Abstract
The production of bacterial ghosts from Escherichia coli is accomplished by the controlled expression of phage φX174 lysis gene E and, in contrast to other gram-negative bacterial species, is accompanied by the rare detection of nonlysed, reproductive cells within the ghost preparation. To overcome this problem, the expression of a secondary killing gene was suggested to give rise to the complete genetic inactivation of the bacterial samples. The expression of staphylococcal nuclease A in E. coli resulted in intracellular accumulation of the protein and degradation of the host DNA into fragments shorter than 100 bp. Two expression systems for the nuclease are presented and were combined with the protein E-mediated lysis system. Under optimized conditions for the coexpression of gene E and the staphylococcal nuclease, the concentration of viable cells fell below the lower limit of detection, whereas the rates of ghost formation were not affected. With regard to the absence of reproductive cells from the ghost fractions, the reduction of viability could be determined as being at least 7 to 8 orders of magnitude. The lysis process was characterized by electrophoretic analysis and absolute quantification of the genetic material within the cells and the culture supernatant via real-time PCR. The ongoing degradation of the bacterial nucleic acids resulted in a continuous quantitative clearance of the genetic material associated with the lysing cells until the concentrations fell below the detection limits of either assay. No functional, released genetic units (genes) were detected within the supernatant during the lysis process, including nuclease expression.
Controlled expression of the cloned φX174 lysis gene E results in the formation of empty bacterial cell envelopes (ghosts) (28, 33). Applicable in a broad range of gram-negative bacterial species, the protein E-mediated lysis procedure gives rise to a new class of genetically inactivated candidate vaccines (11, 16).
In contrast to the majority of tested bacterial species, which are completely inactivated by the lysis process (9, 16), the production of bacterial ghosts from Escherichia coli K-12 species is accompanied by the rare detection of nonlysed inactivated or reproductive cells within the ghost preparation. In previous studies, when green fluorescent protein-derived fluorescence was used for microscopic discrimination between ghosts and nonlysed cells, the percentage of nonlysed reproductive and inactivated E. coli cells was determined as 1.3% and 1.2%, respectively (13). The population of nonlysed cells was cytometrically subdivided into polarized and depolarized cells, and the detected populations were successfully quantified during the time course of lysis (14). The minimum ratios of nonlysed cells within the ghost preparations were cytometrically ascertained to be 4% for the polarized and 1% for the depolarized population, which correlated well with the results derived from classical microbiological procedures, indicating a minimum of 1% reproductive bacteria within the ghost preparations.
It should be emphasized that the ghost preparations which were used in the model studies mentioned above were not produced under optimal lysis conditions. The maximum rate of protein E-mediated inactivation of E. coli, which raises a killing efficiency of 99.99% (equal to a decrease in the viable cell number by 4 orders of magnitude), was intentionally not exploited to the full extent to provide samples which met the requirements for quantitative cytometric analyses so as to gain statistically accurate data within acquisition periods suitable for online monitoring experiments.
As the spectrum of the ghost system is expanded through the utilization of bacterial envelopes as carriers of foreign antigens (10, 24) or as vehicles for delivery for pharmaceutically relevant compounds (18), E. coli K-12 becomes a relevant target for the production of candidate vaccines. Additionally, laboratory strains of E. coli may serve as model organisms for the production of ghost vaccines from their enteropathogenic counterparts. To use E. coli ghosts as vaccines, additional methods which guarantee the total inactivation of the formulations and the preservation of the excellent immunogenic features of the bacterial envelopes have to be identified. Recently, a protocol for the flow cytometric separation of all nonlysed cells within the ghost preparation was established (13). The increase in purity, in terms of absence of viable cells, was determined as being more than three orders of magnitude; however, complete inactivation was not achieved.
As an alternative strategy, the intracellular synthesis of a secondary lethal protein is desired to be combined with the protein E-mediated lysis system to result in ghost preparations that are devoid of any reproductive cells. Intracellular degradation of DNA or RNA was shown to limit the reproductivity of the target cells as a consequence of nuclease expression. Previously, staphylococcal nuclease (26, 27) and the extracellular nuclease of Serratia marcescens (1) were investigated for their killing activities in E. coli after expression from a cloned state lacking the signal sequences for extracellular release. The systems led to a quantitative reduction in the number of viable bacteria within the range of two to five orders of magnitude.
In this work the expression of cloned staphylococcal nuclease during ghost production was tested in E. coli to elucidate its potential to minimize or totally abolish the viability of the lysed culture. The intracellular degradation of bacterial DNA as the result of nuclease activity should further restrict the number of reproductive bacteria within the lysing culture. Additionally, the clearance of residual genetic information is regarded as a beneficial “side effect” resulting in further improved characteristics of bacterial ghosts with respect to minimizing the risk of lateral spread (gene transfer) of critical genetic elements, such as antibiotic resistance genes or genetic determinants for pathogenicity.
The thermostable nuclease (EC 3.1.4.7) of Staphylococcus aureus, hereafter referred to as SNUC, is fully dependent on Ca2+ to act as a phosphodiesterase (3), which cleaves either single- or double-stranded DNA or RNA (15) to produce 3′-phosphomononucleotides, dinucleotides, and 3′,5′-nucleoside diphosphates (2, 3). Additional supplementation with Mg2+ was shown to have a stimulatory effect on the DNase activity of the enzyme (7). The initial phase of digestion is characterized as predominantly endonucleolytic, with only slight site preferences, followed by exonucleolytic degradation (17). Apart from its natural host, the staphylococcal nuclease was expressed in an enzymatic active form in a diverse spectrum of gram-positive and gram-negative bacterial species (5, 8, 23, 31).
MATERIALS AND METHODS
Bacterial strain, plasmids, and the production of ghosts.
E. coli strain NM522 (12; obtained from Stratagene, Amsterdam, The Netherlands), encoding the lacIq gene on an F′ episome, as well as the lysis plasmids pML1 (30) and pAW12 (32) were described previously. The lysis gene E is carried under the transcriptional control of the phage lambda pR/cI857 system encoded by both vectors, which differ in strategies of replication and copy numbers per cell, as the p15A origin of pML1 is derived from the low-copy-number plasmid pACYC174 (6), whereas the ColE1 origin of pAW12 originates from the intermediate-copy-number plasmid pBR322 (4).
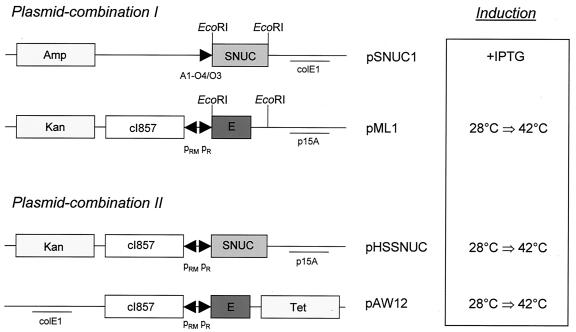
Plasmid pSNUC1 (27) carries the structural gene for staphylococcal nuclease A devoid of the original signal sequence and the amino terminus of nuclease B (29) under the control of the strong synthetic promoter A1-O4/O3 (22). Expression of the nuclease was initiated by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG; obtained from Sigma-Aldrich, Vienna, Austria), which was used at a final concentrations of 5 mM. Plasmid pHSSNUC was constructed via the exchange of the smaller EcoRI fragment of pML1 with the smaller EcoRI fragment of pSNUC1 (Fig. 1). The portion of pML1 which encoded gene E was thus replaced with the structural gene of the nuclease including the ribosome-binding site originating from pSNUC1. The expression of SNUC is therefore controlled by the lambda pR/cI857 system, encoded on a pACYC174-derived vector backbone.
FIG. 1.
Expression plasmids for lysis gene E and the staphylococcal nuclease (SNUC). The vectors are presented as plasmid combinations utilized for the coexpression of gene E and SNUC in E. coli NM522. The expression of the killing genes was triggered by either the addition of an inducer (+IPTG) or a thermal upshift from 28°C to 42°C (28°C ⇒ 42°C). Amp, ampicillin resistance gene; Kan, kanamycin resistance gene; Tet, tetracycline resistance gene; colE1 and p15A, origins of replication; A1-O4/O3, synthetic, chemically inducible promoter; pRM/pR, rightward “maintenance” and rightward promoters of bacteriophage lambda; cI857, gene encoding the thermosensitive repressor for the lambda pR promoter; EcoRI, restrictions sites used for the construction of pHSSNUC.
E. coli NM522 was transformed with the different plasmids and routinely grown in Luria-Bertani (LB) broth (25) containing 2% agar for solid media and supplemented with kanamycin (50 μg/ml), ampicillin (200 μg/ml), or tetracycline (10 μg/ml) where appropriate (antibiotics were obtained from Sigma-Aldrich). For the production of ghosts, the transformed bacteria were grown under selective pressure at an incubation temperature of 28°C to ensure tight repression of the lysis gene E. After reaching mid-log phase, the culture was shifted to 42°C to induce protein E-mediated lysis. The growth and lysis of the bacteria were monitored by measuring the optical density at 600 nm (OD600). Periodically samples were taken and analyzed through a standard automated procedure to determine the actual number of reproductive cells per volume (the procedure is described below). Aliquots of the samples were shock-frozen and stored at −70°C until they were analyzed for their DNA content. Cultures which harbored plasmid pSNUC1 were supplemented with 1% glucose during overnight incubations and subsequently washed twice with LB prior to further experiments. The control experiments and the production of ghosts from cells which harbored an expression plasmid for SNUC were performed by a modified version of the presented protocol, which was extended by additional supplementation with IPTG and/or Mg2+ and Ca2+ at various time points. Mg2+ and Ca2+ were added as MgCl2 and CaCl2, respectively, at final concentrations of 1 mM and 10 mM, respectively.
Determination of the viability of the culture.
The number of CFU within the bacterial samples was determined by a standard automated plating procedure. A spiral plater (WASP system; Don Whitley Scientific Limited, West Yorkshire, UK) was used to ascertain the absolute numbers of reproductive bacteria by plating the pure culture or serial dilutions (in 0.9% NaCl solution) on agar plates. The spiral plater was used as specified by the supplier. For bacterial growth, LB agar plates without added antibiotics or IPTG were used and were incubated at 28°C for at least 20 h prior to colony counting by direct observation. For these assays, the lower limit of detection was 5 or 10 CFU/ml, respectively, referring to the analyzed volume of 200 μl or 100 μl, respectively, of the undiluted culture.
Preparation of total DNA and electrophoretic analysis.
To extract the total DNA content of the cells (or ghosts) of E. coli, 1 ml of the thawed culture was harvested by centrifugation (10 min, 4°C, 10,000 × g). Then 500 μl of the supernatant was collected, and the residual liquid was removed carefully from the pellet. The pellet was subjected to DNA extraction with the Easy-DNA kit (Invitrogen, Groningen, The Netherlands) as specified by the supplier. Finally, the prepared DNA was suspended in 100 μl of TE buffer (Invitrogen) which contained RNase (certified free of DNase contamination; Invitrogen) at a concentration of 40 μg/ml and then incubated at 37°C for 30 min and stored at 4°C. The collected supernatant was extracted twice with phenol-chloroform and precipitated with ethanol. The prepared nucleic acids were suspended in 50 μl of TE buffer supplemented with RNase. After incubation at 37°C for 30 min, the samples were also stored at 4°C. Then 15 μl of the DNA preparations was subjected to electrophoresis, which was performed at constant voltage (120 V) applied to 0.6% or 2% agarose gels. BstEII-digested lambda DNA was purchased from New England Biolabs (Schwalbach, Germany) and served as the molecular size standard.
Real-time PCR and the preparation of DNA standards.
Liquid cultures of E. coli NM522 harboring either plasmid pML1 or pSNUC1 were subjected to DNA extraction via the alkaline lysis procedure to end in DNA standards for SYBR Green I-based real-time PCR (34). The precipitated DNA was suspended in TE buffer containing RNase at a final concentration of 40 μg/ml and incubated at 37°C for 30 min. Tenfold dilution series were prepared in TE buffer to cover the range of dilution factors from 101 to 107. The DNA concentrations of the undiluted standards were determined fluorimetrically via calibration with calf thymus DNA (Sigma-Aldrich) of given concentration and staining with the fluorescent dye Hoechst 33342 (Molecular Probes, Leiden, The Netherlands). A nontemplate control and the dilutions of the quantified standard DNA were used as templates for real-time PCR to determine the linear dynamic range of the assays.
For the quantitative analysis of the DNA preparations, a real-time PCR assay was established with the Rotor-Gene 2000 (Corbett Research, Mortlake, Australia). Specific primers for the genes encoding the β-lactamase (ampr) on pSNUC1 (Amp-Start, 5′-ATGAGTATTCAACATTTCCGTGTC-3′, and Amp-Stop, 5′-TTACCAATGCTTAATCAGTGAGG-3′) or the aminoglycoside 3′-phosphotransferase (kanr) on pML1 (Kan-Start, 5′-ATGAGCCATATTCAACGGGAAA-3′, and Kan-Stop, 5′-TTAGAAAAACTCATCGAGCATCA-3′) were obtained from VBC-Genomics Bioscience Research GmbH (Vienna, Austria). The oligonucleotides were designed for exclusive amplification of the coding region of the corresponding genes.
A single run of quantitative PCR included up to 10 DNA samples of unknown concentrations, three dilutions of the appropriate DNA standard, and one nontemplate control, which were all prepared and analyzed in duplicate. For calibration experiments, seven dilutions of the DNA standard and one nontemplate control were prepared and analyzed in duplicate. The components for PCR in a final volume of 25 μl included 0.1 μl of each primer (100 pmol/μl), 0.5 μl of deoxynucleoside triphosphate s (10 mM each; Roche, Vienna, Austria), 0.25 μl of a freshly prepared aqueous solution (10×) of SYBR-Green I (10,000-fold stock; Molecular Probes), 0.25 μl of Dynazyme DNA polymerase (Finnzymes, Espoo, Finland), 2.5 μl of polymerase buffer (10×; Finnzymes), 20.8 μl of sterile, ultrapure water (ACS-grade reagent; Sigma-Aldrich), and 0.5 μl of the template. For the nontemplate controls, 0.5 μl of TE buffer was added instead of a DNA template. After an initial denaturation step at 94°C for 2 min, the conditions for 40 cycles were 94°C for 28 s, 60°C (kanr) or 62°C (ampr) for 60 s, and 72°C for 60 s, with monitoring of fluorescence after the extension step at 72°C. The PCR products were validated via melting curves (50°C to 94°C in steps of 1°C) and electrophoresis.
RESULTS
Expression of staphylococcal nuclease in E. coli.
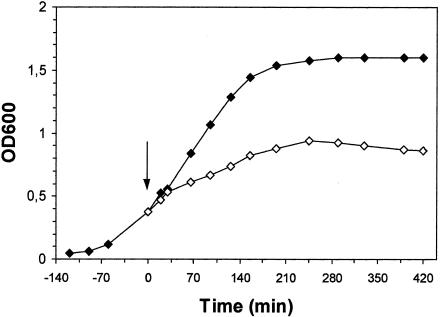
Staphylococcal nuclease A (SNUC) was investigated for its in vivo effects on bacteria after expression in E. coli. Plasmid pSNUC1 was used to direct high levels of nuclease expression in E. coli NM522. As the structural gene of the nuclease was controlled by the strong, synthetic promoter A1-O4/O3, induction with IPTG was predicted to result in intracellular accumulation of the nuclease. E coli NM522(pSNUC1) grown in the presence of Mg2+ and Ca2+ reflected altered growth kinetics after induction with IPTG in comparison to the noninduced control (Fig. 2). The determination of viable counts indicated the presence of a lethal gene product, as the viability of the culture dropped below 1% (106 CFU/ml) 7 h after induction with regard to the initial number of reproductive bacteria (3 × 108 CFU/ml) determined at the time point of IPTG addition.
FIG. 2.
Growth kinetics of E. coli NM522 harboring pSNUC1. The cells were grown at 37°C in the presence of added Mg and Ca until mid-log phase. The culture was split, and one part was induced by the addition of IPTG at a final concentration of 5 mM (◊), whereas the other part served as a noninduced control (⧫). OD600, optical density measured at 600 nm; the arrow indicates the time of induction.
To investigate the intracellular effects of the nuclease on the nucleic acids of the host, total DNA was extracted from samples taken periodically during the time course of expression and investigated by electrophoretic analysis (data not shown). In contrast to the noninduced sample, where the prepared bacterial DNA was shown to build up a mixture of large fragments (>20 kbp), the DNA extracted from induced cultures had been subjected to rapid degradation. DNA prepared from the induced culture was decreasing in amount and in the length of the fragments over time. Samples taken later than 40 min after the addition of IPTG mainly contained DNA fragments shorter than 100 bp or were of marginal residual quantum hardly detectable under the chosen conditions. The microscopic examination of the culture taken 7 h after induction of SNUC revealed the intact morphology of the cells, reflecting no obvious differences in comparison to cells of the noninduced sample (data not shown).
Coexpression of lysis gene E and staphylococcal nuclease.
Two different lysis plasmids were chosen to be tested for the production of ghost preparations with strongly decreased numbers of viable cells as the result of coexpression of gene E and the staphylococcal nuclease. The lysis plasmid pML1, encoding a thermoinducible expression cassette for gene E, was used in combination with the compatible plasmid pSNUC1, carrying SNUC under the control of a chemically inducible promoter. To gain an expression vector for the nuclease which was compatible with the lysis plasmid pAW12, vector pHSSNUC was constructed, encoding SNUC within a lambda pR/cI857-controlled expression unit. With this set of plasmid combinations, the expression of genes E and SNUC could either be induced sequentially (pML1 and pSNUC1) by applying a thermal and a chemical induction signal, respectively, for the different expression cassettes, or simultaneously via a temperature upshift to 42°C (pAW12 and pHSSNUC) (Fig. 1).
E. coli strain NM522 was transformed with either one or both plasmids of each vector combination. The effects of gene expression were studied with regard to the resulting growth and lysis kinetics, the efficiency of ghost production, and the viability of the culture during the time course of the experiments. As the enzymatic activity of the nuclease has been described as being absolutely dependent on added Ca2+, it was suggested that the time point of supplementation (Ca2+ plus Mg2+) be varied as an additional parameter between individual runs. The results of these experiments are summarized in Table 1.
TABLE 1.
Expression and coexpression of E and SNUC
| Plasmid(s) | Time of IPTG additiona (min) | Time of Mg/Ca additiona (min) | Ghost formationb | Maximum inactivationc | Δt[maximum inactivation]d (h) |
|---|---|---|---|---|---|
| pAW12, pHSSNUC | — | — | + | 4 | 2.5-4 |
| — | Inoculation | Impaired | 4-5 | 3.5-4 | |
| — | 0 | Impaired | 4-5 | 3.5 | |
| — | +60 | + | 4-6 | 2.5-4 | |
| pAW12 | — | — | + | 4 | 1.3-1.5 |
| pHSSNUC | — | +60 | ND | 1-2 | 4 |
| pML1, pSNUC1 | — | — | + | 3-4 | 5-6 |
| — | +90 | + | 2-3 | 6 | |
| −45 | — | + | 2-3 | 5 | |
| 0 | 0 | Impaired | ≥7-8* | >8 | |
| 0 | +90 | + | ≥7-8* | >8 | |
| −45 | +90 | + | ≥7-8* | 6->8 | |
| +90 | +90 | + | ≥7-8* | >8 | |
| pML1 | −45 | +90 | + | 2-4 | 5-7 |
| — | — | + | 2-4 | 5-7.5 | |
| pSNUC1 | −45 | +90 | ND | 2-3 | 2-2.5 |
Times refer to the time of temperature upshift (0 min). —, no addition; inoculation, Mg and Ca added at the time of culture inoculation.
+, >95% of bacterial particles were ghosts at the completion of lysis; impaired, protein E-mediated lysis impaired, giving elevated numbers of nonlysed cells; ND, no ghosts detected.
Maximum in activation in log units [log(CFUmax) − log(CFUmin]. *, viable cells below detection limit.
Time period from the temperature upshift to 42°C to the timepoint of minimum variability.
With the plasmid combination pAW12 plus pHSSNUC, rates of inactivation were achieved which were observed to be of slightly increased magnitude over that obtained with the exclusive use of lysis plasmid pAW12 with only minor variations dependent on the time point of supplementation with the divalent cations (Table 1). This was contrary to the detected differences in ghost formation, which was impaired by the early addition of Mg and Ca, resulting in only a minor decrease in the turbidity of the lysing culture and in strongly enhanced numbers of nonlysed cells within the ghost fractions. Furthermore, the time required to reach the minimum viability was observed to be prolonged for the combined approaches in comparison to the exclusive use of pAW12. For plasmid pHSSNUC, a reduction in viable counts by up to two orders of magnitude was determined.
Genetic inactivation of E. coli ghost preparations.
The coexpression of genes E and SNUC in E. coli NM522(pML1, pSNUC1) drastically restricted the reproductivity of the bacterial culture, giving rise to ghost preparations devoid of any detectable viable cells (Table 1). As the concentration of viable bacteria dropped below the lower limit of detection (5 to 10 CFU/ml), the degree of inactivation was calculated as being at least seven to eight orders of magnitude with respect to the absence of any viable cells in the analyzed culture volume. The variation of parameters with regard to the time points of SNUC induction or the addition of the cofactors Mg and Ca did not cause any decrease in the degree of inactivation. It has to be emphasized that the determination of the viable counts was performed under noninduced and nonselective conditions for bacterial growth with respect to the expression cassettes for both of the lethal genes.
Maximum inactivation was either achieved 6 h after lysis induction or detected, as in most cases, after overnight incubation under induced conditions. Although the rates of inactivation were not affected, an inhibitory effect on the formation of ghosts was observed after the simultaneous addition of Mg and Ca and the induction of lysis. This observation correlated well with the results obtained with the vector combination pAW12 plus pHSSNUC, where early supplementation with Mg and Ca caused identical effects.
E. coli NM522 was transformed only with either pSNUC1 or pML1 to investigate further the contribution of each expression element to the rate of inactivation. One of the successful protocols for maximum inactivation including both plasmids (IPTG added at −45 min; temperature shifted to 42°C at 0 min; Mg and Ca added at +90 min) was applied in an unmodified manner to E. coli harboring either pSNUC1 or pML1. The viability of cultures harboring pSNUC1 or pML1 was reduced by more than two and by up to four orders of magnitude, respectively. Furthermore, these experiments gave results resembling those obtained via the “standard” lysis protocol for pML1 without the addition of IPTG or Mg and Ca, indicating no adverse effects of the supplements on ghost formation or killing efficiency. Microscopic examination of all samples except for the simultaneous induction of lysis and the addition of Mg and Ca revealed a maximum number of ghosts, representing more than 95% of the bacterial particles within the preparations.
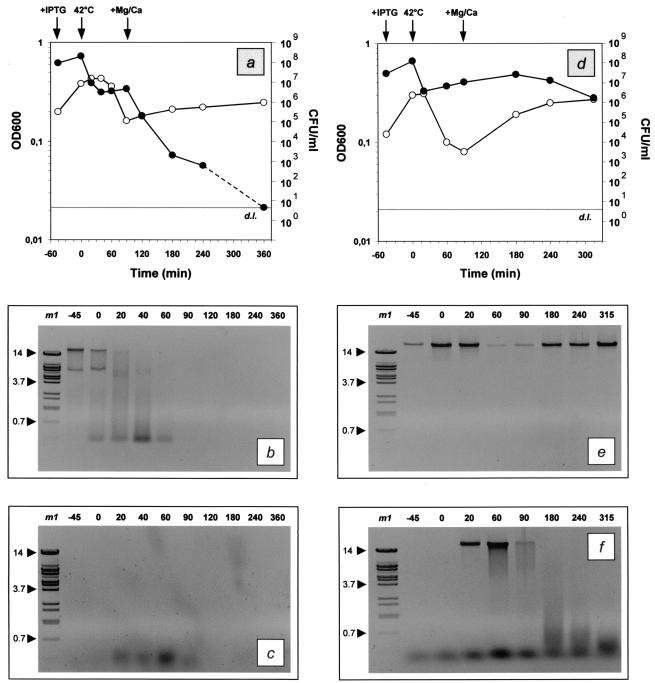
Electrophoretic analysis of genetic content.
Samples of E. coli NM522 harboring either pML1 and pSNUC1 (Fig. 3a) or only pML1 (Fig. 3d) were taken periodically during the time course of growth and lysis (IPTG added at −45 min; temperature shifted to 42°C at 0 min; Mg and Ca added at +90 min), and total DNA was extracted from the cell pellet and the supernatant and analyzed by electrophoretic analysis. For the combinatory approach (pML1 plus pSNUC1), the bacterial DNA within the cells was observed to be degraded soon after the nuclease was induced (Fig. 3b). At the time point of supplementation with Mg and Ca, no residual DNA prepared from the cell pellet could be detected under the chosen conditions. The analysis of the supernatant revealed that no DNA fragments longer than 400 bp could be detected at any time point analyzed (Fig. 3c). These fragments were detected soon after the induction of lysis (time point 20 min) and were no longer visible 30 min after addition of the cations (time point 120 min).
FIG. 3.
Growth and lysis kinetics and electrophoretic analysis of the total DNA content of E. coli NM522 harboring either pML1 plus pSNUC1 (a to c) or only pML1 (d to f). Diagrams a and d focus on the growth and lysis kinetics, which were monitored by measurement of the optical density at 600 nm (OD600; ○) and the determination of the number of CFU per milliliter (•). For practical reasons, the concentration of viable cells that fell below the lower limit of detection (grey lines; d.l.) is illustrated as corresponding to the detection limit, although it does not refer to a certain concentration of cells. The time points of supplementation with IPTG and Mg and Ca and the temperature upshift to 42°C are indicated by arrows. The total DNA contents within the pellets (b, e) and the supernatants (c, f) were analyzed by electrophoretic separation with 0.6% agarose gels. The time points of sampling are given in minutes relative to the time of lysis induction (0 min). Lane m1, BstEII-digested lambda DNA; the sizes of prominent fragments (indicated by arrowheads) are given in kilobases.
These results were contrary to the analysis of the supernatants derived from the control culture harboring only the lysis plasmid (Fig. 3f). Immediately after the induction of lysis, DNA fragments of high molecular weight were expelled from the cells into the supernatant and subsequently degraded. At the same time, the quantity of residual DNA within the cell pellets was decreasing and reached its minimum 1 h after lysis induction (Fig. 3e). The following increase in DNA content corresponded to regrowth of the survivors of the lysis process. The genetic material from cell pellets of the control culture was composed of high-molecular-weight DNA during the entire time course of lysis. Extensive enzymatic fragmentation was never observed after electrophoretic analysis.
Quantitative analysis of genetic content via real-time PCR.
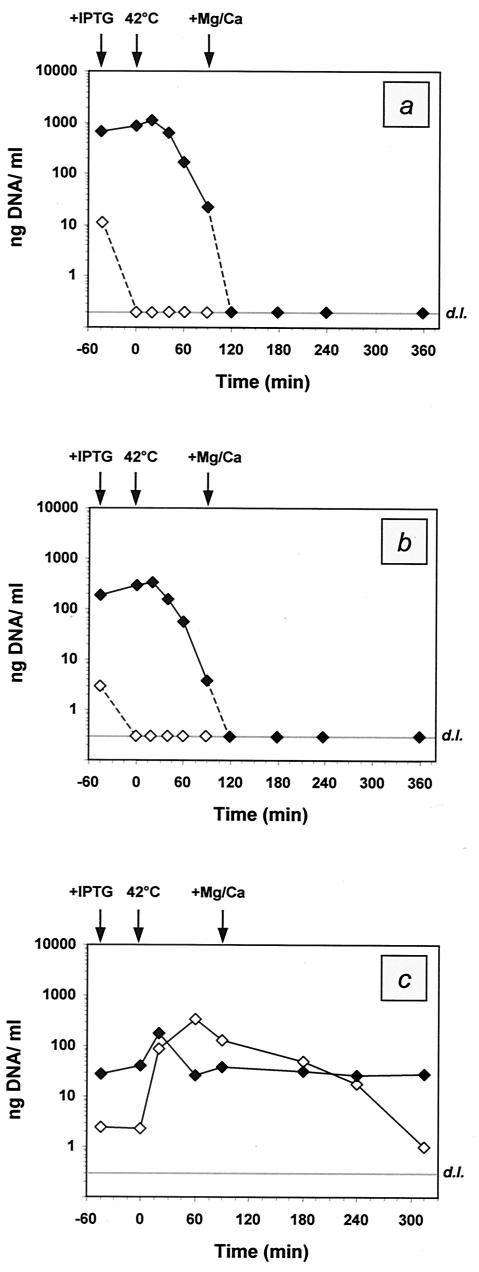
The sets of DNA preparations were subjected to a SYBR Green I-based real-time PCR assay to determine the total genetic content of the genes encoding either β-lactamase (ampr) on pSNUC1 or aminoglycoside 3′-phosphotransferase (kanr) on pML1. After calibration of the systems, the linear dynamic ranges for quantification were determined to span five decades for either approach: for the ampr assay, 950 fg to 9.5 ng per reaction, equivalent to 0.2 ng to 1.9 μg/ml of culture; and for the kanr assay, 1.6 pg to 16 ng per reaction, equivalent to 0.3 ng to 3.2 μg/ml of culture. The detailed results of the quantification via real-time PCR are presented in Fig. 4.
FIG. 4.
Quantitative analysis of the total DNA content within the pellets (⧫) and the supernatants (◊) of E. coli NM522 harboring either pML1 plus pSNUC1 (a, b) or only pML1 (c). The data are presented in nanograms of DNA per milliliter of culture taken at various time points during the production of ghosts. The time points of sampling are given in minutes relative to the induction of lysis (0 min). The quantitative results were obtained via real-time PCR with regard to amplification of either the ampr gene (a) or the kanr gene (b, c) encoded by pSNUC1 and pML1, respectively. For practical reasons, the DNA concentrations that fell below the detection limits (grey lines; d.l.) are illustrated as corresponding to the limits of detection, although they do not refer to certain concentrations. The time points of supplementation with IPTG and Mg and Ca and the temperature upshift to 42°C are indicated by arrows.
For the combinatory approach (pML1 plus pSNUC1), the genetic content (kanr assay; Fig. 4b) within the pellets was determined to be increasing until 20 min after induction of lysis, with the maximum of 330 ng of DNA per ml of culture. Subsequently, a gradual decline in the amount of genetic material was observed before the concentration dropped beyond the detection limit (300 pg/ml) 2 h after lysis induction. The analysis of the supernatants indicated the complete absence of detectable concentrations of DNA during the time course of ghost production except in the initial sample, where 3 ng of residual DNA per ml of the supernatant was determined. Similar results were obtained by quantification of the DNA content via the ampr-based assay (Fig. 4a). However, the absolute concentrations of the quantified DNA were always three- to fourfold higher for the latter approach, reflecting the ratio of copy numbers of pSNUC1 and pML1 per cell.
In correlation with the kanr-based assay, the DNA concentrations were first observed to fall below the detection limit (200 pg/ml) with the sample taken 2 h after induction of lysis. Correspondent to the initial lower cell density of the control culture (pML1), the DNA concentrations within theses pellets (Fig. 4c) determined during the initial phase of the experiment were up to sixfold lower in comparison to the combinatory assay (pML1 plus pSNUC1). After reaching the maximum DNA concentration with 173 ng/ml, the genetic content within the pellet of the control culture was observed to be reduced to a minimum of 26 ng/ml as the result of ongoing lysis. Subsequently, the amount of residual DNA within the pellet remained almost constant at slightly elevated levels. Reciprocal to the DNA content of the cells, the concentration within the supernatant was observed to reach its maximum when the genetic content of the pellet was observed to be minimal. A successive decrease in the DNA concentration of the supernatant was observed till the end of the time course.
DISCUSSION
The coexpression of staphylococcal nuclease and the lysis gene E resulted in the genetic inactivation of E. coli K-12 liquid cultures. The synthesis of lethal protein E gave rise to empty bacterial cell envelopes (ghosts), whereas the expression of SNUC was shown to cause intracellular degradation of the host DNA. Two plasmid combinations were presented, each consisting of two separate expression cassettes for gene E and the nuclease encoded on two compatible vector backbones.
Under optimized conditions, the total inactivation of up to 200 μl of pure ghost preparation could be shown, which corresponded to a decrease in the number of reproductive bacteria by at least seven to eight orders of magnitude. The enzymatic activity of the expressed nuclease was shown to be fully dependent on supplementation with the cofactors Mg2+ and Ca2+. Even with the nuclease expressed at high levels in the absence of the cofactors, the turbidity and the viability of the culture were continuously increasing at rates that were comparable to that of the noninduced controls (data not shown). Whereas the time point of Mg and Ca addition was not critical for the degree of inactivation, protein E-mediated lysis was apparently negatively influenced by supplementation with Mg and Ca at early stage of lysis. The addition of Mg and Ca during the late stage or after lysis was complete had no inhibitory effect on the rate of ghost formation and initiated further reduction in the number of viable cells within the ghost preparations through the activation of the nuclease. The tightly repressed expression systems for gene E and SNUC and the absence of Mg and Ca during the growth phase are suggested to limit the risk of selecting escape mutants, which was shown to be crucial when working on bacterial containment or kill systems (19, 20, 21, 26).
Electrophoretic analysis and real-time PCR were applied for semiquantitative and quantitative characterization of the genetic content within the cells (including ghosts) and the supernatant during the time course of ghost production. During lysis of NM522(pML1, pSNUC1), the concentration of the genetic content within the pellet was first observed to decline four orders of magnitude (real-time PCR) and finally dropped below the detection limit of either assay. Although this study focused on E. coli K-12, it would be feasible to apply this system to other bacterial species to limit the residual genetic content and therefore gain improved characteristics of ghosts to be used as candidate vaccines, carriers of foreign antigens, or vehicles for delivery of pharmaceutical relevant compounds.
Acknowledgments
We are indebted to Jennelle M. Kyd for critical reading of the manuscript.
REFERENCES
- 1.Ahrenholtz, I., M. G. Lorenz, and W. Wackernagel. 1994. A conditional suicide system in Escherichia coli based on the intracellular degradation of DNA. Appl. Environ. Microbiol. 60:3746-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, M., L. A. Heppel, and J. Hurwitz. 1961. The purification and properties of micrococcal nuclease. J. Biol. Chem. 236:3014-3019. [PubMed] [Google Scholar]
- 3.Anfinsen, C. B., P. Cuatrecasas, and H. Taniuchi. 1971. Staphylococcal nuclease: chemical properties and catalysis, p. 177-201. In P. Boyer (ed.), The enzymes, vol. 4. Academic Press, New York, N.Y.
- 4.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 5.Boynton, Z. L., J. J. Koon, E. M. Brennan, J. D. Clouart, D. M. Horowitz, T. U. Gerngross, and G. W. Huisman. 1999. Reduction of cell viscosity during processing of poly(3-hydroxyalkanoates) by chromosomal integration of the staphylococcal nuclease gene in Pseudomonas putida. Appl. Environ. Microbiol. 65:1524-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuatrecasas, P., S. Fuchs, and C. B. Anfinsen. 1967. Catalytic properties and specifity of the extracellular nuclease of Staphylococcus aureus. J. Biol. Chem. 242:1541-1547. [PubMed] [Google Scholar]
- 8.Dutton, E. K., S. A. Ottum, T. C. Bolken, C. A. Franke, and D. E. Hruby. 2000. Expression of active monomeric and dimeric nuclease A from the gram-positive Streptococcus gordonii surface protein expression system. Protein Expr. Purif. 19:158-172. [DOI] [PubMed] [Google Scholar]
- 9.Eko, F. O., M. P. Szostak, G. Wanner, and W. Lubitz. 1994. Production of Vibrio cholerae ghosts (VCG) by expression of a cloned phage lysis gene: potential for vaccine development. Vaccine 12:1231-1237. [DOI] [PubMed] [Google Scholar]
- 10.Eko, F. O., A. Witte, V. Huter, B. Kuen, S. Fürst-Ladani, A. Haslberger, A. Katinger, A. Hensel, M. P. Szostak, S. Resch, H. Mader, P. Raza, E. Brand, J. Marchart, W. Jechlinger, W. Haidinger, and W. Lubitz. 1999. New strategies for combination vaccines based on the extended recombinant bacterial ghost system. Vaccine 17:1643-1649. [DOI] [PubMed] [Google Scholar]
- 11.Eko, F. O., U. B. Mayr, S. R. Attridge, and W. Lubitz. 2000. Characterization and immunogenicity of Vibrio cholerae ghosts expressing toxin-coregulated pili. J. Biotechnol. 83:115-123. [DOI] [PubMed] [Google Scholar]
- 12.Gough, J., and N. Murray. 1983. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J. Mol. Biol. 166:1-19. [DOI] [PubMed] [Google Scholar]
- 13.Haidinger, W., M. P. Szostak, W. Beisker, and W. Lubitz. 2001. Green fluorescent protein (GFP)-dependent separation of bacterial ghosts from intact cells by FACS. Cytometry 44:106-112. [PubMed] [Google Scholar]
- 14.Haidinger, W., M. P. Szostak, and W. Lubitz. 2003. Online monitoring of the Escherichia coli ghost production. Appl. Environ. Microbiol. 69:468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heins, J. N., R. Suriano, H. Taniuchi, and C. B. Anfinsen. 1967. Characterization of a nuclease produced by Staphylococcus aureus. J. Biol. Chem. 242:1016-1020. [PubMed] [Google Scholar]
- 16.Hensel, A., V. Huter, A. Katinger, P. Raza, C. Strnistschie, U. Roesler, E. Brand, and W. Lubitz. 2000. Intramuscular immunization with genetically inactivated (ghosts) Actinobacillus pleuropneumoniae serotype 9 protects against homologous aerosol challenge and prevents carrier state. Vaccine 18:2945-2955. [DOI] [PubMed] [Google Scholar]
- 17.Hörz, W., and W. Altenburger. 1981. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 9:2643-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jechlinger, W., W. Haidinger, S. Paukner, P. Mayrhofer, E. Riedmann, J. Marchart, U. Mayr, C. Haller, G. Kohl, P. Walcher, P. Kudela, J. Bizik, D. Felnerova, E. M. B. Denner, A. Indra, A. Haslberger, M. Szostak, S. Resch, F. Eko, T. Schukovskaya, V. Kutyrev, A. Hensel, S. Friederichs, T. Schlapp, and W. Lubitz. 2002. Bacterial ghosts as carrier and targeting systems for antigen delivery, p. 163-184. In G. Dietrich and W. Goebel (ed.), Vaccine delivery strategies. Horizon Scientific Press, Norfolk, United Kingdom.
- 19.Jensen, L. B., J. L. Ramos, Z. Kaneva, and S. Molin. 1993. A substrate-dependent biological containment system for Pseudomonas putida based on the Escherichia coli gef gene. Appl. Environ. Microbiol. 59:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knudsen, S. M., and O. H. Karström. 1991. Development of efficient mechanisms for biological containment of bacteria. Appl. Environ. Microbiol. 57:85-92.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knudsen, S., P. Saadbye, L. H. Hansen, A. Collier, B. L. Jacobsen, J. Schlundt, and O. H. Karlström. 1995. Development and testing of improved suicide functions for biological containment of bacteria. Appl. Environ. Microbiol. 61:985-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanzer, M., and H. Bujard. 1988. Promoters largely determine the efficiency of repressor action. Proc. Natl. Acad. Sci. 85:8973-8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebl, W., A. J. Sinskey, and K. H. Schleifer. 1992. Expression, secretion and processing of staphylococcal nuclease by Corynebacterium glutamicum. J. Bacteriol. 174:1854-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lubitz, W., A. Witte, F. O. Eko, M. Kamal, W. Jechlinger, E. Brand, J. Marchart, W. Haidinger, V. Huter, D. Felnerova, N. Stralis-Alves, S. Lechleitner, H. Melzer, M. P. Szostak, S. Resch, H. Mader, B. Kuen, B. Mayr, P. Mayrhofer, R. Geretschlager, A. Haslberger, and A. Hensel. 1999. Extended recombinant bacterial ghost system. J. Biotechnol. 73:261-273. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 26.Molin, S., L. Boe, L. B. Jensen, C. S. Kristensen, M. Givskov, J. L. Ramos, and A. K. Bej. 1993. Suicidal genetic elements and their use in biological containment of bacteria. Annu. Rev. Microbiol. 47:139-166. [DOI] [PubMed] [Google Scholar]
- 27.Molin, S., M. Givskov, C. S. Kristensen, A. Bej, and L. Eberl. May1995. A method of limiting the survival of genetically engineered microorganisms in their environment. Patent: international publication number WO 95/10614.
- 28.Schön, P., G. Schrot, G. Wanner, W. Lubitz, and A. Witte. 1995. Two-stage model for integration of lysis protein E of bacteriophage φX174 into the cell envelope of Escherichia coli. FEMS Microbiol. Rev. 17:207-212. [DOI] [PubMed] [Google Scholar]
- 29.Shortle, D. 1983. A genetic system for analysis of staphylococcal nuclease. Gene 22:181-189. [DOI] [PubMed] [Google Scholar]
- 30.Szostak, M. P., A. Hensel, F. O. Eko, R. Klein, T. Auer, H. Mader, A. Haslberger, S. Bunka, G. Wanner, and W. Lubitz. 1996. Bacterial ghosts: non-living candidate vaccines. J. Biotechnol. 44:161-170. [DOI] [PubMed] [Google Scholar]
- 31.Takahara, M., D. W. Hibler, P. J. Barr, J. A. Gerlt, and M. Inouye. 1985. The ompA signal peptide directed secretion of staphylococcal nuclease A by Escherichia coli. J. Biol. Chem. 260:2670-2674. [PubMed] [Google Scholar]
- 32.Witte, A., and W. Lubitz. 1989. Biochemical characterization of φX174-protein-E-mediated lysis of Escherichia coli. Eur. J. Biochem. 180:393-398. [DOI] [PubMed] [Google Scholar]
- 33.Witte, A., G. Wanner, and W. Lubitz. 1992. Dynamics of PhiX174 protein E-mediated lysis of Escherichia coli. Arch. Microbiol. 157:381-388. [DOI] [PubMed] [Google Scholar]
- 34.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-138. [DOI] [PubMed] [Google Scholar]