] [PubMed] [Google Scholar]
- 25.Hamel, C. 1996. Prospects and problems pertaining to the management of arbuscular mycorrhizae in agriculture. Agric. Ecosyst. Environ. 60:197-210. [Google Scholar]
- 26.Hamid, M., I. A. Siddiqui, and S. Shahid Shaukat. 2003. Improvement of Pseudomonas fluorescens CHA0 biocontrol activity against root-knot nematode by the addition of ammonium molybdate. Lett. Appl. Microbiol. 36:239-244. [DOI] [PubMed] [Google Scholar]
- 27.Harrier, L. A. 2001. The arbuscular mycorrhizal symbiosis: a molecular review of the fungal dimension. J. Exp. Bot. 52:469-478. [DOI] [PubMed] [Google Scholar]
- 28.Hewitt, R., J. C. Suit, and D. Billen. 1967. Utilization of 5-bromouracil by thymineless bacteria. J. Bacteriol. 93:86-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobbie, S. E. 1992. Effects of plant species on nutrient cycling. Trends Ecol. E. 7:336-339. [DOI] [PubMed] [Google Scholar]
- 30.Hodge, A. 2000. Microbial ecology of the arbuscular mycorrhiza. FEMS Microbiol. Ecol. 32:91-96. [DOI] [PubMed] [Google Scholar]
- 31.Hongcan, L., Y. Xu, Y. H. Ma, and Z. Peijin. 2000. Characterisation of Micrococcus antarcticus sp. nov., a psychrophilic bacterium from Antarctica. Int. J. Syst. E vol. Microbiol. 50:715-719. [DOI] [PubMed] [Google Scholar]
- 32.Jakobsen, I., and I. L. Rosendahl. 1990. Carbon flow into soil and external hyphae from roots of mycorrhizal cucumber roots. New Phytol. 115:77-83. [Google Scholar]
- 33.Jarstfer, A. G, and D. M. Sylvia. 1992. Inoculum production and inoculation technologies of vesicular arbuscular mycorrhizal fungi, p. 349-377. In B. Metting (ed.), Soil technologies: applications in agriculture, forestry and environmental management. Marcel Dekker, Inc., New York, N.Y.
- 34.Jeffries, P., S. Gianinazzi, S. Perotto, K. Turnau, and J.-M. Barea. 2003. The contribution of arbuscular mycorrhizal fungi in sustainable maintenance of plant health and soil fertility. Biol. Fertil. Soils 37:1-16. [Google Scholar]
- 35.Juteau, P., R. Larocque, D. Rho, and A. LeDuy. 1999. Analysis of the relative abundance of different types of bacteria capable of toluene degradation in a compost biofilter. Appl. Microbiol. Biotechnol. 52:863-868. [DOI] [PubMed] [Google Scholar]
- 36.Kazbekov, E. N., and L. G. Vyacheslavov. 1987. Effects of microwave irradiation on some membrane-related processes in bacteria. Gen. Physiol. Biophys. 6:57-64. [PubMed] [Google Scholar]
- 37.Lark, K. G. 1966. Regulation of chromosome replication and segregation in bacteria. Bacteriol. Rev. 30:3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence, T. S., and W. W. Ford. 1916. Studies on aerobic spore-bearing non-pathogenic bacteria. Spore-bearing bacteria in milk. J. Bacteriol. 1:277-320. [PMC free article] [PubMed] [Google Scholar]
- 39.Mamatha, G., D. J. Bagyaraj, and S. Jaganath. 2002. Inoculation of field-established mulberry and papaya with arbuscular mycorrhizal fungi and a mycorrhiza helper bacterium. Mycorrhiza 12:313-316. [DOI] [PubMed] [Google Scholar]
- 40.McAllister, C. B., I. García-Romera, J. Martin, A. Godeas, and J. A. Ocampo. 1995. Interaction between Aspergillus niger van Tiegh. and Glomus mosseae (Nicol. and Gerd.) Gerd. and Trappe. New Phytol. 129:309-316. [DOI] [PubMed] [Google Scholar]
- 41.McGonigle, T. P., M. H. Miller, D. G. Evans, D. G. Fairchild, and J. A. Swann. 1990. A new method which gives an objective measure of colonisation of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115:495-501. [DOI] [PubMed] [Google Scholar]
- 42.Meselson, M., and F. W. Stahl. 1958. The replication of DNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 44:671-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer, J. R., and R. G. Linderman. 1986. Response of subterranean clover to dual-inoculation with vesicular-arbuscular mycorrhizal fungi and a plant growth-promoting bacterium, Pseudomonas putida. Soil Biol. Biochem. 18:185-190. [Google Scholar]
- 44.Mosse, B. 1962. The establishment of vesicular-arbuscular mycorrhiza under aseptic conditions. J. Gen. Microbiol. 27:509-520. [DOI] [PubMed] [Google Scholar]
- 45.Olsson, P. A., E. Bååth, I. Jakobsen, and B. Söderström. 1996. Soil bacteria respond to presence of roots but not to mycelium of arbuscular mycorrhizal fungi. Soil Biol. Biochem. 4/5:463-470. [Google Scholar]
- 46.Pacovski, R. S. 1989. Influence of inoculation with Azospirillum brasilense and Glomus fasciculatum on sorghum nutrition, p. 235-239. In F. A. Skinner, R. M. Bodden, and I. Fendrik (ed.), Nitrogen fixation with non-legumes. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 47.Poole, E. J., G. D. Bending, J. M. Whipps, and D. J. Read. 2001. Bacteria associated with Pinus sylvestris-Lactarius rufus ectomycorrhizas and their effects on mycorrhiza formation in vitro. New Phytol. 151:743-751. [DOI] [PubMed] [Google Scholar]
- 48.Robarts, R. D., and T. Zohary. 1993. Fact or fiction—bacterial growth rates and production as determined by (methyl-3H)-thymidine? Adv. Microb. Ecol. 13:371-425. [Google Scholar]
- 49.Sacchi, C. T., A. M. Whitney, L. W. Mayer, R. Morey, A. Steigerwalt, A. Boras, R. S. Weyant, and T. Popovic. 2002. Sequencing of 16S rRNA gene: a rapid tool for identification of Bacillus anthracis. Emerg. Infect. Dis. 8:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sen, R., E.-L. Nurmiaho-Lassila, K. Haahtela, and K. Korhonen. 1996. Specificity and mode of primary attachment of Pseudomonas fluorescens strains to the cell walls of ectomycorrhizal fungi, p. 661-664. In D. Azcon-Aguilar and J. M. Barea (ed.), Mycorrhizas in integrated systems: from genes to plant development. ECSC-EC-EAEC Press, Brussels, Belgium.
- 51.Shida, O., H. Takagi, K. Kadowaki, L. K. Nakamura, and K. Komagata. 1997. Emended description of Paenibacillus amylolyticus and description of Paenibacillus illinoisensis sp. nov. and Paenibacillus chibensis sp. nov. Int. J. Syst. Bacteriol. 47:299-306. [DOI] [PubMed] [Google Scholar]
- 52.Smith, S. E., and D. J. Read. 1997. Mycorrhizal symbiosis. Academic Press, New York, N.Y.
- 53.Stone, J. H., M. M. Gabriel, and D. G. Ahearn. 1999. Adherence of Pseudomonas aeruginosa to inanimate polymers including biomaterials. J. Ind. Microbiol. Biotechnol. 23:713-717. [DOI] [PubMed] [Google Scholar]
- 54.Söderström, B. 1992. The ecological potential of the ectomycorrhizal mycelium, p. 77-83. In D. J. Read, D. H. Lewis, A. H. Fitter, and I. J. Alexander (ed.), Mycorrhizas in ecosystems. CAB International, Wallingford, United Kingdom.
- 55.Unge, A., R. Tombolini, M. E. Davey, F. J. de Bruijn, and J. K. Jansson. 1997. GFP as a marker gene, 6.1.13, p. 1-16. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 56.Unge, A., R. Tombolini, L. Molbak, and J. K. Jansson. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl. Environ. Microbiol. 65:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urbach, E., K. L. Vergin, and S. J. Giovannoni. 1999. Immunochemical detection and isolation of DNA from metabolically active bacteria. Appl. Environ. Microbiol. 65:1207-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vyacheslavov, L. G., and M. I. Mosevitsky. 1977. Thymidine uptake in bacteria: the effect of purine nucleosides. Eur. J. Biochem. 74:313-318. [DOI] [PubMed] [Google Scholar]
- 59.Wagner-Dobler, I., R. Pipke, K. N. Timmis, and D. F. Dwyer. 1992. Evaluation of aquatic sediment microcosms and their use in assessing possible effects of introduced microorganisms on ecosystem parameters. Appl. Environ. Microbiol. 58:1249-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyss, P., T. H. Boller, and A. Wiemken. 1992. Testing the effect of biological control agents on the formation of vesicular arbuscular mycorrhiza. Plant Soil 147:159-162. [Google Scholar]