Abstract
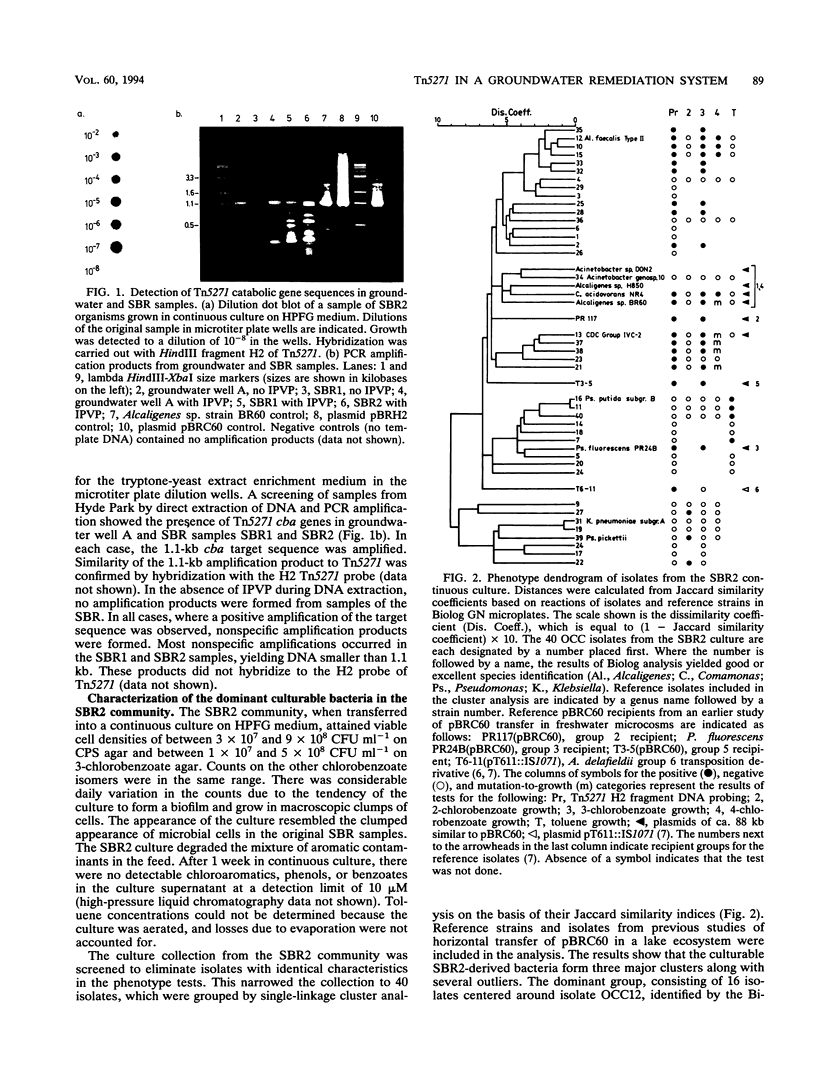
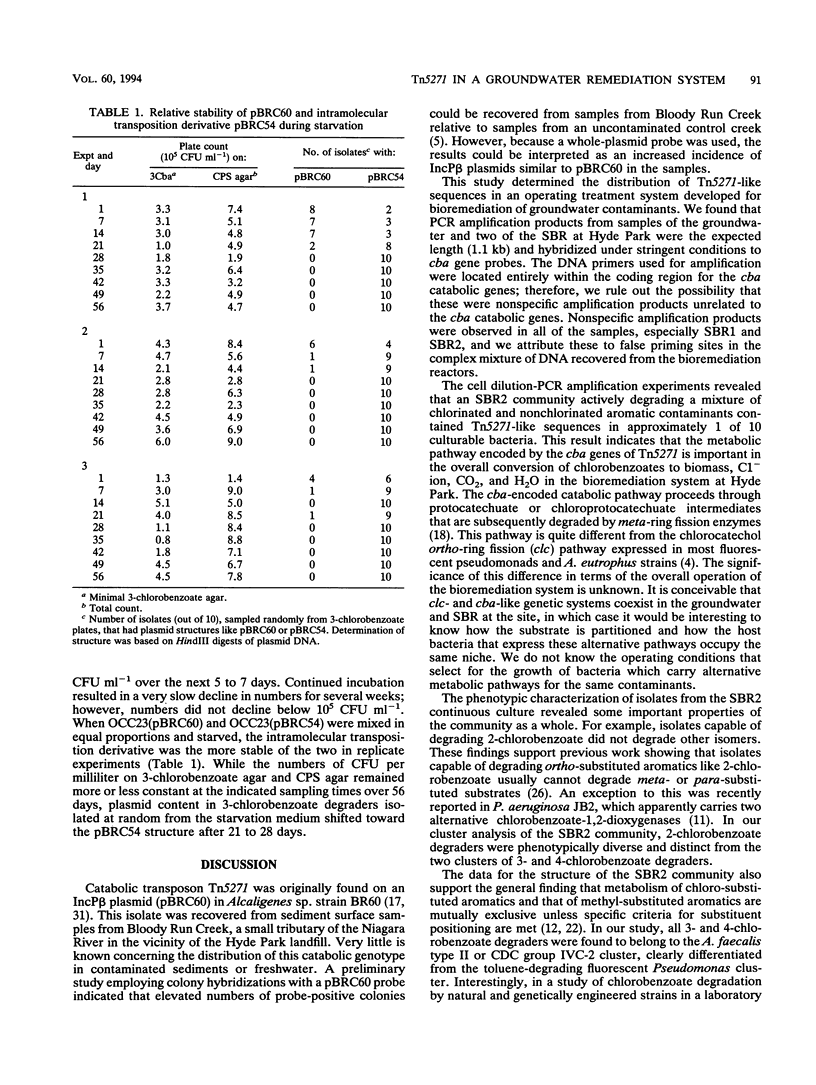
The distribution of Tn5271-related DNA sequences in samples of groundwater and a groundwater bioremediation system at the Hyde Park (Niagara Falls, N.Y.) chemical landfill site was investigated. PCR amplification of target sequences within the cha genes of Tn5271 revealed similar sequences in the groundwater community and in samples from the sequencing batch reactors treating that groundwater. Cell dilution combined with PCR amplification indicated that cha sequences were carried in about 1 of 10 culturable bacteria from the treatment system. Characterization of isolates involved in chlorobenzoate and toluene biodegradation in the treatment system indicated that two phenotypic clusters, Alcaligenes faecalis type 2 and CDC group IVC-2, contained all of the Tn5271 probe-positive isolates from the community. These two groups differed phenotypically from recipient groups isolated following horizontal transfer of pBRC60 (Tn5271) in pristine freshwater microcosms. A genetic rearrangement in Tn5271 attributable to the intramolecular transposition of the flanking element IS1071R was detected in an isolate from the treatment system. Comparison of the structure of the intramolecular transposition derivative from groundwater isolate OCC13(pBRC13) with a laboratory-derived intramolecular transposition derivative of pBRC60 revealed similarities. The rearrangement was shown to increase the stability of the plasmid under starvation conditions.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brenner V., Hernandez B. S., Focht D. D. Variation in chlorobenzoate catabolism by Pseudomonas putida P111 as a consequence of genetic alterations. Appl Environ Microbiol. 1993 Sep;59(9):2790–2794. doi: 10.1128/aem.59.9.2790-2794.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., Chakrabarty A. M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulthorpe R. R., Wyndham R. C. Involvement of a chlorobenzoate-catabolic transposon, Tn5271, in community adaptation to chlorobiphenyl, chloroaniline, and 2,4-dichlorophenoxyacetic acid in a freshwater ecosystem. Appl Environ Microbiol. 1992 Jan;58(1):314–325. doi: 10.1128/aem.58.1.314-325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulthorpe R. R., Wyndham R. C. Transfer and Expression of the Catabolic Plasmid pBRC60 in Wild Bacterial Recipients in a Freshwater Ecosystem. Appl Environ Microbiol. 1991 May;57(5):1546–1553. doi: 10.1128/aem.57.5.1546-1553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinsted J., de la Cruz F., Schmitt R. The Tn21 subgroup of bacterial transposable elements. Plasmid. 1990 Nov;24(3):163–189. doi: 10.1016/0147-619x(90)90001-s. [DOI] [PubMed] [Google Scholar]
- Haugland R. A., Sangodkar U. M., Chakrabarty A. M. Repeated sequences including RS1100 from Pseudomonas cepacia AC1100 function as IS elements. Mol Gen Genet. 1990 Jan;220(2):222–228. doi: 10.1007/BF00260485. [DOI] [PubMed] [Google Scholar]
- Hickey W. J., Focht D. D. Degradation of mono-, di-, and trihalogenated benzoic acids by Pseudomonas aeruginosa JB2. Appl Environ Microbiol. 1990 Dec;56(12):3842–3850. doi: 10.1128/aem.56.12.3842-3850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higson F. K., Focht D. D. Utilization of 3-chloro-2-methylbenzoic acid by Pseudomonas cepacia MB2 through the meta fission pathway. Appl Environ Microbiol. 1992 Aug;58(8):2501–2504. doi: 10.1128/aem.58.8.2501-2504.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Kröckel L., Focht D. D. Construction of chlorobenzene-utilizing recombinants by progenitive manifestation of a rare event. Appl Environ Microbiol. 1987 Oct;53(10):2470–2475. doi: 10.1128/aem.53.10.2470-2475.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure N. C., Fry J. C., Weightman A. J. Survival and catabolic activity of natural and genetically engineered bacteria in a laboratory-scale activated-sludge unit. Appl Environ Microbiol. 1991 Feb;57(2):366–373. doi: 10.1128/aem.57.2.366-373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure N. C., Weightman A. J., Fry J. C. Survival of Pseudomonas putida UWC1 containing cloned catabolic genes in a model activated-sludge unit. Appl Environ Microbiol. 1989 Oct;55(10):2627–2634. doi: 10.1128/aem.55.10.2627-2634.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu C. H., Wyndham R. C. Cloning and expression of the transposable chlorobenzoate-3,4-dioxygenase genes of Alcaligenes sp. strain BR60. Appl Environ Microbiol. 1993 Nov;59(11):3625–3633. doi: 10.1128/aem.59.11.3625-3633.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu C., Ng J., Singh R., Straus N., Wyndham C. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8312–8316. doi: 10.1073/pnas.88.19.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael D., Kreps S., Mergeay M. Identification of a catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J Bacteriol. 1993 Mar;175(6):1674–1681. doi: 10.1128/jb.175.6.1674-1681.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre M., Mailhiot K., Ahmad D., Massé R. Isolation and preliminary characterization of a 2-chlorobenzoate degrading Pseudomonas. Can J Microbiol. 1989 Apr;35(4):439–443. doi: 10.1139/m89-067. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Iino T. Naphthalene degrading genes on plasmid NAH7 are on a defective transposon. Mol Gen Genet. 1990 Aug;223(1):33–39. doi: 10.1007/BF00315794. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Minegishi K., Iino T. Toluene transposons Tn4651 and Tn4653 are class II transposons. J Bacteriol. 1989 Mar;171(3):1386–1393. doi: 10.1128/jb.171.3.1386-1393.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyndham R. C. Evolved aniline catabolism in Acinetobacter calcoaceticus during continuous culture of river water. Appl Environ Microbiol. 1986 Apr;51(4):781–789. doi: 10.1128/aem.51.4.781-789.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyndham R. C., Singh R. K., Straus N. A. Catabolic instability, plasmid gene deletion and recombination in Alcaligenes sp. BR60. Arch Microbiol. 1988;150(3):237–243. doi: 10.1007/BF00407786. [DOI] [PubMed] [Google Scholar]
- van der Meer J. R., Zehnder A. J., de Vos W. M. Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain P51. J Bacteriol. 1991 Nov;173(22):7077–7083. doi: 10.1128/jb.173.22.7077-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]




