Abstract
The MLL gene, the closest human homologue to the Drosophila trithorax gene, undergoes chromosomal translocation with a large number of different partner genes in both acute lymphoid and acute myeloid leukemias. We have identified a new partner gene, EEN, fused to MLL in a case of acute myeloid leukemia. The gene is located on chromosome 19p13, where two other MLL partner genes, ENL and ELL/MEN have also been identified. The deduced protein of 368 aa contains a central α-helical region and a C-terminal Src homology 3 (SH3) domain most similar to the C-terminal SH3 domain found in the Grb2/Sem-5/Drk family of genes. Sequence analysis of the fusion MLL/EEN transcript in our patient reveals that exon 6 of MLL is fused to the N-terminal end of EEN, a fusion that would create a chimeric protein that includes the major functional domain of EEN. EEN is expressed in a variety of tissue types and encodes a protein of approximately 46 kDa. The EEN protein is the human homologue of a member of a recently described murine SH3 domain-containing protein family. It is also highly related to a putative gene identified in Caenorhabditis elegans, and a number of similar sequences are present in the EST databases of several species.
The cloning and characterization of genes localized at nonrandom cytogenetic breakpoints in acute leukemia have facilitated studies of the molecular pathways involved in cellular transformation (1). The human homologue of the Drosophila trithorax gene, MLL (also referred to as HRX, ALL-1, and Htrx) is located on chromosome 11q23 (2–5); this chromosomal band is involved in translocations with at least 25 different chromosome partners, and at least 10 of the partner genes translocated to MLL have been cloned (for reviews, see refs. 6 and 7). MLL consists of 36 coding exons and spans at least 90 kb of DNA (8). The gene has two regions: the SET domain (9) and a cysteine-rich, PHD zinc-finger domain (10–12), highly related to those present in the Drosophila trithorax gene, a genetic regulator of homeobox genes. In mice in which one allele of the MLL is eliminated (knocked out) by homologous recombination, sternal and axial skeletal malformations, reminiscent of bidirectional homeotic transformations, are observed; homozygous deletion of MLL results in an embryonic lethal mutation (13). These data further support the hypothesis that MLL is the functional homologue of the trithorax gene in mammals.
As a result of the reciprocal nature of the translocations that occur between MLL and a partner gene, two fusion genes and, therefore, two potential transcripts can be formed. However, the fusion transcript that is consistently observed is that of the der(11)chromosome (7, 14), where the N-terminal portion of MLL is fused to the C-terminal end of the partner gene. The diversity of the partner genes that fuse to MLL has made it difficult to postulate a common mechanism to explain the biological basis for leukemogenesis of the fusion transcript. The first biological evidence for a leukemogenic role of these translocations has recently been described for the MLL/AF9 fusion (15). MLL rearrangements detected in the absence of chromosomal translocations involving 11q23 led to the description of two other mechanisms of oncogenic involvement. Partial duplication of MLL has been observed in cases of acute myeloid leukemia with normal karyotype or trisomy 11 (16, 17), and deletion of exon 8 has been seen in cases of T-cell acute lymphoblastic leukemia (18).
Identification of MLL rearrangements by Southern blotting or detection of MLL–partner gene fusion transcripts by reverse transcriptase–PCR are increasingly used to identify patients with poor prognostic outcome (19–21). We adopted these procedures as a general screen and identified a patient with rearrangement of MLL but without any of the common fusion transcripts. Thus we undertook the characterization of the consequences of MLL rearrangement in this patient.
MATERIALS AND METHODS
Patient Details.
The patient was a 22-month-old female with a diagnosis of acute myeloid leukemia based on morphological and cytochemical criteria. The patient was identified in a general screen of cases of acute leukemia for novel genes fused to MLL based on two selection criteria: DNA rearrangement detected with an MLL probe and a negative result in reverse transcriptase–PCR analyses of the four more common MLL fusion RNA products. Cytogenetic analysis revealed her karyotype as 46, XX/47, XX +21.
Molecular Screening of Patient Material.
Digests (BamHI, EcoRI, and SacI) of genomic DNA (10 μg) were screened for MLL rearrangement with the probe P/S4 as described (17, 22). Total RNA obtained from the bone marrow of the patient or established cell lines, RS4;11 (23), ML-2 (24), MONO MAC 6 (25), HB1119 (4), was isolated by the single-step method (26). cDNA was reverse transcribed with random hexamers and subjected to PCR amplification using specific primers to amplify chimeric transcripts from MLL/AF4, MLL/AF6, MLL/AF9, or MLL/ENL fusions (27).
Cloning of Fusion mRNA and the Genomic DNA.
Total RNA from the bone marrow of the patient was subjected to rapid amplification of cDNA ends (RACE) strategy using a 3′ RACE kit (Life Technologies, Grand Island, NY). Primers used are available on request (C.W.S.). PCR products were cloned using the UDG and Clonamp systems (Life Technologies). The novel sequence isolated from the RACE clone was used as a probe to screen a human fetal brain cDNA library (a kind gift from A. Ashworth, Institute of Cancer Research, London) constructed in λZAPII. Positive clones were isolated and converted to pBluescript phagemids according to the manufacturer’s instructions (Stratagene). All sequencing was performed using fluorescently labeled dideoxy terminators on 373A or 377 Applied Biosystems automated sequencers. The DNA sequence was assembled and analyzed, and both the nucleotide and predicted ORF were compared with public databases using various online and network facilities, including those of the Science and Engineering Research Council (England)-funded Daresbury’ SEQNET, Switzerland’s ExPASy, the U.K. European Bioinformatics Institute (EBI), and the U.S. government-funded National Center for Biotechnology Information.
The chromosomal fusion was amplified from genomic DNA (100 ng) by long-range PCR using the Expand Long Template PCR System (Boehringer Mannheim). The amplified band was cloned by TA cloning into pGEM-T (Promega) and sequenced and compared with the MLL breakpoint sequence (28).
Chromosomal Localization.
Two pairs of gene-specific primers (sequences available on request) were designed from the novel cDNA and genomic sequence analyses to perform PCR-based mapping on a monochromosomal somatic cell hybrid DNA panel (United Kingdom Human Genome Mapping Project Resource Center, Hixton, U.K.).
Fluorescence in situ hybridization (FISH) was employed to confirm and further define the genomic position of the new gene. The 4.5-kb MLL/EEN genomic fragment obtained from long-range PCR, which contained 3.8 kb of EEN, was used for FISH on normal human lymphocyte chromosome spreads using previously published methodologies (29). Images of metaphase preparations and hybridization signals were captured from a Nikon microscope and merged using cytovision image analysis software (Applied Imaging Company, Sunderland, U.K.).
RNA and Protein Expression Studies.
Multiple-human-tissue Northern blots (CLONTECH) were hybridized with a 2.6-kb EEN riboprobe derived by in vitro transcription of the EEN7 clone.
EEN cDNA clones were linearized, extracted by phenol chloroform, precipitated by ethanol, and resuspended in water. Protein products were expressed using the TNT coupled reticulocyte lysate system (Promega) according to the manufacturer’s instructions. Protein products were analyzed on an SDS/12% polyacrylamide gel. The gel was dried and autoradiographed for 1–3 days at −70°C.
RESULTS
Rearrangement of MLL and Noninvolvement of AF4, AF6, AF9, or ENL.
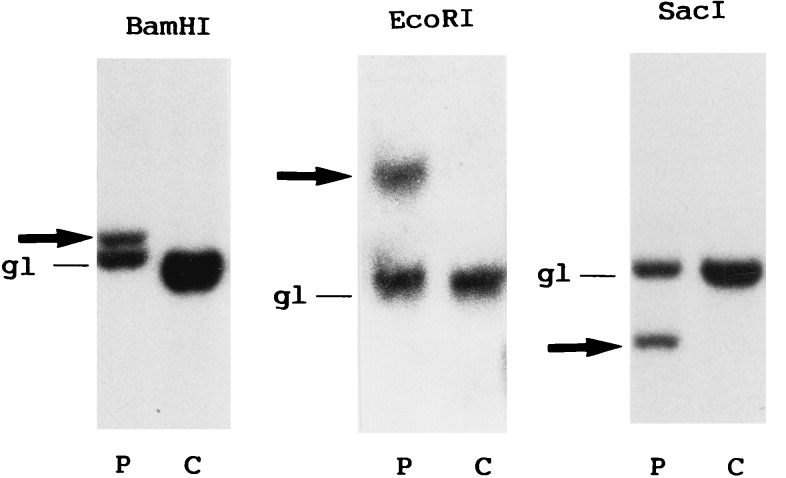
Southern blot analysis of DNA prepared from the leukemic cells of our patient using the probe P/S4 revealed the presence of rearranged bands with the three restrictions enzyme digests employed (Fig. 1). This allowed us to localize the MLL genomic breakpoint between exons 6 and 8. However, no PCR products were amplified from the cDNA prepared from these cells, when MLL/AF4, MLL/AF6, MLL/AF9, and MLL/ENL specific primer pair combinations were used (ref. 30 and data not shown). We inferred that MLL in this patient may be fused to a novel partner gene sequence.
Figure 1.
Genomic blot analysis for MLL rearrangement. Leukemic (P) and control (C) DNAs were digested with the three enzymes shown, blotted, and hybridized with P/S4. gl, germline bands; arrows, rearranged bands.
Cloning and Identification of a Novel MLL Fusion Product.
As all characterized MLL fusion genes to date have been shown to retain the derivative chromosome 11, the 3′ RACE method was chosen to isolate the fusion partner of MLL in this patient’s leukemic cells. The clones generated were screened with MLL primers from exons 6 and 9 upstream and downstream, respectively, of the breakpoint mapped above. Clones that contained exon 6 but not exon 9, and were larger than 250 bp, were sequenced. The sequence obtained from one clone (ES59) contained a 172-bp novel sequence with an ORF fused in-frame with the 3′ end of MLL exon 6 and, thus, could represent a potential new MLL translocation partner gene (Fig. 2). Although, at the time of these analyses (02/96), the sequence did not match any known genes present in the GenBank nucleotide database (January 1996), there were a number of highly related, although distinct, EST sequences with significant homology present. The presence of this fusion transcript was confirmed by reverse transcriptase–PCR in the patient RNA using primers form MLL exon 6 and the 172-bp novel sequence (data not shown).
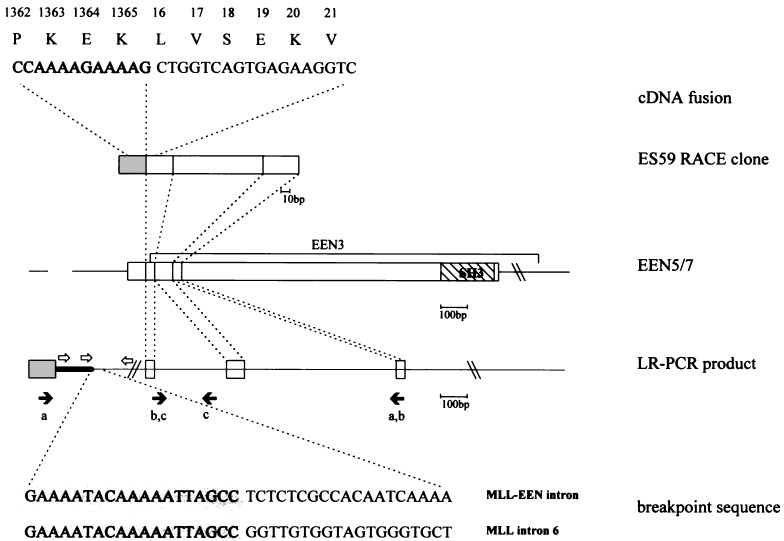
Figure 2.
The cDNA fusion sequence and amino acid translation and breakpoint sequences from the patient are depicted. The stippled areas are derived from the MLL locus. The positions of Alus identified by our sequence analysis, and in refs. 28, 31, and 32, are indicted by open arrows. The origin of the primers used in long-range PCR (pair a) and screening of the somatic cell hybrid DNA (pairs b, c) are indicated by solid arrows. Shaded box, exon 6 of MLL; open boxes, exons of EEN.
Analysis of Genomic Breakpoint.
To determine whether the fusion RACE clone was the result of a DNA translocation, we investigated the genomic DNA from the leukemic cells of this patient using long range PCR with primers from MLL exon 6 and a primer derived from the 3′ end of ES59. A unique 4.5-kb genomic PCR product was amplified from the leukemic DNA but not from control normal DNA. Sequence analysis of the fragment revealed that the MLL-contiguous sequences continued for 650 bp and were identical to those of MLL intron 6, after which they completely diverged (Fig. 2). At the other end of the 4.5 kb fragment, the sequence contained the 172-bp novel cDNA sequence present in ES59 interrupted by two introns spanning about 1.5 kb of genomic sequence (Fig. 2). These data are consistent with a genomic fusion due to the translocation of a partner gene to MLL and would, following splicing, generate the sequence represented in ES59.
The breakpoint in MLL occurs within and interrupts Alu26 repeat in intron 6 (31), between the nucleotides at positions 1257/1258 (numbering according to the 11q23 breakpoint sequence in GenBank accession no. HSU04737; ref. 28). The chromosome 19 breakpoint does not occur in an Alu; thus the rearrangement is not due to homologous recombination. Four Alu sequences occur within intron 6 and may contribute to the recombinogenic nature of this region (31, 32). In the sequence originating from chromosome 19, an Alu is present in the opposite orientation beginning about 250 bp downstream of the breakpoint (GenBank accession no. U66000U66000). The MLL breakpoint occurs in Region I as defined by Broeker et al., the region in which de novo patients often map (32). Although there are numerous sequences that are similar to the topoisomerase II consensus binding site (33), the nearest (matching 15 of the 18 of the consensus) lies at least 30 bases upstream.
Chromosomal Localization of the New MLL Partner Gene.
Using gene-specific primers designed from the 172-bp novel sequence, a 1.5-kb band was localized to the chromosome 19-containing member of a somatic cell hybrid panel. In addition, a 162-bp band was amplified from the monochromosomal panel members containing chromosomes 11, 15 (also containing 11q), and 17. All amplified bands were purified and subjected to sequence analysis. The 1.5-kb band on chromosome 19 was identical to that found in the 3′ end of the genomic breakpoint sequenced above. Thus, chromosome 19 is the genomic localization of the novel gene. In contrast, sequence analysis of the products from the other three hybrids each contained a sequence with a 10-bp deletion (GenBank accession nos. U66001U66001 and U66002U66002) but otherwise showed greater than 85% identity to the 172-bp cDNA (data not shown). We assume that these are representative of one or more processed pseudogenes (34, 35). To confirm the localization of the new gene, PCR-based chromosome mapping was performed with an additional pair of primers, one of which was intronic and therefore specific to the intron-containing gene. A unique 190-bp fragment was amplified from the DNA of the hybrid containing chromosome 19, but in no other members of the panel.
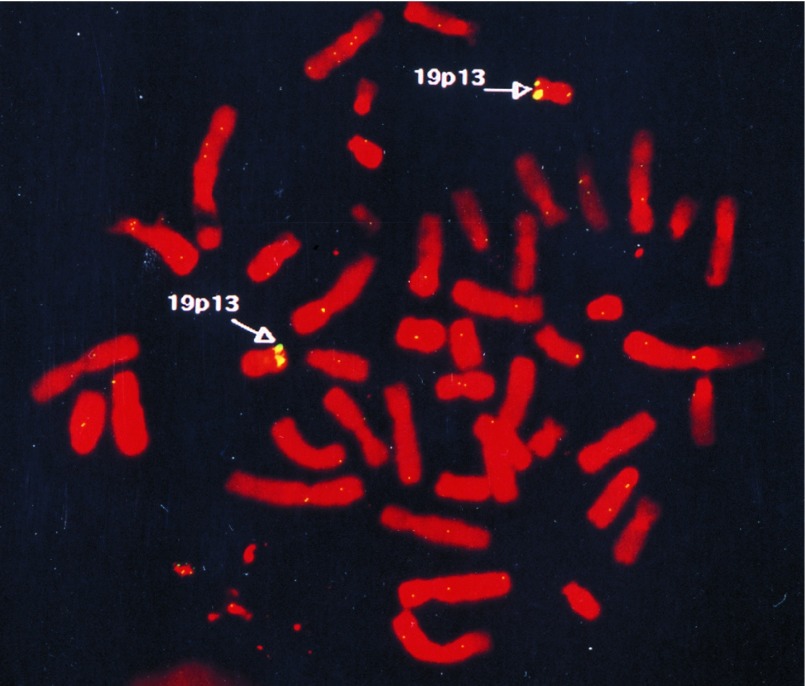
To define the chromosomal localization of the new gene more precisely, the 4.5-kb genomic breakpoint fragment from the patient was labeled for FISH on lymphocyte metaphases from a normal individual. The probe was found to hybridize specifically to chromosome 19p13 (Fig. 3). Out of 40 metaphases analyzed, detectable hybridization signals of two chromosome 19s at the p13 region were noted in 28 (70%), and only one of the two chromosome 19s was labeled at the same region in 10 (25%). Interestingly, two previously identified, but distinct, MLL partner genes, ENL (4, 36) and ELL/MEN (37–39) have been localized. We therefore designated the new gene as EEN (Extra Eleven Nineteen).
Figure 3.
Chromosomal localization of EEN by FISH. Normal metaphase spreads were hybridized with the fluorescently labeled 4.5-kb MLL/EEN genomic fragment.
Isolation and Sequencing of a Novel cDNA Containing ES59.
The 172-bp novel sequence identified in RACE clone ES59 was used as a probe to screen a human fetal brain cDNA library. Three positive clones containing the full 172-bp sequence were identified. Two clones (EEN5 and EEN7) of 2.6 kb appeared to be identical. The third clone, EEN3, was smaller with an insert size of 2.1 kb. When EEN3 and EEN7 were sequenced, they were found to be identical in the overlapping region, except for a G in EEN7 and an A in EEN3 in the first position of the codon of amino acid 331, which would result in a Asn/Asp difference at this position. The EEN7 clone contains an ORF (Fig. 4a) that spans 1107 nt (GenBank accession no. U65999U65999). The predicted initiation methionine codon is present within a Kozak consensus sequence (40). This ORF was followed by a termination codon and at least 1.3 kb of 3′ untranslated sequence and is predicted to encode a protein of 368 aa. This frame is the same as that identified in the ES59 fusion transcript that showed a join between MLL exon 6 and EEN at amino acid 16 (Fig. 2).
Figure 4.
Predicted coding sequence of EEN compared with other related sequences. (a) A multiple-sequence alignment of proteins found to be most closely related to the following GenBank accession numbers: EEN (U65999U65999), SH3p8 (U58885U58885), SH3p4 (U58886U58886), SH3p13 (U58887U58887), and Ce-SH3 (C. elegans CELF35A5.8, U46675U46675). Dark and light shading represent identical residues and conserved residues, respectively. Arrowheads indicate the point of MLL fusion to the predicted EEN amino acid sequence. (b) The sequence alignment of EEN (amino acids 312–360) to Src homology 3 (SH3) domain-containing proteins. The SwissProt accession numbers for the sequences shown are as follows: GRB2 (Human Grb2 C-terminal SH3 domain, P29354P29354), Sem5 (C. elegans Sem5 C-terminal domain, P29355P29355), Drk (Drosophila Drk C-terminal SH3 domain, Q0812), Myos1c (Myosin 1c heavy chain, P10569) and SRC (Human Src, P12931).
Sequence comparisons with the public databases revealed that the predicted EEN protein contains an SH3 domain found in many proteins involved in signal transduction as well as in cytoskeletal components (for review, see ref. 41). The EEN SH3 domain is similar to the C-terminal SH3 domain identified in the members of the myosin heavy chain family, but the closest identity is to the SH3 domain present in the Sem-5/Grb-2/Drk protein family members (Fig. 4b). While this paper was in preparation, the mouse homologue of EEN was cloned and reported as a result of the screening of a phage expression library with SH3 ligand peptides (Fig. 4a); the EEN SH3 domain can functionally bind to a class II type proline-rich sequence (42). In addition, the EEN protein has a high degree of sequence homology (30% identity, 50% similarity over 350 aa) to the protein encoded by a putative gene identified in C. elegans (GenBank accession no. U46675U46675: ref. 43 and direct submission to database) and to another encoded by a C. elegans EST sequence (accession no. M75843M75843; ref 44). Finally, several human EST sequences encode for a highly similar, although distinct, sequence (GenBank accession nos. T50023T50023, H22727H22727, HSC1HH051, and HSC1XE051). During the course of our library screening for EEN, we also identified an EEN-related gene (unpublished data) that represents the human counterpart to the mouse SH3p13, a member of the same SH3 domain-containing family present from C. elegans to human.
Expression Pattern of EEN.
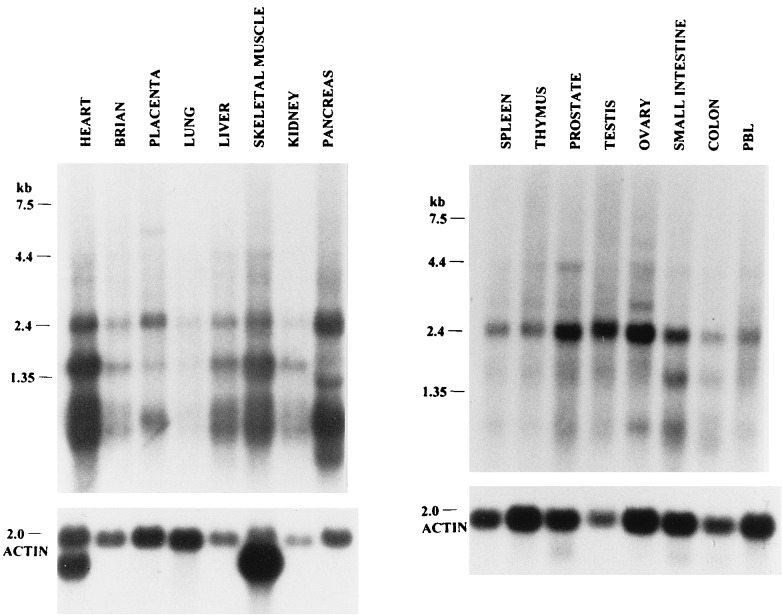
To investigate the predominant sites of expression of the EEN transcript, Northern blots containing RNA isolated from various tissues were hybridized to a riboprobe derived from the 2.6-kb EEN7 cDNA clone. Multiple transcript sizes were expressed in all tissue types examined (Fig. 5), with the highest expression observed in the pancreas and a low level of expression in the lung. The 2.6-kb transcript is the major transcript observed in most tissues, including spleen, thymus, prostate, testis, ovary, small intestine, colon, peripheral blood leukocytes, placenta, and pancreas, whereas the 2.0- and 1.2-kb transcripts were also strongly expressed in heart, liver, skeletal muscle, placenta, and kidney. Transcript sizes of 1.7 and 1 kb were also expressed in the pancreas.
Figure 5.
Northern blot analysis of EEN in multiple tissues. The blots were hybridized to a 2.6-kb EEN riboprobe. Molecular weights are given in kilobases. The blots were then stripped and rehybridized with an actin probe to control for RNA quality.
Protein Encoded by EEN cDNA.
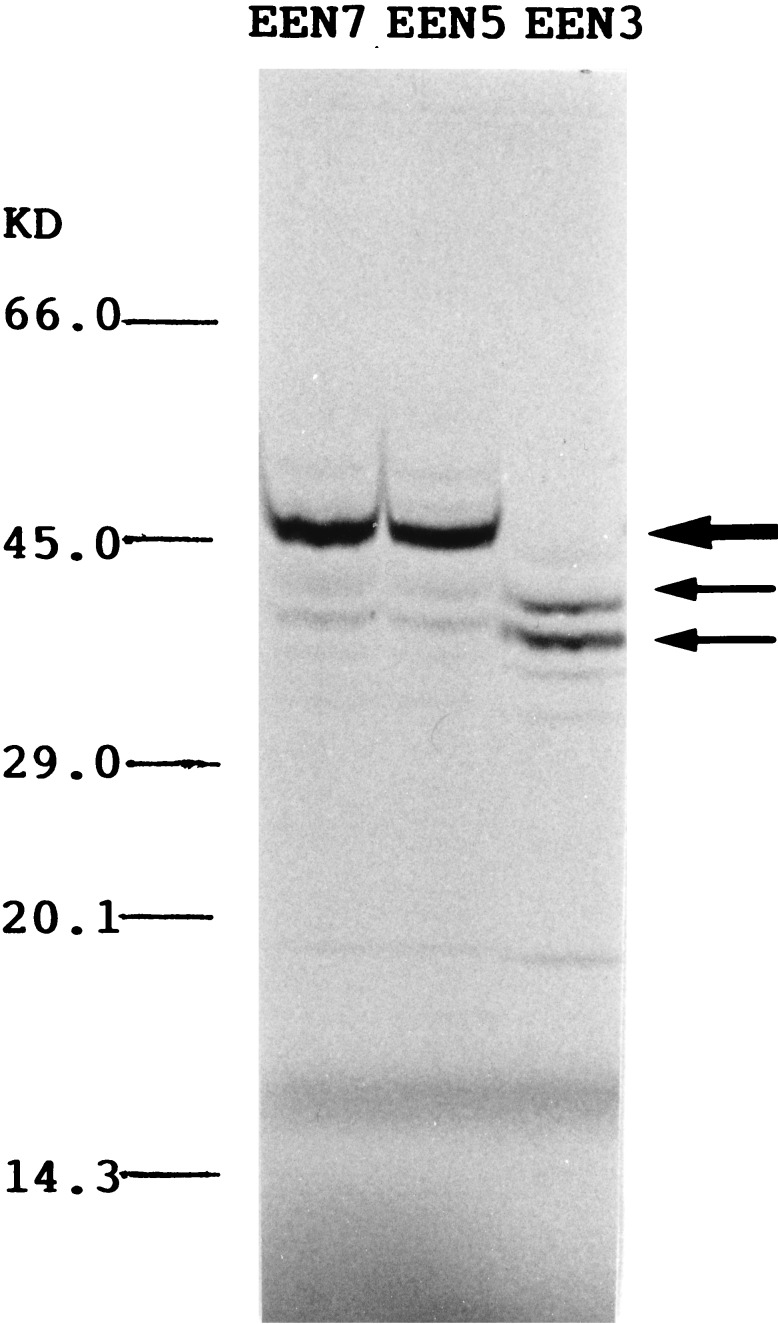
Several protein products of EEN were expressed by in vitro transcription and translation studies using the different EEN clones (Fig. 6). The EEN5 and EEN7 clones both encoded a major protein product of 46 kDa, probably encoded by the 1107-bp ORF contained in these two clones. The 46-kDa protein product was absent in EEN3, which lacks the ATG potential start codon present in EEN5/EEN7. The two major protein products of about 40 and 35 kDa observed in EEN3 were also weakly expressed in EEN5/EEN7 transcription/translation products.
Figure 6.
Analysis of proteins encoded by the EEN cDNA clones. The major 46-kDa band observed in EEN5 and EEN7 is indicated by a large arrow; the smaller products of 36 and 40 kDa expressed in all transcription/translation reactions are indicated by small arrows.
DISCUSSION
The t(11;19)(q23;p13) is a recurring chromosomal abnormality found in different types of acute leukemia, although heterogeneity in the breakpoints of chromosome 19p13 has been reported (45–47). Two genes on 19p13 [ENL (4, 36), involved in acute lymphoid leukemia, and ELL/MEN (37–39), involved in acute myeloid leukemia] have been identified in fusions with MLL on chromosome 11q23. Here, we report the identification of a third novel gene localized to 19p13 and found in a translocation with MLL in a case of acute myeloid leukemia. FISH analysis reveals that EEN is in the close vicinity of ENL and ELL/MEN. At the cytogenetic level, it may be difficult to distinguish MLL/ENL, MLL/ELL, and MLL/EEN fusions, and it is likely that other cases of t(11;19) may result in this gene fusion.
Northern blot analyses show that EEN is expressed as multiple transcripts in different tissues with a major transcript of 2.6 kb. We have cloned and sequenced a 2.6-kb cDNA from a fetal brain library, with an ORF encoding 368 aa. Comparisons with sequences in the databases revealed the presence of an SH3 domain; this SH3 domain has also been isolated independently and, like the Sem-5/Grb2/Drk SH3 domain (41, 48, 49) to which it is most closely related, will recognize a class II SH3 ligand (42). SH3 domains are conserved protein modules of 55–75 aa found in many proteins involved in signal transduction, but also in cytoskeletal elements and subunits of a neutrophilic cytochrome oxidase (41). Murine EEN (SH3p8 in Fig. 4a) is one of three related proteins isolated in the functional screen performed by Sparks et al. (42), and thus forms a new family of genes. Protein prediction programs suggest the presence of a helical structure in the central portion of the protein followed by the SH3 domain located at the C terminus. As MLL is fused to EEN at amino acid 16, the fusion product of der(11) transcript contains the AT hooks and methyltransferase homology regions of MLL, joined to the α-helical region and SH3 domain of EEN.
The nature of the molecular events resulting from the fusion of MLL to the various partner genes is unknown. There are no obvious features that are shared by the partner genes identified to date (for review, see ref. 50), and in this respect, EEN is no different. SH3 domains have been shown to mediate protein–protein interactions by binding proline-rich motifs in ligand proteins (41, 51, 52). As MLL contains several proline-rich sequences, one could postulate that the SH3 domain of EEN may interact with this region of MLL in either the fusion or the normal MLL protein. Formation of chimeric protein dimers has been suggested for MLL/AF1p, MLL/AF6, MLL/AF10, and MLL/AF17 fusions via protein dimerization domains present in the partner genes (7). Experiments investigating the possible interaction between the EEN SH3 domain and MLL will address this possibility.
Acknowledgments
We thank Cary So and Stanley Ko for technical assistance; David Cain and Harry King (Institute of Cancer Research, London) and Johnson Lau (Sequencing Core, Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, FL) for performing sequencing; Mel Greaves, Zhu Chen, and Cathy Price for helpful discussions; and Peter Rice (Sanger Center, Cambridge, U.K.) for the help with gcg program prettybox. C.W.S. is supported by a Croucher Foundation Scholarship, and C.C. is funded by a Human Capital and Mobility European Union Fellowship. M.-M.L., Q.-H.H., and S.-J.C. were supported in part by a grant from the National Natural Science Foundation of China. The work is funded in part by the University of Hong Kong Committee on Research and Conference Grants 337/046/0028 and 335/046/0060, and The Leukaemia Research Fund of Great Britain.
ABBREVIATIONS
- RACE
rapid amplification of cDNA ends
- FISH
fluorescence in situ hybridization
- SH3
Src homology 3
Footnotes
References
- 1.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Djabali M, Selleri L, Parry P, Bower M, Young B D, Evans G A. Nat Genet. 1992;2:112–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 3.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 4.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 5.Zieman-van der Poel S, McCabe N R, Gill H J, Espinosa R, Patel Y, Harden A, Rubinelli P, Smith S D, LeBeau M M, Rowley J D, Diaz M O. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greaves M F. Leukemia. 1996;10:372–377. [PubMed] [Google Scholar]
- 7.Bernard O A, Berger R. Genes Chromosomes Cancer. 1995;13:75–85. doi: 10.1002/gcc.2870130202. [DOI] [PubMed] [Google Scholar]
- 8.Rasio D, Schichman S A, Negrini M, Canaani E, Croce C M. Cancer Res. 1996;56:176–1769. [PubMed] [Google Scholar]
- 9.Chen J D, Chan C S, Pirrotta V. Mol Cell Biol. 1992;12:598–608. doi: 10.1128/mcb.12.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aasland R, Gibson T J, Stewart A F. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 11.Kokan M H M, Saibm A, de The H. Mol Biol Genet. 1995;318:733–739. [Google Scholar]
- 12.Saha V, Chaplin T, Gregorini A, Ayton P, Young B D. Proc Natl Acad Sci USA. 1995;92:9737–9741. doi: 10.1073/pnas.92.21.9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu B D, Hess J L, Horning S E, Brown G A J, Korsmeyer S J. Nature (London) 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 14.Rowley J. Genes Chromosomes Cancer. 1992;5:264–266. doi: 10.1002/gcc.2870050316. [DOI] [PubMed] [Google Scholar]
- 15.Corral J, Lavenir I, Impey H, Warren A J, Forster A, Larson T A, Bell S, McKenzie A N J, King G, Rabbitts T H. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. ‘. [DOI] [PubMed] [Google Scholar]
- 16.Schichman S A, Canaani E, Croce C M. J Am Med Assoc. 1995;273:571–576. [PubMed] [Google Scholar]
- 17.So C W, Ma Z G, Price C M, Dong S, Chen S J, Gu L J, So C K C, Wiedeman L M, Chan L C. Cancer Res. 1997;57:117–122. [PubMed] [Google Scholar]
- 18.Lochner K, Siegler G, Fuhrer M, Greil J, Beck J D, Fey G H, Marchalek R. Cancer Res. 1996;56:2171–2188. [PubMed] [Google Scholar]
- 19.Biondi A, Rambaldi A, Rossi V, Elia L, Caslini C, Basso G, Battista R, Barbui T, Mandelli F, Masera G, Croce C, Canaani E, Cimino G. Blood. 1993;82:2943–2947. [PubMed] [Google Scholar]
- 20.Griesinger F, Elfers H, Ludwig W-D, Falk M, Rieder H, Harbott J, Lampert F, Heinze B, Hoelzer D, Thiel E, Riehm H, Worman B, Fonatsch C, Hiddenmann W. Leukemia. 1994;8:542–548. [PubMed] [Google Scholar]
- 21.Stock W, Thirman M J, Dodge R K, Rowley J D, Diaz M O, Wurster-Hill D, Sobol R E, Davey F R, Larson R A, Westbrook C A, Bloomfield C D. Leukemia. 1994;8:1918–1922. [PubMed] [Google Scholar]
- 22.Morgan G J, Cotter F, Katz F E, Ridge S A, Domer P, Korsmeyer S, Wiedemann L M. Blood. 1992;80:2172–2175. [PubMed] [Google Scholar]
- 23.Stong R C, Korsmeyer S J, Parkin J L, Arthur D C, Kersey J H. Blood. 1985;65:21–31. [PubMed] [Google Scholar]
- 24.Drexler H G, MacLeod R A F, Quentmeier H, Steube H. DSM Catalogue of Human and Animal Cell Lines. 4th Ed. Germany: Braunschweig; 1994. [Google Scholar]
- 25.MacLeod R A F, Voges M, Drexler V G. Blood. 1993;82:3221–3222. [PubMed] [Google Scholar]
- 26.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 27.Repp R, Borkhardt A, Haupt E, Kreuder J, Brettreich S, Hammermann J, Nishida K, Harbott J, Lampert F. Leukemia. 1995;9:210–215. [PubMed] [Google Scholar]
- 28.Gu Y, Alder H, Nakamura T, Schichman S A, Prasad R, Canaani O, Saito H, Croce C M, Canaani E. Cancer Res. 1994;54:2327–2330. [PubMed] [Google Scholar]
- 29.Mao M, Yu M, Tong J H, Ye J, Zhu J, Huang Q H, Fu G, Yu L, Zhao S Y, Waxman S, Lanotte M, Wang Z Y, Tan J Z, Chen S J, Chen Z. Proc Natl Acad Sci USA. 1996;93:5910–5914. doi: 10.1073/pnas.93.12.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Z G, Huang Q H, Dong S, Cao Q, Su X Y, Huang W, Wang Z Y, Gu L J, Chen S J, Chen Z. Chin J Hematol. 1996;17:3–6. [Google Scholar]
- 31.Marschalek R, Greil J, Lochner K, Nilson I, Siedgler G, Zweckbronner I, Beck J D, Fey G H. Br J Haematol. 1995;90:308–320. doi: 10.1111/j.1365-2141.1995.tb05151.x. [DOI] [PubMed] [Google Scholar]
- 32.Broeker P L S, Super H G, Thirman M J, Pomykala H, Yonebayashi Y, Tanabe S, Zeleznik-Le N, Rowley J D. Blood. 1996;87:1912–1922. [PubMed] [Google Scholar]
- 33.Spitzner J R, Muller M T. Nucleic Acids Res. 1988;16:5533–5556. doi: 10.1093/nar/16.12.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanin E F. Annu Rev Genet. 1985;19:253–372. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- 35.Wiedemann L M, Perry R P. Mol Cell Biol. 1984;4:2518–2528. doi: 10.1128/mcb.4.11.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura T, Alder H, Gu Y, Prasad R, Canaani O, Kamada N, Gale R P, Lange B, Crist W M, Nowell P C, Croce C M, Canaani E. Proc Natl Acad Sci USA. 1993;90:4631–4635. doi: 10.1073/pnas.90.10.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thirman M J, Levitan D A, Kobayashi H, Simon M C, Rowley J D. Proc Natl Acad Sci USA. 1995;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitani K, Kanda Y, Ogawa S, Tanaka T, Inazawa J, Yazaki Y, Hirai H. Blood. 1995;85:2017–2024. [PubMed] [Google Scholar]
- 39.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 40.Kozak M. Cell. 1996;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 41.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 42.Sparks A B, Hoffman N G, McConn S J, Fowlkes D M, Kay B K. Nat Biotech. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 43.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 44.Waterston R, Martin C, Craxton M, Hynh C, Coulson A, Hillier L, Durbin R K, Green P, Shownkeen R, Halloran N, Hawkins T, Wilson R, Berks M, Du Z, Thomas K, Thierry-Mieg J, Sulston J. Nat Genet. 1992;1:114–123. doi: 10.1038/ng0592-114. [DOI] [PubMed] [Google Scholar]
- 45.Huret J L, Brizard A, Slater R, Charrin C, Bertheas M F, Guihot F, Hahlen K, Kroes W, van Leeuwen E, Schoot E V D, Beishuizen A, Tanzer J, Hagemeijer A. Leukemia. 1993;7:152–160. [PubMed] [Google Scholar]
- 46.Mitani K, Sato Y, Kobayashi Y, Shibasaki Y, Kasuga M, Inaba T, Hayashi Y, Miura Y, Miyazono K, Hirai H, Urabe A, Takaku F. Am J Hematol. 1989;31:253–257. doi: 10.1002/ajh.2830310407. [DOI] [PubMed] [Google Scholar]
- 47.Poirel H, Rack K, Delabesse E, Raadford-Weiss I, Troussard X, Debert C, Leboeuf D, Bastard C, Picard F, Veil-Buzyn A, Flandrin G, Bernard O, Macintyre E. Blood. 1996;87:2496–2505. [PubMed] [Google Scholar]
- 48.Lim W A, Richards F M, Fox R O. Nature (London) 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 49.Feng S, Chen J K, Yu H, Simon J A, Schreiber S L. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 50.Canaani E, Nowell P C, Croce C M. Adv Cancer Res. 1995;66:213–234. doi: 10.1016/s0065-230x(08)60255-9. [DOI] [PubMed] [Google Scholar]
- 51.Shokat K M. Chem Biol. 1995;2:509–514. doi: 10.1016/1074-5521(95)90183-3. [DOI] [PubMed] [Google Scholar]
- 52.Lowenstein E J, Daly R J, Batzer A G, Li W, Margolis B, Lammers R, Ulrich A, Skolnik E Y, Bar-Sagi D, Schlessinger J. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]