Abstract
Highly efficient production of a Thermomyces lanuginosus IOC-4145 β-1,4-xylanase was achieved in Pichia pastoris under the control of the AOX1 promoter. P. pastoris colonies expressing recombinant xylanase were selected by enzymatic activity plate assay, and their ability to secrete high levels of the enzyme was evaluated in small-scale cultures. Furthermore, an optimization of enzyme production was carried out with a 23 factorial design. The influence of initial cell density, methanol, and yeast nitrogen base concentration was evaluated, and initial cell density was found to be the most important parameter. A time course profile of recombinant xylanase production in 1-liter flasks with the optimized conditions was performed and 148 mg of xylanase per liter was achieved. Native and recombinant xylanases were purified by gel filtration and characterized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, circular dichroism spectroscopy, matrix-assisted laser desorption ionization-time of flight-mass spectrometry and physicochemical behavior. Three recombinant protein species of 21.9, 22.1, and 22.3 kDa were detected in the mass spectrum due to variability in the amino terminus. The optimum temperature, thermostability, and circular dichroic spectra of the recombinant and native xylanases were identical. For both enzymes, the optimum temperature was 75°C, and they retained 60% of their original activity after 80 min at 70°C or 40 min at 80°C. The high level of fully active recombinant xylanase obtained in P. pastoris makes this expression system attractive for fermentor growth and industrial applications.
β-1,4-Xylanases present great potential in several biotechnological applications. They catalyze the hydrolysis of internal β-1,4-d-xylose units and are used in bread making, clarification of beer and juices, and the conversion of xylan-containing lignocellulosic materials to d-xylose, which can be converted to a variety of bioproducts with high aggregate value (3). Xylanases are also believed to be essential in improving the nutritional quality of animal feed and in the recovery of textile fibers (15, 35). In addition, increasing concern over preserving the environment from industrial wastes has raised interest in applying microbial xylanases in the pulp and paper industry. The aim of these biotechnological processes is to reduce or replace the harmful alkaline extraction of hemicellulose and the need for chlorine in the bleaching process without affecting the cellulose fiber strength of paper products. Xylanase treatment breaks up the cell wall structure, thereby enabling lignin removal in subsequent stages. The fragmentation of the xylan polymer allows free diffusion of the portions of residual lignin that are covalently attached to xylan (35).
For the commercial realization and economic viability of xylanase production, it is necessary to identify organisms that produce high levels of this enzyme. Several strains of the thermophilic fungus Thermomyces lanuginosus secrete high levels of xylanases, which are very active and stable at elevated temperatures (9, 27, 31). Purified and unpurified extracellular xylanase preparations from this fungus have been successfully used for the bleaching of pulps (4, 11, 20, 21).
Purkarthofer et al. (28) were the first to report that the T. lanuginosus strain DSM 5826 secreted a xylanase during submerged cultivation in a complex medium. Accordingly, we demonstrated that T. lanuginosus strain IOC4145, isolated from Brazilian soil, secreted cellulase-free xylanase activity in submerged and semisolid fermentation with corncob as the substrate (9). Xylanase activity in the semisolid fermentation culture medium was high, displayed considerable thermostability, and was stable over a broad pH range (4.0 to 11.0). However, the scale-up of semisolid fermentation is still a challenge because controlling the heat and mass transfer and dissolved oxygen is very complex. To facilitate enzyme production and the industrial use of the thermophilic xylanase preparations, one should develop a heterologous expression system that produces large amounts of secreted protein with an organism that can be grown in industrial-scale fermentors.
In recent years, several industrial yeasts have been developed as recombinant host systems for the commercial production of heterologous proteins. These organisms combine ease of genetic manipulation with the ability to perform many eukaryotic posttranslational modifications (6). One of the most commonly used systems is the methylotrophic yeast Pichia pastoris, in which expression is driven by one of the strongest known regulated promoters, the alcohol oxidase I (AOX1) promoter, which is induced by methanol and repressed by other carbon sources such as glucose, glycerol, and ethanol (23). Another important feature of this system is its ability to achieve extremely high cell densities, enabling efficient protein production and secretion (7, 34).
For a broad application, the cost of enzymes is one of the main factors determining the economics of a process. Cost reduction of enzyme production by optimizing the fermentation medium is a basic principle for industrial applications. Medium optimization was traditionally done by varying one factor while keeping the other factors at a constant level. This technique is time-consuming and incapable of detecting the true optimum due, especially, to interactions among factors. One of the most useful techniques to identify the relative significance of factors and find optimal conditions that give maximal results is statistically based experimental designs, such as the 23 factorial design (24).
In this work we describe the cDNA cloning, heterologous expression, and characterization of a recombinant T. lanuginosus xylanase. We optimized growth conditions of the Pichia pastoris system for high-level laboratory-scale xylanase expression, determining the most relevant medium components for enzyme production by the use of a 23 factorial experimental design involving two concentrations of each factor. This enabled us to obtain the secretion of a large amount of pure recombinant xylanase in P. pastoris, avoiding the use of semisolid media for the production of the native enzyme. In addition, we compared the recombinant enzyme to the native xylanase with respect to structure and activity and found them to be identical. To our knowledge this is the first report on the expression of a thermostable xylanase from a thermophilic fungus in P. pastoris.
MATERIALS AND METHODS
Materials.
Oligonucleotides were purchased from DNAgency (Malvern, Pa.). Plasmids were from Invitrogen (Carlsbad, Calif.; pPIC9) and from Stratagene (La Jolla, Calif.; pSK+). Restriction enzymes were from Amersham Biosciences (Uppsala, Sweden) and Pfu DNA polymerase was from Stratagene. Yeast nitrogen base (YNB) without amino acids was from BD/Difco Laboratories (Franklin Lakes, N.J.). Biotin, methanol, glycerol, and mono- and dibasic potassium phosphate were from Merck (Haar, Germany). Dextrose and other chemicals were purchased from Sigma (St. Louis, Mo.). Escherichia coli strain XL1Blue (Stratagene) was used for propagation of plasmids, and P. pastoris strain GS115 (his4) (Invitrogen) was used for protein expression. Thermomyces lanuginosus was isolated from Brazilian soil and identified by Fundação Instituto Oswaldo Cruz, Rio de Janeiro, Brazil, under the code IOC-4145 (9).
Reverse transcription-PCR and cloning of T. lanuginosus xylanase cDNA.
T. lanuginosus was grown in 1% birchwood xylan (9), and total RNA was extracted with Trizol reagent (Invitrogen). First-strand cDNA synthesis was performed with 2 μg of total RNA, 200 ng of random hexamers (Amersham Biosciences), 1× reverse transcription buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2), 10 mM dithiothreitol, 0.5 mM deoxynucleoside triphosphate mix, 5 U of RNase inhibitor (Amersham Biosciences), and 200 U of Superscript II reverse transcriptase (Invitrogen) in a total volume of 20 μl at 42°C for 50 min. The cDNA encoding XynA was amplified by PCR with P9XILfor (5′-GCTTACGTAGAATTCCAGACAACCCCCAACTCGGAG-3′) as the sense primer. The underlined sequence corresponds to the tetrapeptide TyrValGluPhe encoded by pPIC9, containing an EcoRI restriction site (Fig. 1). The antisense primer was P9XILrev (5′-TTAATTCGCGGCCGCTTAGCCCACGTCAGCAACGGT-3′). The underlined sequence corresponds to part of the pPIC9 plasmid and a NotI restriction site (Fig. 1).
FIG. 1.
Plasmid construction for the expression of T. lanuginosus XynA in P. pastoris. (A) 5′ region of the recombinant XynA/pPIC9 construct, showing the last eight amino acids of the S. cerevisiae α-factor secretion signal and the tetrapeptide TyrValGluPhe (bold characters) of the pPIC9 plasmid placed ahead of the first three amino acids of the mature XynA sequence. The arrowheads indicate the KEX2 and STE13 protease cleavage sites necessary for the proteolytic processing of the secretion signal. The EcoRI restriction site used to digest the PCR product for subcloning into the pPIC9 plasmid is indicated by an arrow and underlined. (B) 3′ region of the recombinant XynA/pPIC9 construct, corresponding to the last six amino acids of the mature XynA sequence and a stop codon followed by (bold characters) 24 nucleotides of the pPIC9 expression plasmid. The NotI restriction site used to digest the PCR product for subcloning into the pPIC9 plasmid is indicated by an arrow and underlined.
The primers were designed based on the T. lanuginosus XynA gene sequence deposited in GenBank (accession number U35436) (30). The PCR was performed with 3 μl of the reverse transcription reaction mix in a total volume of 100 μl with 2.5 U of Pfu DNA polymerase (Stratagene), 200 ng of each primer, 0.2 mM deoxynucleoside triphosphate mix, and 1× PCR buffer (20 mM Tris-HCl [pH 8.8], 2 mM MgSO4, 10 mM KCl, 10 mM [NH4]2SO4, 0.1% Triton X-100, 0.1 mg of bovine serum albumin per ml). PCR cycling conditions consisted of an initial step of 5 min at 95°C, followed by 30 cycles of 45 s at 95°C, 1 min at 60°C, and 2 min at 72°C, and a final extension step of 10 min at 72°C.
A 605-bp PCR product consisting of the entire XynA coding region without its original signal sequence was excised from an agarose gel after electrophoresis and purified by adsorption to silica. This product was digested with EcoRI and NotI and cloned into pSK+ and the XynA sequence was checked by DNA sequencing with dideoxy chain terminator chemistry (29). The sequence of the cloned PCR product was identical to the XynA sequence deposited by Schlacher et al. (30).
Construction of the expression plasmid.
Cloned XynA was excised from pSK+ by digestion with EcoRI and NotI and ligated into pPIC9 previously digested with the same enzymes. This allowed in-frame cloning into the Saccharomyces cerevisiae α-factor secretion signal of pPIC9 with the KEX2 and STE13 protease cleavage sites placed ahead of XynA (Fig. 1). The plasmid (reXynA/pPIC9) was linearized with SalI in order to favor integration at the P. pastoris his4 locus, and 5 μg was used to electroporate P. pastoris cells under the conditions specified by the supplier.
Screening of His+ transformants for methanol utilization phenotype.
Selection of His+ transformants was done on minimal selective MD medium (1.34% YNB, 4 × 10−5% biotin, 1% dextrose, and 1.5% agar). The Mut (methanol utilization) phenotype of recombinant clones was determined by patching His+ transformants on MD and MM (1.34% YNB, 4 × 10−5% biotin, 0.5% methanol, and 1.5% agar) plates.
Selection of a high-secreting clone.
The clones that produced the highest levels of xylanase activity were chosen based on the Congo red-polysaccharide interactions (36). Clones were plated on MM agar (1.5%) containing 1% xylan, and after 96 h at 30°C the agar was stained with Congo red (1 mg/ml) for 15 min. The zones of xylan hydrolysis were seen as large haloes after flooding the agar with 1 M HCl. The selected positive colonies were cultured in 4 ml of BMG medium (100 mM potassium phosphate [pH 6.0], 1.34% YNB, 4 × 10−5% biotin, and 1% glycerol) in 50-ml plastic tubes. When an optical density at 600 nm (OD600) of 13 to 15 (6.5 × 108 to 7.5 × 108 cells/ml) was reached, the cultures were centrifuged (1,500 × g for 5 min), and the cell mass was resuspended to an OD600 of 1 (0.5 × 108 cells/ml) in BMM medium (100 mM potassium phosphate, pH 6.0, 1.34% YNB, 4 × 10−5% biotin, 0.5% methanol) to induce recombinant xylanase (recombinant XynA) expression. These cultures were grown for 72 h at 30°C at 250 rpm in a total volume of 5 to 7 ml in 50-ml plastic tubes or 50 ml in 500-ml shake flasks and supplemented with 0.5% methanol every 24 h. The experiment was carried out in duplicates.
Experimental design.
Three factors were studied for xylanase production: methanol and YNB concentrations and initial cell density. A 23 factorial design involving two concentrations for each factor (Table 1) was effective in searching for an optimum condition. Xylanase production for each triplicate of shake flask cultures was determined by enzyme activity assays after a growth period of 72 h. The software Statistica '99 for Windows version 5.5 (StatSoft, Tulsa, Okla.) was used to evaluate and to compare the amount of xylanase produced by each medium. The Pareto chart was constructed with this software and shows the values of the Student's t test (24) for each component of the medium. The dotted line indicates the minimal magnitude of statistically significant effects to a confidence level of 95%.
TABLE 1.
Assigned concentrations of variables for different levels of the 23 factorial design
| Factor | Levela
|
|
|---|---|---|
| −1 | +1 | |
| Methanol (%, vol/vol) | 0.5 | 1.0 |
| YNB (%, wt/vol) | 0.9 | 1.8 |
| Initial cell density (108 cells/ml) | 0.5 | 5 |
The numbers indicate the maximum (+1) and the minimum (−1) concentrations of the factors.
Recombinant XynA expression in P. pastoris.
The P. pastoris clone secreting the highest levels of recombinant XynA (clone 7) was cultured in 100 ml of BMG medium at 30°C with constant shaking in 500-ml shake flasks. When cultures reached an OD600 of about 13 to 15 (6.5 × 108 to 7.5 × 108 cells/ml), they were centrifuged (4,000 × g for 5 min) and the cell mass was resuspended in 250 ml of BMM to an OD600 of 10 (5 × 108 cells/ml). Cultures were grown at 30°C with constant shaking on a rotary shaker in 1-liter shake flasks for 96 h. The cultures were supplemented daily with 0.5% methanol, and the pH was adjusted to 6.0 every 24 h. The experiment was carried out in duplicate.
XynA production in semisolid fermentation.
A 20% (vol/vol) mycelium suspension (9) was used to inoculate 500-ml shake flasks, containing 15 g of corncob as a carbon source and 22.5 ml of production medium (0.2% peptone, 0.2% meat extract, 0.2% NaCl, and 0.1% KH2PO4). After inoculation, the shake flasks were incubated at 45°C for 3 days.
Purification of XynA and recombinant XynA.
T. lanuginosus or P. pastoris cells were removed from the culture media by filtration under vacuum with filter paper (Whatman, no. 4, fast flow rate) or by centrifugation at 1,500 × g for 5 min, respectively. Cell-free media were dialyzed against water at 4°C for 2 days, lyophilized, and reconstituted in 6 or 3 ml of water, respectively. XynA and recombinant XynA were purified by gel filtration on a Superdex G-75 HR column (30 by 1.0 cm) on a fast protein liquid chromatography system (Amersham Biosciences, Uppsala, Sweden). The column was equilibrated and eluted in 20 mM Tris (pH 7.5)-150 mM NaCl. The flow rate was 0.5 ml/min, and the absorbance of the eluant was monitored at 214 and 280 nm. The fractions (1.5 ml) were analyzed for protein content and xylanase activity as described below. The active fractions were pooled and referred to as the purified enzyme preparation.
Enzyme assays.
Xylanase activity was assayed with birchwood xylan as the substrate (9). One hundred microliters of the reaction mixture containing 10 μl of diluted enzyme solution and 90 μl of a 1% suspension of xylan in universal buffer (pH 6.0) was incubated at 75°C for 3 min. In the initial time course and growth experiments, the reducing sugars were determined by the dinitrosalicylic acid (DNS) procedure (22), and xylose was used as a standard. Xylanase activity was also determined by the Somogyi-Nelson procedure, which is more accurate in the determination of reducing sugars, to measure specific activities and to quantify the total amount of xylanase produced in the optimized growth conditions (5, 33). Released xylose was measured spectrophotometrically at 540 nm. One unit of enzyme activity was defined as the amount of enzyme capable of releasing 1 μmol of reducing sugar per minute under the assay conditions. Protein concentration was measured by the method of Lowry et al. (19) with bovine serum albumin as a standard.
All experiments were performed in duplicate or triplicate, and the analytical measurements were performed at least in triplicate. All statistical analyses of the experimental data were done in Microsoft Excel (Office 2000).
Physicochemical parameters.
The effect of temperature on xylanase activity and stability was determined as described by Damaso et al. (9). The residual activity was measured as stated above.
Polyacrylamide gel electrophoresis.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was carried out on a 16% (wt/vol) polyacrylamide gel in the Laemmli system (17), and proteins were visualized by Coomassie blue staining.
Circular dichroism.
Circular dichroism measurements were obtained on a Jasco J-715 spectropolarimeter at 25°C in a quartz cell of 0.1-cm path length, with two scans per spectrum, a scan speed of 50 nm/min, and an 8-s integration time.
MALDI-TOF analysis.
Recombinant XynA (100 ng) was analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry with a Voyager DE Pro mass spectrometer (Applied Biosystems, Foster City, Calif.). The sample was mixed with a saturated solution of α-cyano-4-hydroxycinnamic acid containing 50% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid. The spectrum was acquired in direct mode and interpreted with the Voyager software version 5.0.
RESULTS
Construction of the expression vector and selection of the clones.
The nucleotide sequence encoding the entire T. lanuginosus XynA without its original signal sequence was cloned in frame with the S. cerevisiae α-factor secretion signal into the expression vector pPIC9 (Fig. 1), and the resulting plasmid was used to transform P. pastoris. A total of 500 yeast colonies were randomly picked and grown in MM agar containing 1% xylan. After a 72-h growth period, the plates were stained to detect substrate hydrolysis, and 13 colonies which produced the largest haloes were chosen (data not shown).
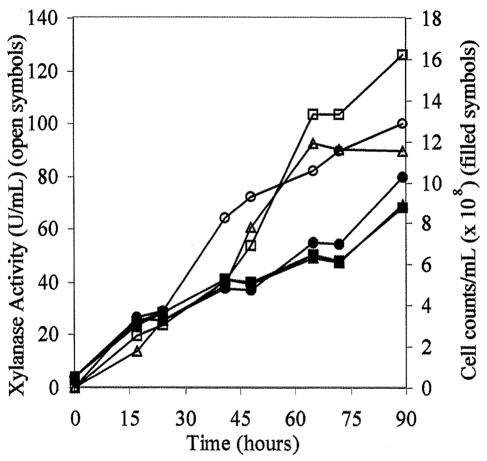
The presence of xylanase activity was also tested in the 13 positive colonies in 5-ml cultures. The highest level of enzyme activity was obtained after 72 h of methanol induction (data not shown). Three colonies producing the highest levels of xylanase activity were grown in 500-ml shake flasks with an initial cell density of 0.5 × 108 cells/ml. As shown in Fig. 2, the secretion level of recombinant XynA was in the range of 90 to 126 U/ml after 90 h of induction. The clone expressing the highest level of activity, named reXynA7, was used in the experiments that follow to optimize enzyme production.
FIG. 2.
Time course of recombinant XynA expression in 50 ml of BMM medium inoculated with the three highest-expressing clones. Cell counts (filled symbols) and xylanase activity (open symbols) were monitored throughout the experiment for clone 5 (•,○), clone 7 (▪,□), and clone 13 (▴,▵). Xylanase activity was measured by the DNS procedure as described in Materials and Methods. The values correspond to the averages of five measurements, and the error was below 10%.
For these experiments and in the next section, xylanase activity was measured by the DNS procedure, which, although simple and fast, can give values that are not proportional to the actual number of hemiacetal reducing groups (5). Therefore, these values are included mostly for comparative purposes.
Optimization of medium constituents.
Different statistical experimental designs have been employed successfully for native xylanase production (8, 10, 14); however, they have not been used for recombinant systems.
Experiments with a 23 factorial design (24) were carried out with two values for each factor to determine the best medium composition and initial cell density for recombinant XynA production (Table 1). We found that the concentrations of methanol and YNB as well as the initial cell density exert distinct effects on xylanase production. The maximum and minimum xylanase activities, measured by the DNS procedure, were obtained in media M6 and M2, respectively (Table 2). As shown in Table 2, each medium has a different composition, varying the levels of each factor. M1 has the lowest values of each factor, while M8 has the highest.
TABLE 2.
Effect of medium (M1 to M8) composition on the production of recombinant XynA by P. pastoris after 48 h of induction in 5-ml cultures
| Medium | Composition
|
Xylanase activity (U/ml)a | ||
|---|---|---|---|---|
| Methanol (%, vol/vol) | YNB (%, wt/vol) | Initial cell density (108 cells/ml) | ||
| M1 | 0.5 | 0.9 | 0.5 | 157 |
| M2 | 1.0 | 0.9 | 0.5 | 152 |
| M3 | 0.5 | 1.8 | 0.5 | 156 |
| M4 | 1.0 | 1.8 | 0.5 | 168 |
| M5 | 0.5 | 0.9 | 5.0 | 305 |
| M6 | 1.0 | 0.9 | 5.0 | 360 |
| M7 | 0.5 | 1.8 | 5.0 | 340 |
| M8 | 1.0 | 1.8 | 5.0 | 264 |
Xylanase activity is the average of three experiments (DNS procedure).
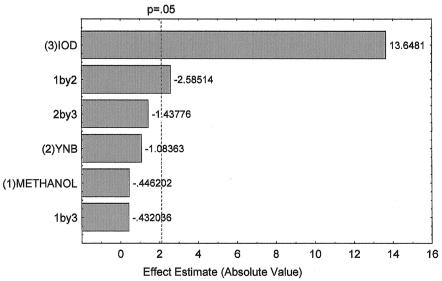
The Pareto chart was used to compare the amount of recombinant XynA produced in all media. The chart shows the values of the Student t test for each component of the BMM medium, and the dotted line indicates the minimal magnitude of statistically significant effects to a confidence level of 95% (Fig. 3). The Pareto chart clearly shows that methanol and YNB did not have any significant effect on recombinant XynA production. On the other hand, the initial cell density of the culture was shown to be the most important factor that affected enzyme production. While for other microorganisms the main impact on growth and cost of culture is generally the nitrogen source, for P. pastoris we could reduce this factor without being detrimental to protein production. Finally, as shown in Table 2, medium M5 (0.9% YNB, 0.5% methanol, and initial cell density of 5 × 108 cells/ml) was the best one, considering the yield in xylanase activity and the amount of YNB and methanol used for enzyme production. This optimized medium was named BMM(M5) and used for the next set of experiments.
FIG. 3.
Pareto chart of the effect of medium composition on recombinant XynA expression by P. pastoris. The Pareto chart shows the values of the Student's t test for each medium component. The dotted line indicates the confidence level of 95% (i.e., P = 0.05). Values to the right of this line are statistically significant. Initial cell density (IOD) is the influence of initial cell density on xylanase production. 1by2 means the influence of both methanol and YNB concentration on xylanase production, and 2by3 stands for the influence of both YNB concentration and initial cell density on xylanase production. 1by3 stands for the influence of both methanol concentration and initial cell density on xylanase production. Xylanase activity was measured by the DNS procedure as described in Materials and Methods. The experiment was carried out in triplicate, and the values correspond to the averages of five measurements, with an error below 10%.
Optimized expression of recombinant XynA in P. pastoris.
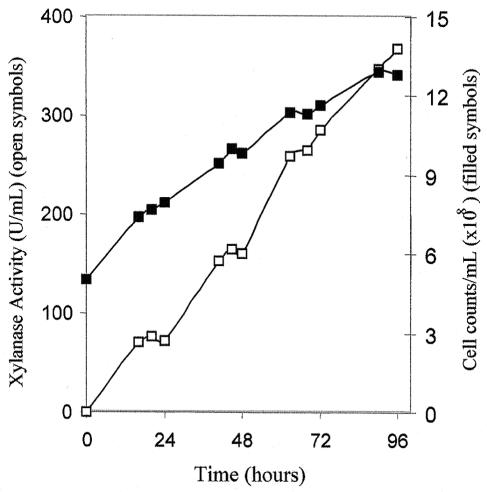
The kinetic profile of recombinant XynA expression was studied in 250 ml of BMM(M5). Every 12 h of induction, a sample was collected and the cell density and enzyme activity were monitored (Fig. 4). The maximum expression of recombinant XynA occurred after 96 h of induction. Comparing the results obtained in 250-ml cultures (Fig. 4) with those found in 5-ml BMM(M5) cultures (Table 2), it can be verified that the level of enzyme expression did not change significantly by the use of a larger culture.
FIG. 4.
Time course of recombinant XynA expression in 250 ml of optimized medium BMM(M5) inoculated with P. pastoris clone 7. Cell counts (▪) and xylanase activity (□) were monitored throughout the experiment. Xylanase activity was measured by the DNS procedure as described in Materials and Methods. The values correspond to the averages of five measurements, and the error was below 10%.
Purification of XynA and recombinant XynA.
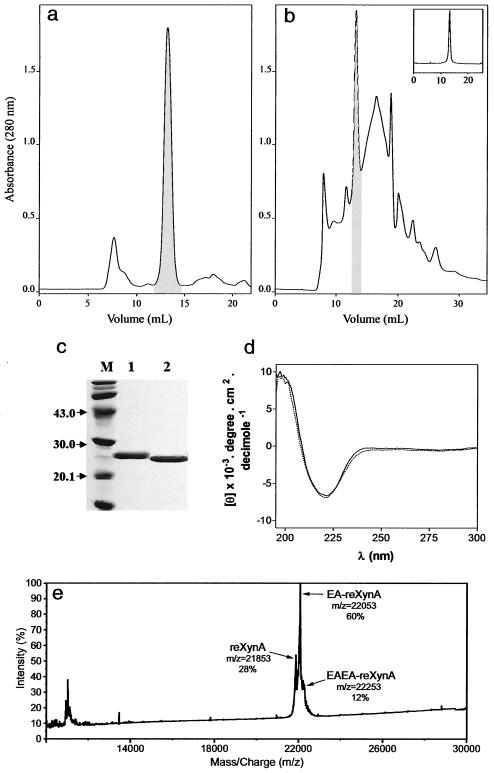
Recombinant XynA was purified to homogeneity from P. pastoris culture medium after 4 days of induction, while native XynA was purified from T. lanuginosus culture medium after 3 days of semisolid fermentation. The recombinant enzyme was purified by a single gel filtration chromatographic step (Fig. 5a), but the native enzyme had to be subjected to two gel filtration steps (Fig. 5b). One significant advantage in the use of P. pastoris for xylanase production in relation to the native fungus is that the main secreted protein found in the medium is recombinant XynA, while several contaminant proteins are present in the semisolid fermentation medium (Fig. 5a and b).
FIG. 5.
Purification of XynA and recombinant XynA. Culture media from P. pastoris (a) and T. lanuginosus (b) were processed and applied to a Superdex G-75 HR column as described in Materials and Methods. All fractions were assayed for xylanase activity, and active fractions are in the hatched areas. The active fractions in b were pooled and subjected to another gel filtration step (inset) as described in Materials and Methods. (c) SDS-PAGE of the purified enzymes. Lanes: M, molecular mass markers; 1, purified recombinant XynA (8 μg); 2, purified XynA (8 μg). (d) circular dichroic spectra of recombinant XynA (solid line) and XynA (doted line) were measured as described in Materials and Methods. (e) MALDI-TOF mass spectrum of purified recombinant XynA. Three forms of the protein were detected, and the corresponding peaks, molecular mass, and relative abundance are indicated.
Purified recombinant XynA migrated as a single homogeneous band on an SDS-PAGE gel (16% acrylamide) with an apparent molecular mass of 26.9 kDa, which is similar to that of the native enzyme, 25.5 kDa (Fig. 5c, lanes 1 and 2, respectively). MALDI-TOF analysis of recombinant XynA revealed a mixture of three differentially processed isoforms (Fig. 5e). The calculated molecular masses were 22.1 kDa for the most abundant protein species (60%), 21.9 kDa for the second highest peak (28%), and 22.3 kDa for the least abundant isoform (12%). Native XynA has a theoretical molecular mass of 21.3 kDa, and our construct, if fully processed, would give a molecular mass of 21.9 kDa due to the addition of the tetrapeptide TyrValGluPhe to the recombinant XynA sequence (Fig. 1). This value completely agrees with the one found for the second most abundant protein species detected in the mass spectrum. The molecular mass of the main protein species is consistent with the GluAla dipeptide being left at the amino terminus of recombinant XynA. The experimental molecular mass found for the least abundant species agrees with a form in which the GluAlaGluAla tetrapeptide is left at the amino terminus. These observations would fully account for the size difference observed in the SDS-PAGE gel between the native and recombinant enzymes. This type of amino-terminal variability is commonly observed when heterologous proteins are secreted by P. pastoris with the α-factor preprotein signal sequence (7).
As activity values measured by the DNS procedure can give higher values due to overoxidation of sugars, we measured the specific activity by the more accurate method of Somogyi-Nelson (5, 32). We obtained 323 U/mg and 272 U/mg for the native and recombinant enzymes, respectively. The activities present in the culture media were also analyzed by the Somogyi-Nelson procedure, enabling us to quantify the amount of active protein made in the T. lanuginosus IOC-4145 and in the P. pastoris media. In semisolid medium, T. lanuginosus IOC-4145 produced 88.5 U/ml after 120 h, which corresponded to 270 mg of XynA per liter. P. pastoris, after 96 h of methanol induction, secreted 40.2 U/ml (360 U/ml measured by DNS procedure), representing 148 mg of virtually pure and active recombinant XynA per liter of culture.
To determine if the native and recombinant proteins had a similar secondary structure content, we performed circular dichroism spectra for both enzymes (Fig. 5d). Absorption maxima at 197 nm and minima at 221 nm clearly indicated that they were identical in relation to β-strand and α-helical structures.
Effect of temperature on xylanase activity and stability.
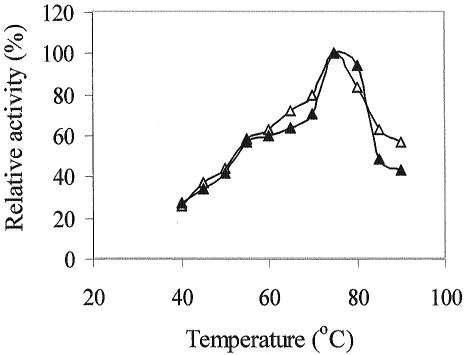
The optimal temperature for xylanase activity was determined for the purified native and recombinant enzymes, and both showed an identical profile, with maximal activity at 75°C (Fig. 6).
FIG. 6.
Effect of temperature on the activity of recombinant XynA (▵) and XynA (▴). Xylanase activity was measured by the DNS procedure at different temperatures as described in Materials and Methods. The values correspond to the averages of five measurements, and the error was below 10%.
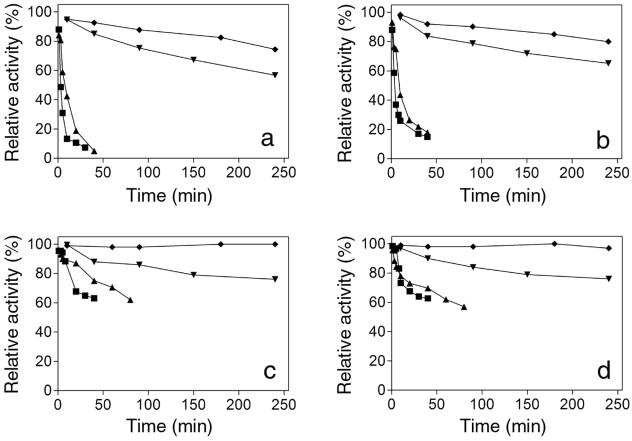
An investigation of thermostability revealed that the purified recombinant XynA and XynA enzymes (Fig. 7c and d, respectively) showed identical stability. Furthermore, they were more stable in the pure form than in the crude culture medium (Fig. 7a and b). At 50°C, the enzymes retained full activity after 4 h of incubation, while the crude culture medium lost almost 20% of its original activity. Purified recombinant XynA and XynA also retained 60% of their activity after 40 min at 80°C or 80 min at 70°C, while the crude medium lost 95% of the initial activity before 40 min of incubation at these temperatures.
FIG. 7.
Thermostability of recombinant XynA and XynA. Crude culture medium of P. pastoris expressing recombinant XynA (a), purified recombinant XynA (c), crude culture medium of T. lanuginosus (b), and purified XynA (d) were incubated at 50°C (♦), 60°C (▾), 70°C (▴), and 80°C (▪). Relative xylanase activity is shown, and the values correspond to the averages of five measurements, with an error below 10%.
DISCUSSION
Currently, the best medium for the production of native T. lanuginosus xylanase, irrespective of the strain used, is semisolid medium, with corncob as the substrate (9). However, the scale-up of this type of fermentation is not yet efficient. Therefore, we chose to produce a recombinant xylanase with the P. pastoris expression system. The crude supernatant obtained after centrifugation of the culture medium contains fewer proteins than T. lanuginosus semisolid fermentation supernatant (Fig. 5a and b), enabling the use of the cell-free P. pastoris medium in industrial processes without the need for further enzyme purification.
The first characterization and cDNA cloning of a xylanase from T. lanuginosus DSM 5826 was reported by Schlacher et al. (30), who expressed the enzyme as a LacZ fusion protein in E. coli. Our work is the first to report the expression of a highly thermophilic fungal xylanase in P. pastoris and, additionally, the first to optimize medium conditions by an experimental design protocol. The optimization of production medium with experimental design is a very useful tool to attain high levels of enzyme activity, with a lower cost. In this study, after medium optimization, the yield of xylanase expression was increased approximately threefold from the screening of positive clones to the 48-h induction experiment with BMM(M5) (Table 2).
A comparison of xylanase activities found in the culture medium of several T. lanuginosus strains grown in submerged fermentation for 6 days with corncob as the substrate revealed a great variability of enzyme production, ranging from 3,590 U/ml to 361 U/ml (31). The xylanase activity found in semisolid medium (850 U/ml) and in submerged medium (500 U/ml) of T. lanuginosus IOC-4145 cultures (unpublished observations) is similar to what was found by Singh et al. (31) for some of the lowest-expressing strains and is not very different from what we obtained with the recombinant system (360 U/ml). Even though the values cited above were all obtained with the DNS procedure to measure xylanase activity, they are useful for the purpose of comparison.
Various bacterial and fungal xylanases have been expressed in different eukaryotic systems, and, in all cases, enzyme expression levels as well as specific activities were much lower than what we obtained in this work (2, 13, 16, 18, 25, 26, 37, 38).
The characterization of recombinant XynA by SDS-PAGE and MALDI-TOF analysis showed that its relative molecular mass was higher than that of the native enzyme due to the addition of the tetrapeptide TyrValGluPhe in our construct and to heterogeneity in its amino-terminal processing, which could include up to four extra amino acids. The primary sequence of XynA does not display any N- or O-glycosylation signals (not shown), and, accordingly, no evidence for glycosylated isoforms was observed in the mass spectrum of recombinant XynA (Fig. 5e). Structural data obtained by protein crystallography for the T. lanuginosus DSM 5826 xylanase revealed that the amino terminus points outward and does not participate in the folding of the polypeptide chain, indicating that the extra amino acids may not be relevant for enzyme activity or conformation (12). However, as we observed a 16% lower xylanase activity with recombinant XynA, it may be that one of the alternative forms has a lower activity or is inactive. For the most part, though, the native and recombinant enzymes seemed to be identical as shown by the optimum temperature, thermostability, and content of β-strand and α-helical structures.
The recombinant and native xylanases showed similar thermostability and were as stable at high temperatures as the xylanases secreted by other T. lanuginosus strains (1, 32). Surprisingly, in the culture medium, both enzymes were less thermostable than in the purified form (Fig. 7a to d). The data were plotted as the log10 of the percentage of the remaining activity against time for the different temperatures (data not shown), and we observed a nonlinear decay in temperatures above 70°C, indicating that other factors such as proteolysis or protein aggregation may play a role in the inactivation of the unpurified enzymes.
In conclusion, recombinant XynA was efficiently secreted by P. pastoris, producing a recombinant protein with properties similar to the native one. With this system, detailed structure-function analysis of the xylanase can be carried out by means of site-directed mutagenesis. In addition, the recombinant xylanase showed enough thermostability to be used as a bleaching aid in the pulp and paper industry, as the native xylanases from T. lanuginosus strains IOC-4145 and SSBP have been used in the treatment of sugar cane bagasse pulp (11) and in bagasse soda pulp (4), respectively. Our next step will be to produce recombinant XynA in fermentor cultures, as we have observed through 23 factorial design that initial cell density is a crucial factor for a high level of protein expression. P. pastoris is particularly suitable for growth in fermentors, where parameters such as pH, aeration, and carbon source feed rate can be easily controlled (6), so very high cell densities can be achieved, which will probably enable us to increase the protein yield even further.
Acknowledgments
This work was supported by research grants given by Faperj, Finep/Pronex (014/98), and CNPq.
REFERENCES
- 1.Bennet, N. A., J. Ryan, P. Biely, M. Vrsanska, M. L. kremnicky, B. J. Macris, C. Dimitris, P Kekos, P. Katapodis, M. Claeyssens, W. Nerinckx, P. Ntauma, and M. K. Bhat. 1998. Biochemical and catalytic properties of an endoxylanase purified, from the culture filtrate of Thermomyces lanuginosus ATTC46882. Carbohydr. Res. 306:445-455. [DOI] [PubMed] [Google Scholar]
- 2.Berrin, J., G. Williamson, A. Puigserver, J. Chaix, W. R. McLauchlan, and N. Juge. 2000. High-level production of recombinant fungal endo-β-1, 4-xylanase in the methylotrophic yeast Pichia pastoris. Protein Expr. Purif. 19:170-187. [DOI] [PubMed] [Google Scholar]
- 3.Biely, P. 1985. Microbial xylanolytic systems. Trends Biotechnol. 3:286-290. [Google Scholar]
- 4.Bissoon, S., L. Christov, and S. Singh. 2002. Bleach boosting effects of purified xylanase from Thermomyces lanuginosus SSBP on bagasse pulp. Process Biochem. 37:567-572. [Google Scholar]
- 5.Breuil, C., and J. N. Saddler. 1985. Comparison of the 3, 5-dinitrosalicylic acid and Nelson-Somogyi methods of assaying for reducing sugars and determining cellulase activity. Enzyme Microb. Technol. 7:327-332. [Google Scholar]
- 6.Cereghino, G., J. Cereghino, C. Ilgen, and J. Cregg. 2002. Production of recombinant proteins in fermentor cultures of the yeast Pichia pastoris. Curr. Opin. Biotechnol. 13:329-332. [DOI] [PubMed] [Google Scholar]
- 7.Cereghino, J. L., and J. M. Cregg. 2000. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 24:45-66. [DOI] [PubMed] [Google Scholar]
- 8.Couri, S., S. C. Terzi, Pinto, G. A. S., S. P. Freitas, and A. C. A.Costa. 2000. Hydrolytic enzyme production in solid-state fermentation by Aspergillus niger 3T5B8. Process Biochem. 36:255-261. [Google Scholar]
- 9.Damaso, M. C. T., C. M. M. C. Andrade, and N. Pereira Jr. 2000. Use of corncob for endoxylanase production by thermophilic fungus Thermomyces lanuginosus IOC-4145. Appl. Biochem. Biotechnol. 84-86:821-834. [DOI] [PubMed] [Google Scholar]
- 10.Ghanem, N. B., H. H. Yusef, and H. K. Mahrouse. 2000. Production of Aspergillus terreus xylanase in solid-state cultures: application of the Plackett-Burman experimental design to evaluate nutritional requirements. Bioresource Technol. 73:113-121. [Google Scholar]
- 11.Gonçalves, A. R., D. S. Ruzene, C. M. Andrade, M. C. Damaso, and N. Pereira, Jr. 2001. Bleaching of ethanol/water-bagasse pulps by with xylanase from Thermomyces lanuginosus, p. 205-206. In Proceedings of the 8th International Conference on Biotechnology in the Pulp and Paper Industry. Helsinki, Finland.
- 12.Gruber, K., G. Klintschar, M. Hayn, A. Schlacher, W. Steiner, and C. Kratky. 1998. Thermophilic xylanase from Thermomyces lanuginosus: high-resolution X-ray structure and modeling studies. Biochemistry 37:13475-13485. [DOI] [PubMed] [Google Scholar]
- 13.Haan, R. D., and W. H. Z. Zyl. 2001. Differential expression of the Trichiderma reesei β-xylanase II (xyn2) gene in the xylose-fermenting yeast Pichia stipitis. Appl. Microbiol. Biotechnol. 57:521-527. [DOI] [PubMed] [Google Scholar]
- 14.Kadowaki, M. K., C. G. M. Souza, R. C. G. Simão, and R. M. Peralta. 1997. Xylanase production by Aspergillus tamarii, Appl. Biochem. Biotechnol. 66:97-106. [Google Scholar]
- 15.Kalogeris, E., P. Christakopoulos, D. Kekos, and B. J. Macris. 1998. Studies on the solid-state production of thermostable endoxylanases from Thermoascus aurantiacus: characterization of two isozymes. J. Biotechnol. 60:155-163. [Google Scholar]
- 16.La Grange, D. C., I. S. Pretorius, M. Claeyssens, and W. H. van Zyl. 2001. Degradation of xylan to D-xylose by recombinant Saccharomyces cerevisiae coexpressing the Aspergillus niger β-xylosidase (xlnD) and the Trichoderma reesei xylanase II (xyn2) genes. Appl. Environ. Microbiol. 67:5512-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:681-685. [DOI] [PubMed] [Google Scholar]
- 18.Li, X., and L. G. Ljungdahl. 1996. Expression of Aureobasidium pullulans XynA in, and secretion of the xylanase from Saccharomyces cerevisiae. Appl. Environ. Microbiol. 62:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1975. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 20.Madlala, A. M., S. Bissoon, S. Singh, and L. Christov. 2001. Xylanase-induced reduction of chlorine dioxide consumption during elemental chlorine-free bleaching of different pulp types. Biotechnol. Lett. 23:345-351. [Google Scholar]
- 21.Medeiros, R., G. Silva Jr., B. C. Salles, R. S. Estelles, and E. X. F. Filho. 2002. The performance of fungal xylan-degrading enzyme preparations in elemental chlorine-free bleaching for Eucalyptus pulp. J. Ind. Microbiol. Biotechnol. 28:204-206. [DOI] [PubMed] [Google Scholar]
- 22.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal. Chem. 31:426-428. [Google Scholar]
- 23.Minning, S., A. Serrano, P. Ferrer, C. Sola, R. D. Schmid, and F. Valero. 2001. Optimization of the high-level production of Rhizopus oryzae lipase in Pichia pastoris. J. Biotechnol. 86:59-70. [DOI] [PubMed] [Google Scholar]
- 24.Montgomery, D. C. 1997. Design and analysis of experiments, 4th ed. John Wiley & Sons, New York, N.Y
- 25.Passoth, V., and B. Hahn-Hägerdal. 2000. Production of a heterologous endo-1, 4-β-xylanase in the yeast Pichia stipitis with an O2-regulated promoter. Enzyme Microb. Technol. 26:781-784. [DOI] [PubMed] [Google Scholar]
- 26.Pérez-González, J. A., L. H. Graaff, J. Visser, and D. Ramón. 1996. Molecular cloning and expression in Saccharomyces cerevisiae of two Aspergillus nidulans xylanase genes. Appl. Environ. Microbiol. 62:2179-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puchart, V., P. Katapodis, P. Biely, L. Kremnický, P. Christakopoulos, M. Vršanská, D. Kekos, B. J. Macris, and M. K. Bhat. 1999. Production of xylanases, mannanases, and pectinases by the thermophilic fungus Thermomyces lanuginosus. Enzyme Microb. Technol. 24:355-361. [Google Scholar]
- 28.Purkarthofer, H., M. Sinner, and W. Steiner. 1993. Cellulase-free xylanase from Thermomyces lanuginosus: optimization of production in submerged and solid-state culture. Enzyme Microb. Technol. 15:677-682. [Google Scholar]
- 29.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlacher, A., K. Holzmann, M. Hayn, W. Steiner, and H. Schwab. 1996. Cloning and characterization of the gene for the thermostable xylanase XynA from Thermomyces lanuginosus. J. Biotechnol. 49:211-218. [DOI] [PubMed] [Google Scholar]
- 31.Singh, S., P. Reddy, J. Haarhoff, P. Biely, B. Janse, B. Pillay, D. Pillay, and B. A. Prior. 2000. Relatedness of Thermomyces lanuginosus strains producing a thermostable xylanase. J. Biotechnol. 81:119-128. [DOI] [PubMed] [Google Scholar]
- 32.Singh, S., B. Pillay, and A. Orior. 2000. Thermal stability of β-xylanases produced by different Thermomyces lanuginosus strains. Enzyme Microb. Technol. 26:502-508. [DOI] [PubMed] [Google Scholar]
- 33.Somogyi, M. 1952. Notes on sugar determination. J. Biol. Chem. 195:19-23. [PubMed] [Google Scholar]
- 34.Sreekrishna, K., R. G. Brankamp, K. E. Kropp, et al. 1997. Strategies for optimal synthesis and secretion of heterologous proteins in the methylotrophic yeast Pichia pastoris. Gene 190:55-62. [DOI] [PubMed] [Google Scholar]
- 35.Subramaniyan, S., and P. Prema. 2002. Bio/Technology of microbial xylanases: enzymology, molecular biology, and application. Crit. Rev. Biotechnol. 22:33-64. [DOI] [PubMed] [Google Scholar]
- 36.Teather, R. M., and P. J. Wood. 1982. Use of Congo Red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43:777-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsujibo, H., T. Ohtsuki, T. Ho, I. Yamazaki, K. Miyamoto, M. Sugiyama, and Y. Inamori. 1997. Cloning and sequence analysis of genes encoding xylanases and acetyl xylan esterase from Streptomyces thermoviolaceus OPC-520. Appl. Environ. Microbiol. 63:661-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh, D. J., and P. L. Bergquist. 1997. Expression and secretion of a thermostable bacterial xylanase in Kluyveromyces lactis. Appl. Environ. Microbiol. 63:3297-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]