Abstract
The remodelling of the extracellular matrix (ECM) has been shown to be highly upregulated in cancer and inflammation and is critically linked to the processes of invasion and metastasis. One of the key enzymes involved in specifically degrading the heparan sulphate (HS) component of the ECM is the endo-β-glucuronidase enzyme heparanase. Processing of HS by heparanase releases both a host of bioactive growth factors anchored within the mesh of the ECM as well as defined fragments of HS capable of promoting cellular proliferation. The finding that heparanase is elevated in a wide variety of tumor types and is subsequently linked to the development of pathological processes has led to an explosion of therapeutic strategies to inhibit its enzyme activity. So far only one compound, the sulphated oligosaccharide PI88, which both inhibits heparanase activity and has effects on growth factor binding has reached clinical trials where it has shown to have promising efficacy. The scene has clearly been set however for a new generation of compounds, either specific to the enzyme or with dual roles, to emerge from the lab and enter the clinic. The aim of this review is to describe the current drug discovery status of small molecule, sugar and neutralising antibody inhibitors of heparanase enzyme activity. Potential strategies will also be discussed on the selection of suitable biomarker strategies for specific monitoring of in vivo heparanase inhibition which will be crucial for both animal model and clinical trial testing.
Keywords: heparanase, heparan sulphate, inhibitor, inflammation, cancer, angiogenesis
Introduction
The extracellular matrix (ECM) plays a key role in both normal and disease processes as diverse as angiogenesis, inflammation, wound healing and tumour cell invasion. In the ECM, heparan sulphate proteolglycans (HSPGs) interact with fibronectin, laminin, collagen and growth factors to help maintain cellular architecture. HSPGs themselves act as a storage depot for a number of cytokines and growth factors such as basic fibroblast growth factor (bFGF), vascular endothelial growth factor (VEGF), keratin growth factor (KGF), hepatocyte growth factor (HGF) and transforming growth factor β (TGFβ), which bind specifically to the heparan sulphate (HS) glycosaminoglycan chains (Bernfield et al., 1999; Kreuger et al., 2006). The synthesis of HS on the core HSPG protein begins within the Golgi apparatus and requires the sequential action of a HS synthase enzyme followed by modification brought about by a series of additional enzymes: sulfotransferases, N-deacetylase/N-sulfotransferase, C-5 epimerase, 2-O-sulfotransferase, 6-O-sulfotransferase and 3-O-sulfotransferase. This postglycosylation modification produces the diverse repertoire of HS sulphated species, which is important for generating protein-binding sites and cleavage sites for endoglucuronidase enzymes such as heparanase. The complete HS molecule has a domain structure organization (Lyon and Gallagher, 1998; Merry et al., 2001; Murphy et al., 2004) with N- and O-sulphate groups clustered into iduronic acid-rich S-domains separated by the NA domains, which contain N-acetylglucosamine-glucuronic acid disaccharide repeats with low levels of O-sulphation. Heparanase cleavage sites are spaced within these regions of low sulphation. The fact that HS is so widely distributed and evolutionarily conserved is testament to the vital importance of this molecule in cell development and function. Remodelling of the ECM following heparanase cleavage of HS results both in the liberation of glycosaminoglycan-anchored bioactive molecules and HS fragments of approximately 5–7 kDa in size that modulate growth factor binding to their receptors (Sanderson et al., 2004). The actual amount of heparanase required for HS remodelling appears to be of key importance as low levels of the enzyme enhances FGF2 binding and subsequent downstream activation of ERK/FAK signalling in human metastatic melanoma cells (70W), whereas very high levels are inhibitory (Reiland et al., 2006).
Heparanase activity has been detected in activated immune cells including T and B cells, macrophages, neutrophils, mast cells mediating extravasation and traffic to inflammatory sites (Vaday and Lider, 2000), and the enzyme is highly expressed in placental tissue where it is involved in trophoblast cell implantation (Dempsey et al., 2000). Platelets are also a very rich source of heparanase and their aggregation with tumour cells is believed to facilitate tumour cell metastases and ECM disassembly following platelet degranulation (Freeman and Parish, 1998). The key role played by heparanase in malignancy was confirmed by antisense cDNA transfection studies that showed a significant reduction in the invasive and metastatic properties of tumour cells (Uno et al., 2001). Expression of an anti-heparanase ribozyme construct in human MDA-MB-435 breast carcinoma cells reduced significantly heparanase enzyme activity and also invasion through a matrigel basement membrane (Edovitsky et al., 2004). A large number of publications now clearly link heparanase expression to the process of tumourigenesis in a wide number of cancers, including bladder (Gohji et al., 2001), colon (Friedmann et al., 2000; Sato et al., 2004), gastric (Tang et al., 2002; Takaoka et al., 2003), breast (Maxhimer et al., 2002), oral (Ikuta et al., 2001), oesophageal (Mikami et al., 2001), pancreatic (Koliopanos et al., 2001; Rohloff et al., 2002; Kim et al., 2002), brain (Marchetti and Nicolson, 2001), thyroid (Xu et al., 2003), prostate (Ogishima et al., 2005a, 2005b) and acute myeloid leukemia (Vlodavsky et al., 2002). Collectively, this evidence suggests that heparanase plays a fundamental role in sustaining the pathology of malignant diseases and therefore that it may provide a potential target for anti-cancer therapy. An exciting development in the understanding of heparanase function was the finding that the protein has a role in cell adhesion. Overexpression of heparanase in Eb T-cell lymphomas induced cell adhesion via β1 integrin and Rac activation (Goldshmidt et al., 2003). Inactive variants of the protein produced the same pro-adhesive effects suggesting, for the first time, that heparanase has functions over and above its enzymatic activity. The as yet elusive pro-adhesive domain of heparanase was also shown to be responsible for the phosphorylation of Akt in primary endothelial cells (Gingis-Velitski et al., 2004) and it has been postulated that this may provide anti-apoptotic protection in tumour cells (Cohen et al., 2005).
In the absence of a 3D structure of the enzyme, a number of groups have taken a structure prediction strategy to carry out a rational design of new high-affinity inhibitors (Ishida et al., 2004; Zhou et al., 2006). Heparanase has been the subject of a number of excellent reviews in recent years examining its mode of action, disease association and efficacy of specific inhibitory molecules (Parish et al., 2001; Vlodavsky et al., 2002; Simizu et al., 2004b; Ferro et al., 2004; Ilan et al., 2006).
This review will focus on some of the latest heparanase inhibitory compounds and antibodies to have emerged, and discuss new therapeutic strategies to downregulate the actions of this important protein (Figure 1). The development of the Oxford GlycoSciences (OGS) heparanase inhibitor series of compounds is presented as a case study example of the structure–activity relationship (SAR) involved in the progression of this class of drugs through animal model testing. It is envisaged that these inhibitors will serve as valuable research tools for both in vitro and in vivo studies into the functioning of the enzyme in both normal and disease settings. Progression of heparanase inhibitors towards clinical testing is critically dependent on obtaining biomarkers to study the efficiency of heparanase knockdown, and hence for the first time a discussion is given on potential strategies that may be useful to assess heparanase inhibitor efficacy.
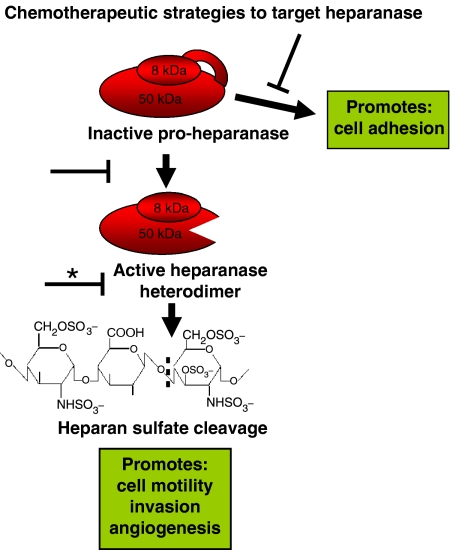
Figure 1.
The latent form of heparanase (pro-heparanase) is schematically represented as a contiguous fusion of 8 and 50 kDa subunits joined by an intervening linker region. Proteolytic digestion of the linker is thought to induce a conformation change that opens up the active site and allow HS cleavage. The minimum HS epitope required for heparanase digestion is indicated. The most direct route to inhibit heparanase action and consequently most heavily investigated (*) is to inhibit the enzymatic activity. Other potential targets implicated from recent studies on the enzyme include the linker peptide proteases or the putative pro-adhesive domains of the enzyme.
Heparanase enzyme
In 1999, five groups published papers describing the cloning of the heparanase gene (Fairbanks et al., 1999; Kussie et al., 1999; Hulett et al., 1999; Toyoshima and Nakajima, 1999; Vlodavsky et al., 1999). Up until that time there was considerable controversy in the literature regarding the actual molecular weight of active heparanase with sizes ranging from 8 to 137 kDa all having been reported in Oosta et al. (1982), Hoogewerf et al. (1995) and Freeman and Parish (1998). Recombinant chemokine connective tissue-activating peptide (CTAP)-III (Rechter et al., 1999) was shown to digest HS from a metabolically labelled naturally produced ECM substrate. It seems likely, however, that the CTAP III proteins used in this study contained contaminating activities, whereas a later study showed that highly purified native CTAP III from platelets was completely devoid of heparanase activity. In addition, immunoaffinity depletion of CTAP III from platelets left behind an active HS degrading species of ∼55 kDa consistent with that expected of the HPSE1 gene product (Castor et al., 2002). Confirmation that heparanase enzyme activities from both natural and tumour sources were similar came from the finding that both platelet and carcinoma cell heparanase activity (Freeman et al., 1999) degraded heparin and HS to equivalent size fragments and Western blotted positive against a HPSE1 antibody.
The human heparanase gene is located on chromosome 4q21.3 (Baker et al., 1999; Dong et al., 2000) and encodes a pre pro-polypeptide of 543 amino acids. The protein is N-glycosylated at six putative sites and tunicamycin inhibitor studies have shown that these are important for the kinetics of oestrogen receptor (ER) to Golgi transport and for enzyme secretion (Simizu et al., 2004a). Signal peptidase cleavage of the pre pro-enzyme within the ER (position A35) generates the latent 65 kDa (as judged by its mobility on SDS PAGE gels) pro-enzyme form of the enzyme. The active heterodimer form of the enzyme consists of an N-terminal 8 kDa subunit (Q36–E109) non-covalently associated with a C-terminal 50 kDa subunit (K158–I543) that is subsequently produced by proteolysis of the pro-enzyme and excision of the intervening 6 kDa linker region (S110–Q157) (Fairbanks et al., 1999; Levy-Adam et al., 2003; McKenzie et al., 2003). As the active residues (E225 and E343) required for HS cleavage are both contained within the 50 kDa subunit (Hulett et al., 2000), it is presumed that the 8 kDa subunit binding produces some conformation change within the active site that facilitates catalysis. The precise nature of the proteases responsible for generating both subunits is an area of intensive investigation. Extensive mutagenesis studies indicate that the bulky hydrophobic group Y156 is essential for correct heparanase processing and activation at the P2 position of the cathepsin L-like linker cleavage site 2 (i.e. between amino acids Q157 and K158) (Abboud-Jarrous et al., 2005). In contrast, mutations around site 1 (i.e. between amino acids E109and S110) had no effect, suggesting that this region contains a novel protease recognition motif. The proteolytic processing of heparanase occurs intracellularly within the late endosomes and lysosomes (Nadav et al., 2002; Goldshmidt et al., 2002a, 2002b; Gingis-Velitski et al., 2004; Vreys et al., 2005; Zetser et al., 2004). The requirement for a low-pH environment for efficient processing is consistent with this reaction being carried out in acidic intracellular organelles. For example, pH increasing agents such as chloroquine and bafilomycin A1 inhibit heparanase processing (Zetser et al., 2004) and a pH range between 4 and 5 was found to be optimal in vitro processing of heparanase using endosome/lysosome cell fractions (Cohen et al., 2005).
Regulation of heparanase gene transcription
Numerous studies using quantitative RNA analysis methods have shown a striking correlation between tumour progression and the increase in heparanase mRNA expression. Upregulation of heparanase transcription by modulating the levels of transcription factors able to bind to its promoter appears to be a common factor of most malignant cells studied to date. Hypomethylation of the heparanase promoter in benign prostatic hyperplasia and normal bladder correlated with increased levels of heparanase expression (Ogishima et al., 2005a, 2005b). Treatment of C6 rat glioma or human choriocarcinoma cells that have higher levels of promoter methylation with demethylating agents, such as 5-azacytidine, restored heparanase activity and potentiated metastasis capacity in vivo (Shteper et al., 2003). The transcription factors SP1 and Ets are associated with basal level heparanase regulation (Jiang et al., 2002), whereas the early growth response 1 transcription factor is involved in the inducible regulation of heparanase transcription (de Mestre et al., 2003, 2005). The presence of functional oestrogen response elements within the heparanase promoter suggested a hormonal control mechanism that in particular could have a role in the development and progression of breast cancer (Elkin et al., 2003). Induction of heparanase expression in ER positive MCF7 human breast carcinoma cells was shown to be inhibited by treatment of by the oestrogen antagonist ICI 182,780 (Elkin et al., 2003). Two interferon-stimulated response elements consensus sequences are also present in the heparanase promoter and may mediate the effect of interferon-γ on heparanase gene expression in endothelial cells (Edovitsky et al., 2006) and T lymphocytes (Sotnikov et al., 2004) upregulated heparanase gene expression and enzymatic activity. Under normal cellular conditions, the heparanase promoter needs to be tightly regulated and transcriptional repression at least in part is produced by the binding of the p53 tumour suppressor via recruitment of histone deacetylases. Tumour variants of p53 were unable to bind to the promoter and hence failed to repress transcription (Baraz et al., 2006).
Promoter study analysis show that heparanase by itself can act to promote further the expression of other proteins involved in tumour malignancy and angiogenesis such as Cox-2 (Ohtawa et al., 2006) and HIF1α (Naomoto et al., 2007). It is unclear whether this involves the direct translocation of heparanase into the nucleus, heparanase-mediated signalling or activation as a consequence of increased HS turnover. Other studies have shown that heparanase is upregulated together with matrix metalloproteinase 2 (MMP2) in vascular smooth muscle cells (Fitzgerald et al., 1999) and in an animal model of cardiac hypertrophy (Kizaki et al., 2005). It would be interesting to take these observations further and analyse global transcription changes (e.g. microarray transcriptome profiling of a regulatable heparanase cell line) brought upon by heparanase expression to see whether heparanase expression triggers a downstream expression cascade of other key disease-associated proteins.
Heparanase cellular localization
It is clear that heparanase has an important housekeeping function within the late endosome and lysosomal organelles where it is involved in the processing and recycling of HS from internalized HSPGs. It is not restricted to these cellular locations, however, as immunofluorescence and cell fractionation studies clearly show the presence of the protein in the nucleus, perinuclear regions and, plasma membrane (Marchetti et al., 2000; Goldshmidt et al., 2001, 2002a, 2002b, 2003). Intracellular pools of heparanase can be mobilized by demand and relocated to the cell surface or secreted where it may degrade cell surface HS. In this context it has been shown that immature dendritic cells contain a resting pool of nuclear and cytoplasmic heparanase, which can be actively translocated to cell surface membrane extensions upon lipopolysaccharide stimulation. This cell surface heparanase degrades ECM HS and is thought to play a role in cellular transmigration (Benhamron et al., 2006). The nuclear role of heparanase is still undefined but is thought to play a role in nuclear HS degradation and stimulation of gene expression. Heparanase protein contains two potential nuclear localization sequences (amino acid positions 271–277 and 427–430), which may mediate its nuclear translocation as evidenced in cancer cell lines (Schubert et al., 2004) and human gastric and oesophagus cancer sections (Takaoka et al., 2003). Strong heparanase expression was localized to the invasion front of head and neck squamos cell carcinomas and in disseminated tumour cells where it correlated with poor prognosis (Beckhove et al., 2005). Promonocytic leukaemia U937 and THP-1 cells have been used as models to study the spatial regulation of heparanase in macrophages. Activation of the cells with PMA induced HS degradation of endothelium-derived ECM and re-distribution of heparanase to the cell surface (but not secreted) (Sasaki et al., 2004). Treatment of human microvascular endothelial cells with TNFα and Il-1β (Chen et al., 2004) and human peripheral T cells with TNFα (Sotnikov et al., 2004) increased secretion dramatically. Tumour-derived cells, on the other hand, seem to secrete heparanase more efficiently via agonist stimulation with nucleotides such as adenosine triphosphate, adenosine diphosphate and adenosine. These nucleotides act through the P2Y G-protein coupled receptor (GPCR) signal transduction cascade via protein kinase C to effect the movement of intracellular endosome/lysosome heparanase pools towards the cell surface (Shafat et al., 2006a, 2006b). The P2Y nucleotide receptor is found on the surface of platelets where it plays a major role in platelet aggregation and thrombus stabilization and is a well-established target in its own right for anti-thrombotic drugs (Kahner et al., 2006). Gene expression profiling data across a number of primary human cancers also indicates that many GPCRs including P2Y are overexpressed in tumours (Li et al., 2005) that may initiate a re-routing of intracellular heparanase to cell surface locations during disease progression.
A number of cell types, for example, primary fibroblasts, endothelial cells and tumour cell lines, have been shown to be able to internalize extracellular latent 65 kDa heparanase via binding to cell surface HSPGs or the low-density lipoprotein receptor (LDLR)-related protein and mannose 6-phosphate receptor (Vreys et al., 2005). This offers yet another mechanism for normal cells to regulate their local levels of heparanase and offers the potential for cancer cells to acquire more heparanase enzyme from the extracellular space to increase their tumourigenicity (Figure 2).
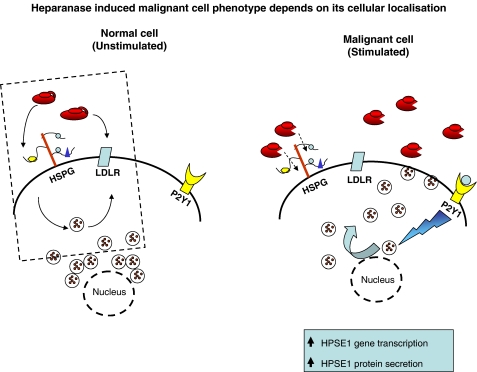
Figure 2.
Heparanase has an important housekeeping role in degrading recycled glycosaminoglycan chains of internalized HSPG molecules, which accounts for its endosomal/lysosomal roles in normal/unstimulated cells. Latent enzyme can be internalized by binding to membrane-bound co-receptors such as HSPGs and LDLR. A recent study has described a route where intracellular heparanase can be actively secreted via the P2Y1 cell surface receptor. In the scenario of a malignant tumour cell it can be envisaged that a combination of increased heparanase expression at the mRNA level coupled with appropriate signals forcing pools of heparanase protein to the cell surface could rapidly mobilize ECM components required for proliferation and metastases (adopted from Shafat et al., 2006a).
Small molecule inhibitors
Case study: development of OGS series of heparanase inhibitors
Between 1999 and 2003, OGS undertook a programme aimed to develop small-molecule inhibitors against heparanase enzyme activity.
High-throughput screening of a 50 000 compound library using a modified HS/bFGF 96-well plate assay (Figure 3a) identified the 2,3-dihydro-1,3-dioxo-1H-isoindole-5-carboxylic acid compound (Figure 4, molecule 2a) as a inhibitor of heparanase activity (IC50 of 8 μM) and with modest cell-based anti-angiogenic activity (IC50 40 μM). This compound satisfied a number of favourable drug selection criteria including molecular weight, lipophilicity, polar surface area and synthetic accessibility of related structures. An SAR programme directed to identify more potent and selective inhibitors using this compound as a scaffold was therefore initiated (Courtney et al., 2004). Early studies identified key structural features that could not be modified without the loss of inhibitory activity, namely the carboxylic acid, isoindole-5-carboxylic acid moiety and pendant benzoxazole ring system. Instead substitutions were made on the central aromatic core and benzoxazole moiety to study their effect and the efficacy of some of these molecules (Figure 4). This series of compounds led to the selection of derivative 2 h (OGT 2492) with its much-improved anti-angiogenic potency (IC50, 2 μM) and enzyme inhibitory potency (IC50, 3.0 μM) for further optimization (Figure 3b). A number of substitutions were then made on the pendant phenyl ring of OGT 2492 (Figure 4). Introduction of fluoro to the 4-position (compound 6b) led to a significant increase in heparanase inhibitory activity. A loss of activity was observed with the introduction of an ortho-methoxy group (6i). Translocating the methoxy substituent on the central phenyl ring to the 4-position produced the most potent compound in the series (6p) with an IC50 of 0.2 μM against the enzyme and an IC50 of 3 μM in the angiogenesis assay. There was a poor correlation between the IC50 data from both assays for each compound probably due to solubility issues, poor cellular uptake or an off-target component to the activity. One clear exception however was the 3′,5′-diflouro derivative (6i), which had close matching potencies (IC50 of 0.9 μM against the enzyme and IC50 of 0.75 μM in the angiogenesis assay).
Figure 3.
(a) OGS heparanase high-throughput assay. The assay is based on the use of the specific binding of bFGF (shown as triangles) to HS pre-adsorbed onto plates (in red). Following cleavage of high molecular weight HS by heparanase, the smaller material generated no longer adheres to the surface of the six-well plate and hence heparanase activity can be followed by as a reduction in bFGF binding (as measured by a HRP-conjugated bFGF antibody). (b) Angiogenesis assay. Human endothelial cells were co-cultured with fibroblast cells and left for 12 days to form tubule structures. Tubules are stained positive with anti-CD31 and secondary alkaline phosphatase conjugate and quantified using an image analysis programme. Plates show controls plates incubated with dimethylsulphoxide, VEGF or recombinant HPSE1 protein. The anti-angiogenic activity of 1 μM OGT 2492 is indicated (adopted from Courtney et al., 2004).
Figure 4.
Structure and efficacy of a select number of 2,3-dihydro-1,3-dioxo-1H-isoindole-5-carboxylic acid OGS heparanase inhibitors (adopted from Courtney et al., 2004). SAR derivatives of each class are shown.
A series of solubilizing substitutions were introduced to improve compound solubility with the aim of narrowing the gap between IC50 data in both assays (Figure 4c). These molecules were potent inhibitors in both primary and angiogenesis assays and had more comparable IC50 values. A two-fold variation was observed with compounds 9a (0.5 μM IC50 against the enzyme vs 0.25 μM in the angiogenesis assay) and 9e (2.0 μM IC50 enzyme vs 1.0 μM angiogenesis), suggesting that these compounds should be more amenable for cellular assays including angiogenesis, cell invasion and cell motility studies.
A second-phase research programme at OGS centred on the furanylthiazole acetic acid compound as a new SAR scaffold (Courtney et al., 2005). This compound inhibited heparanase enzyme activity relatively weakly at IC50 of 25 μM, but was deemed a good candidate for optimization because of the scope for introducing a variety of structural changes within the molecule. In a similar fashion to the 2,3-dihydro-1, 3-dioxo-1H-isoindole-5-carboxylic acid series, all compounds were tested for efficacy in the primary enzyme assay and secondary cell-based angiogenesis assay. A table of some selected molecules showing structures and IC50 data is shown in Figure 5. The 2-chloro substituent was found to be essential for inhibition activity and so modifications were introduced at the 4-chloro position (compound 4a). The most potent compound in that series was the 4-chlorobenzamide variant, which had an IC50 of 2.5 μM against the enzyme and IC50 of 2 μM in the angiogenesis model. Extending the amide moiety to a cinnamoyl group increased dramatically the inhibition potency towards the enzyme by a factor of 10–20-fold, but the molecules had poor bioavailability and short half-life (0.5 h in mice). As the synthetic access to the furanylthiazole acetic acids was difficult, it was decided to seek replacements for this unit. Using a technique of ‘structural morphing', it was shown that bicyclic heteroaromatic acids could be structurally overlayed onto the furanylthiazole acetic acids and so these compounds were put forward as effective surrogates. Indeed, compound 11i displayed a comparable heparanase inhibitory activity (IC50 of 0.4 μM) to that of compound 5b. The IC50 in the angiogenesis assay however was five times less potent; nevertheless, this compound was selected for further SAR analysis.
Figure 5.
Structure and efficacy of a select number of furanyl-1,3-thiazol-2-yl and benzoaxazol-5-yl acetic acid OGS heparanase inhibitors (adopted from Courtney et al., 2005).
Compounds 12d and 15b were shown to have much more favourable Absorption, Distribution, Metabolism, and Excretion (ADME) characteristics compared with the original 5b compound. Oral dosing studies in mice models (at 20 mg kg−1) showed that the plasma concentration remained high at almost 10 times the IC50 values. In addition, the compounds showed no major inhibition of human cytochrome P450 isoenzymes (IC50 >30 μM) and displayed modest cell penetration in a Caco 2 cell model of intestinal drug transport (Courtney et al., 2005). Overall, it is likely that these compounds will be useful as biological tools to dissect the function of heparanase enzyme in both normal and disease settings. Unfortunately there is no published data on the efficacy of any of the OGS small molecule inhibitors in animal studies, hence it remains to be seen whether these compounds will actually have efficacy in vivo.
Small molecule inhibitors: Imclone Systems Incorporated
A novel class of N-(4-{[4-(1H-benzoimidazol-2-yl)-arylamino]-methyl}-phenyl)-benzamides were identified as potent inhibitors of heparanase enzyme activity. The N-(4-{[5-(1H-benzoimidazol-2-yl)-pyridin-2-ylamino]-methyl}-phenyl)-3-bromo-4-methoxy-benzamide derivative had an IC50 value of 0.29 μM against the platelet enzyme. The highest plasma concentration achieved in mice was only 5.5 μM, however, this compound series were not deemed suitable for ‘proof-of-concept' evaluation in animal models (Xu et al., 2006).
A second series of heparanase inhibitors namely the 1-[4-(1H-benzoimidazol-2-yl)-phenyl]-3-[4-(1H-benzoimidazol-2-yl)-phenyl]-ureas were developed by the same company and further evaluated for their inhibition potency and suitability for in vivo animal model studies. Among them, the 1, 3-bis-[4-(1H-benzoimidazol-2-yl)-phenyl]-urea (compound 7a) displayed good heparanase inhibitory activity (IC50 of 0.27 μM). Compound 7a inhibited the proliferation of B16-BL6 melanoma cells in vitro by <50% at 100 μM. The plasma concentration of the compound was significantly improved over the first series of compounds with values of 31 μM at 1 h and 23 μM by 4 h. The inhibitor was dosed intraperitoneal (i.p.) to C57 mice at 30 mg kg−1 in a B16-BL6 melanoma tail vein model and mice killed at day 21. Results showed an inhibition of lung metastasis by approximately 50% as compared to the vehicle-treated group indicating the potential of these drugs as therapeutic agents (Pan et al., 2006).
Sugar inhibitors
PI-88 Progen Industries Limited
PI-88 is a mixture of highly sulfonated mannan oligosaccharides, consisting of predominantly penta- and tetra-sized species isolated from the yeast pichia pastoris. It is the only heparanase inhibitor to date to have reached the clinical trial testing stage and is currently being evaluated in multiple-phase II clinical trials as a monotherapy or in combination with standard chemotherapy for metastatic melanoma, non-small cell lung cancer, prostate cancer, post-resection hepatocellular carcinoma and multiple myeloma (http://www.progen.com.au/in/Aug06FactSheet.pdf). It is increasingly clear that PI-88 has a dual mode of action, inhibiting heparanase enzyme activity and interfering with the binding or action of HS-bound growth factors.
In an RIP1-Tag2 mouse model of multistage pancreatic islet, carcinogenesis PI-88 was shown to reduce the number of early progenitor lesions and impair tumour growth at later stages. Strikingly, the reduction in tumour angiogenesis correlated with a reduced association of VEGF-A with its receptor VEGF-R2 on tumour epithelium (Joyce et al., 2005). The compound also inhibits in vitro angiogenesis of human placental blood vessels (Brown et al., 1996) and tumour growth and metastases of highly invasive mammary adenocarcinoma 13762 MAT cells (Parish et al., 1999).
A family of anomerically pure, pentasaccharide analogues of PI-88 were synthesized to allow a combinatorial chemistry approach to be carried out and to facilitate structural characterization (Karoli et al., 2005). All the analogues were still able to inhibit heparanase activity (IC50, 1 μM) and to bind the proangiogenic growth factors FGF-1, FGF-2 and VEGF with high affinity. The n-octyl derivative had improved pharmacokinetic properties in that it was cleared three times mores slowly than PI-88, and hence may provide increased systemic exposure. Replacement of the terminal phosphate group of PI-88 oligosaccharide mixture with a sulpho moiety showed that only the tetra and pentasaccharide species inhibited heparanase activity (IC50, 1 μM), whereas smaller size species only partially inhibited even at saturating concentrations. In rat and rabbit models of balloon angioplasty, PI-88 inhibits vascular smooth muscle cell proliferation and reduces intimal thickening. PI-88 directly blocks the interaction of HS with FGF-2 and thus blocks the downstream ERK1/2 activation and cellular signalling cascades (Francis et al., 2003). A series of HS mimetic compounds called sulphated-linked cyclitols were shown to inhibit heparanase activity, albeit less than PI-88 and to bind with varying selectivity to a number of HS-binding proteins such as FGF-1, FGF-2, VEGF-2 and interleukin-8 (Ferro et al., 2004). It is anticipated that the study of these compounds will eventually lead to the design of heparanase inhibitory PI-88 analogues that also specifically target particular HS-binding proteins.
Heparins
Heparins, in particular the low-molecular-weight species (LMWH), have been shown to increase the survival of patients with advanced cancer (Klerk et al., 2005; Rickles, 2006). A recent, in-depth retrospective analyses review of clinical trial data over a 45-year period from 1960 to 2005 in addition to prospective clinical studies has suggested that heparins have a beneficial effect on cancer survival. Data were collated on the type and dose of heparin used, exposure time, interval between drug administration and cancer cell inoculation, and the animal tumour model used. Importantly, a distinction was also made in the analysis between heparin effects on the primary tumour or on established metastases and effects on the metastatic potential of infused cells. Results show that heparins affect the formation of metastasis rather than the growth of primary tumours due to either effects on blood coagulation, inhibition of cancer cell-platelet and -endothelial interactions by selectin inhibition or inhibition of cell invasion and angiogenesis (reviewed in Niers et al., 2006). The anti-coagulant properties of heparin however limit the extensive use of these compounds as anti-tumour agents. Chemically modified non-anti-coagulant species of heparin can be given at higher efficacious doses and are safer to use when there are concerns of bleeding complications. Glycol splitting of heparin, which involves periodate oxidation/borohydride reduction of naturally or partially 2-O-sulfated heparins, abolishes the anti-coagulant activity through a loss of anti-thrombin binding and increase of in vitro heparanase inhibitory activity (Casu et al., 2002; Naggi et al., 2005). Glycol-split heparins have a significantly reduced ability to displace bFGF from ECM, making them more specific anti-heparanase drugs.
Size-fractionated heparin oligosaccharides have been analysed for their anti-angiogenic properties in a number of growth factor-dependent angiogenesis animal models. Both octa- and deca-oligosaccharide species (all doses given at 20 mg kg−1 day−1) given to mice bearing subcutaneously (s.c.) sponges impregnated with FGF-2 reduced the micro vessel density to levels below control over a 14-day period. Octasaccharides also significantly reduced micro vessel density over a similar 2-week period in a HEC-FGF2 human endometrial cancer cells that were implanted in a hollow fibre placed s.c. in vivo and of implanted H460 lung carcinoma cells in hollow fibres over 3 weeks. Crucially, clinical levels of octasaccharides failed to show any significant in vitro anti-coagulant effects, showing the potential of these compounds as therapeutic agents. It is not yet clear however whether the ability of these glycosaminoglycans to act as anti-angiogenesis inhibitors is via an affect on heparanase inhibition in addition to them acting as HS competitor inhibitors for growth factor binding (Hasan et al., 2005).
Suramin
Suramin is a polysulphonated napthylurea that was originally developed to combat the early stages of trypanosomiasis (sleeping sickness), but more recently shown to have promising anti-tumouregenic properties in treating human malignancies. The clinical use of this drug has been hampered, however, by its severe side effects, which include neuro and renal toxicity (Stein et al., 1989). Various suramin analogues have been described that still retain the anti-tumour effect with significantly less toxicity (Firsching et al., 1995; Braddock et al., 1994). Three analogues, NF127, NF145 and NF 171, were shown to inhibit heparanase purified from brain metastatic melanoma (70W) cells with IC50 values of 20–30 μM, twice as potent as suramin itself. The compounds also inhibited completely the invasion of human 70W cells at 10 μM and of heparanase-induced angiogenesis with the most potent compound NF145 having an IC50 of being 236 μM (Marchetti et al., 2003). In addition to the anti-heparanase activity, it has also been postulated that suramin may have a dual role in binding growth factors in such a way as to block subsequent binding to growth factor receptors (Pesenti et al., 1992).
Natural product inhibitors
The metabolite (+)−trachyspic acid was isolated from yeast culture broths and both enantiomeric forms shown to inhibit heparanase with a similar IC50 value of 36 μM (Shiozawa et al., 1995). The inhibitor RK-682 (IC50, 17 μM) was also identified by screening 10 000 microbial culture broths but lacks selectivity as it also inhibits other enzymes such as phospholipase A2, HIV-1 protease and the protein phosphatases CD45 and VHR (Ishida et al., 2004). RK-682 was selected as the lead compound in a combined combinatorial chemistry and protein ligand docking analysis to try and improve on the target selectivity. Benzyl, methyl and isopropyl variants of RK-682 inhibited heparanase activity to varying degrees (IC50 of 17 μM, >100 μM, >100 μM, respectively). Encouragingly, all derivatives poorly inhibited various glycosidases and VHR phosphatase enzyme demonstrating that selectivity had been achieved. The benzyl derivative of RK-682 also inhibited migration (IC50, 3.0 μM) and invasion (IC50, 1.5 μM) of HT1080 cells (Ishida et al., 2004).
Two fungal glucuronide metabolites with a dimeric 2,4-dihydroxy-6-alkylbenzoic acid (orcinol p-depside) aglycone have shown significant heparanase inhibition with an IC50 of 10 μM (Wang et al., 2005) and inhibition of B16-F10 melanoma cell invasion. These compounds are promiscuous however in that they also inhibit bacterial heparinase and telomerase enzyme activities, and hence any future use would warrant detailed SAR studies to try to overcome this lack of target selectivity.
Neutralising antibodies
Heparanase has been shown to play a role in the development of restenosis following vascular injury by releasing bFGF and by initiating the bFGF-mediated proliferation of smooth muscle cells. A neutralizing rabbit heparanase antibody raised against the active site region G215–D234 of the 50 kDa subunit (Myler and West, 2002) was evaluated for efficacy in a rat carotid balloon injury model. Local delivery of 200 μg anti-heparanase IgG was found to inhibit bFGF release by approximately 60% at 4 days in comparison with a non-specific rabbit IgG sham control. Heparanase neutralization was also shown to reduce significantly neointima hyperplasia as judged by the thickness of the intimal cell layer at 14 days (Sham: 81±17 μM, anti-heparanase IgG: 34±9 μM) (Myler et al., 2006). The effect of neutralizing antibody on inhibiting complete bFGF release in this model system is not absolute, however potentially due to the release of bFGF via other mechanisms, for example platelet factor-4 (Myler and West, 2002). This study nevertheless indicates clearly that the strategy for targeting heparanase via an antibody directed towards the putative enzyme active site cleft may well be of therapeutic benefit in the treatment of restenosis.
A role for heparanase has also been implicated in the pathogenesis of proteinuria where it degrades the HS side chains of HSPGs within the glomerular basement membrane, and in so doing alters their perm selective properties. A rabbit peptide antibody against the heparanase 50 kDa subunit (R382–F398) was raised and tested for its ability to inhibit urinary protein secretion in an animal model of passive Heymann nephritis. A significant reduction was observed at day 5 of disease with the heparanase neutralizing antibody as compared with normal rabbit serum control (62±11 mg day−1 compared with 203±43 mg day−1, respectively) (Levidiotis et al., 2004). The efficacy of this antibody on protein excretion was also tested in a later study by the same group using a model of accelerated anti-glomerular basement disease. An increase in glomerular heparanase was shown using immunohistochemical analysis at day 10 of disease and Western-blot analysis of kidney cortex confirmed that the active 58 kDa heparanase species was restricted to the diseased kidney at this time period. Proteinuria at day 10 of disease was significantly reduced by the intravenous injection 24 h before disease induction of anti-heparanase antiserum compared to injection of normal rabbit serum control (126±25 and 216±17.8 mg day−1, respectively; Levidiotis et al., 2005). Although the precise role of heparanase in human glomerunonephritis is still unclear, these studies indicate that inhibition of the enzyme activity using a neutralizing antibody approach may be beneficial in reducing proteinuria.
In addition to the strategy of targeting specific regions of heparanase using peptide antibodies, the full-length recombinant active 58 kDa enzyme has also been used to immunize both rabbits and chickens for the generation of polyclonal antibodies. Addition of 100 μg ml−1 anti-heparanase antiserum dramatically inhibited the invasion of human ovarian cancer line OC-MZ-6 through matrigel by 95%, whereas addition of control IgG at a similar concentration had no effect (He et al., 2004). It remains to be seen whether other regions of heparanase protein such as the heparin-binding domains, active-site region around E343 and small 8 kDa subunit can also be selectively targeted by antibodies. Once the protein adhesion moieties on heparanase have been delineated fully, it will be interesting to speculate on the potential therapeutic benefit of an antibody combination cocktail consisting of enzyme neutralizing and anti-adhesion heparanase effects.
Inhibitory proteins
A common theme to currently recognized naturally occurring endogenous protein inhibitors of heparanase-mediated HS digestion is that they are all highly basic proteins, which are presumed to bind to negatively charged HS via electrostatic interactions. So far three such proteins have been identified and these are described below.
Histidine-rich glycoprotein
Histidine-rich glycoprotein (HRG) is a relatively abundant 75 kDa plasma glycoprotein (1–2 μM in humans) that binds to and masks the heparanase cleavage site on HS. It has also been proposed to prevent the heparanase-induced release of FGF from the ECM and so act as an inhibitor to the pro-angiogenic activity of heparanase (Freeman and Parish, 1997). Studies using pgsA-745 Chinese hamster ovary cell lines which are deficient in cell surface glycosaminoglycans and with cells pre-treated with mammalian heparanase have shown that HRG binds predominantly to cell surface HS specifically and that this interaction occurs via the N-terminal domain of HRG (Jones et al., 2004).
Eosinophil major basic protein
A natural heparanase inhibitor protein, major basic protein (MBP), has been identified in activated eosinophils. MBP is a 14 kDa (117-amino acid residues) highly basic protein, whose complement of basic residues consists entirely of arginine residues. Eosinophil lysates and MBP were shown to completely inhibit recombinant heparanase activity in a concentration-dependent manner at 0.2 μM. Two other eosinophil proteins, peroxidase and eosinophil cationic proteins, also inhibited partially heparanase activity at similar concentrations. Poly-L-arginine alone inhibited only at very high concentrations, however, suggesting that the inhibition of heparanase by MBP is structurally specific and does not solely depend on the presence of any highly positively charged protein. It is proposed that active secretion of a heparanase inhibitor protein by eosinophils may confer a degree of protection to these cells, and thus help to hinder the progression of inflammation and cancer (Temkin et al., 2004).
HS-interacting protein
Heparanase and a synthetic peptide of HS-interacting protein (HIP/RPL29) recognize common sites on cell surface and ECM HS. Incubation of heparanase-containing melanoma cellular extracts or partially purified heparanase preparations with cell surface- or ECM-derived HS and HIP peptide, but not a scrambled sequence of this peptide or other HS-binding proteins present in ECM, inhibited heparanase action completely (Marchetti et al., 1997).
LMWH endogenous inhibitor
An LMWH (2–10 kDa), heat stable, natural endogenous inhibitor of heparanase has also been identified in butanol extracted B16-F10 cells, although the nature of this species remains uncharacterized (Keren and LeGrue, 1989).
The analysis of these natural inhibitors and their regulation warrants further investigation as they may well provide a mechanism by which cells can rapidly modulate heparanase activity. In addition, it will be important to study whether the levels of these natural inhibitors are altered in malignancies, for example overexpression of one of the above HS-binding proteins or downregulation of the non-proteinaceous LMWH inhibitor could in effect produce a local increase in heparanase activity by removing the natural regulator molecule.
Surrogate markers: efficacy of heparanase inhibition
The development of selective markers of drug efficacy or biomarkers during the drug discovery process is vital for making preclinical evaluations through each phase of clinical trials. Biomarkers predict a patient's response to a particular compound and act as an effective surrogate end point. With the potential of an increasing number of heparanase inhibitors entering the clinic, it is increasingly important to develop minimally invasive tests to assess drug efficacy. Below is a compilation of a number of strategies that could be further developed into a diagnostic product for use in heparanase inhibitor clinical trials.
Serum VEGF levels
Heparanase cleavage of the ECM HS chain attached to HSPGs liberates a number of growth factors. In a recent study of over 114 patients with cutaneous melanoma, the serum levels of VEGFR1, TGFβ1 and VEGF were found to be significantly higher in patients compared with controls. Positive correlations were also found with advanced stages of disease (Tas et al., 2006). Two of the OGS heparanase inhibitors (OGT 2115 and OGT 2492) have been evaluated in an MDA435 xenograft tumour model to study their effect on serum VEGF levels (Figure 6). Drug OGT 2492 had the most dramatic effect with an approximate 50% reduction, whereas OGT 2115 reduced serum VEGF levels by a more modest 20%. Whilst this data suggests that monitoring serum VEGF levels may be a useful indicator of heparanase drug efficacy, this test is unlikely to be specific for the target as a number of other enzymes in particular MMPs are known to have an effect on serum VEGF levels. In this context, the MMP inhibitor batimastat (BB-94) inhibited the MMP9- and MMP2-induced release of VEGF in SKOV3 and OVCAR3 ovarian carcinoma cells in a dose- and time-dependent manner (Belotti et al., 2003).
Figure 6.
Animals were dosed with OGT compounds, OGT 2492 and OGT 2115, at 25 mg Kg−1 by i.p. injection (mean of four animals) and serum samples analysed by VEGF ELISA at 24 days (adopted from Courtney et al., 2004).
Cellular heparan sulphate levels
Monoclonal anti-HS antibodies have proved to be useful tools to evaluate the presence or absence of HS in various cell types. High levels of cellular heparanase activity have been shown to correlate with a reduction of HS staining using the commercially available Seigagaku 10E4 antibody, which specifically binds to mixed HS domains containing both N-acetylated and N-sulphated disaccharide units (Van den Born et al., 2005). Two colorectal cell lines, HT29 and Lovo, were assessed for their heparanase mRNA status by quantitative analysis (Figure 7a) and shown to have low and high levels, respectively, of heparanase mRNA. Subsequent immunofluorescence staining of these cell types using the anti-HS antibody 10E4 confirmed that high levels of heparanase enzyme observed in the Lovo cell line corresponds to poor staining with the HS antibody, whereas the opposite is observed for the low enzyme expressing HT29 cell line (Figure 7b).
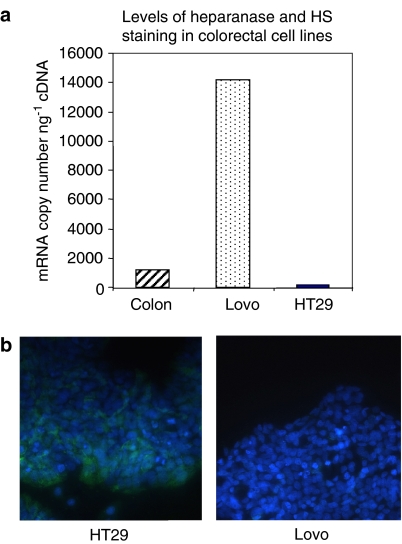
Figure 7.
(a) Quantitative profiling of the level of heparanase mRNA in HT29 and Lovo colorectal cell lines. Normal colon mRNA levels were measured as a reference standard (adopted from McKenzie et al., 2003). (b) Immunofluorescence staining of HT29 and Lovo cell lines with anti-HS antibody 10E4 (shown in green), nuclear staining is shown in blue.
A more quantitative study was undertaken at OGS to investigate whether this inverse relationship between heparanase enzyme levels and HS staining was observed over a range of measured heparanase enzyme concentrations (McKenzie et al., 2003). MDA-435 cells in culture were incubated with increasing amounts of pro-HPSE1 enzyme (from 0 to 200 ng ml−1) and left for 24 h to internalize the enzyme and convert it intracellularly into the active heterodimer enzyme. Cells were permeabilized, incubated with anti-HPSE1 antibody or HS antibody and measured by fluorescence-activated cell sorter analysis (Figure 8a). Quantification of the fluorescence (Figure 8b) showed an almost linear inverse correlation demonstrating the potential of using cell HS staining as a prognostic marker of heparanase enzyme activity. An inverse relationship between heparanase levels and HS staining has also been described previously using histochemical analysis of paraffin-embedded thyroid tumour sections (Xu et al., 2003).
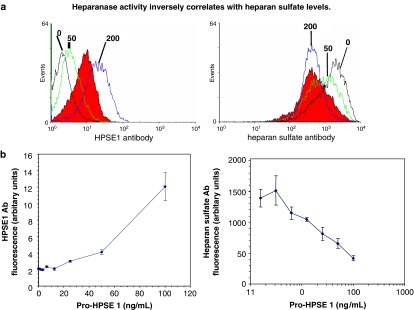
Figure 8.
(a) Recombinant pro-heparanase protein (0–200 ng ml−1) was added to MDA435 cells and left for 24 h to internalize protein fully. Permeabilized cells were incubated with HPSE1 antibody or anti-HS antibody (10E4) and measured by FACS. The ovarian cancer line OV90, a known high heparanase expresser, is shown in red as a reference standard (taken from McKenzie et al., 2003). (b) Quantification of the levels of HS and heparanase staining at each dose of exogenously added pro-heparanase protein (taken from McKenzie et al., 2003).
As many immune cells (Vlodavsky et al., 1992) have been shown to contain high levels of heparanase enzyme, it is possible that a simple FACS-based blood test, such as the one just described, could be employed to monitor heparanase levels and in particular heparanase inhibition during a patient–drug treatment programme. The progress of anti-HS antibody development also lends itself to the exciting possibility of producing a HS antibody specific to the HS ‘stub' produced upon heparanase cleavage.
Urinary levels of heparanase protein
A highly sensitive enzyme-linked immunosorbent assay (ELISA) method for quantitating the level of heparanase as low as 200 pg ml−1 has been developed. A study of urine samples from normal individuals fell in the range of 69±20 pg ml−1 heparanase, whereas those from primary transitional bladder cell carcinoma patients (TCC) showed a statistically significant increase to 286±100 pg ml−1. Interestingly, monitoring of heparanase levels in TCC patients following surgical resection of the primary tumour showed a significant reduction in urinary heparanase protein levels down to 107±23 pg ml−1 which is consistent with the tumour tissue being the original source of the excreted protein. A similar study on urine from leukaemia patients showed a statistically significant increase up to 345±97 pg ml−1 (Shafat et al., 2006a, 2006b). The ELISA assay could thus serve as a diagnostic and prognostic marker for bladder cancer and may be useful in monitoring any effect on tumour growth shrinkage following treatment with heparanase inhibitors.
Conclusions
Heparanase has an important housekeeping role in the metabolic turnover of HS chains in normal cells. A wide number of studies demonstrate that the enzyme is upregulated in a number of malignant diseases and that this overexpression correlates directly with an increase in processes such as cellular invasion, angiogenesis and metastasis. Increasingly, a substantial body of literature has supported the potential of heparanase as a viable therapeutic target and this has accelerated the number of selective drug development strategies. These have taken the form of small molecule, sugar, natural product inhibitors and of neutralising antibodies which have all shown promising efficacy at the pre-clinical stage. The compound PI88 is the only heparanase inhibitor to date to reach human clinical trials testing. Classically, the main target of heparanase inhibitors has been aimed at the enzymatic function of the protein; however, recent studies have indicated the future potential of targeting the pro-adhesion activity and inhibiting the proteases that cleave the linker region separating the 8 and 50 kDa subunits. Increasingly, there is a need to develop surrogate markers for in vivo drug efficacy to keep up with the variety of heparanase inhibitors emerging from academic and industrial pre-clinical research. The most promising strategies may involve a combination analysis of serum growth factors, excreted heparanase and antibody staining of cellular HS. Heparanase represents an excellent therapeutic target and the exciting challenge ahead will be to see how the encouraging slew of drugs performs in more advanced animal model studies.
Acknowledgments
I would like to thank Professor Israel Vlodavsky and Professor John Gallagher for critical reading of this article and for their continued support. Thank you also to Dr Philip Woodman and Dr Rebecca Tyler for proof reading and to Alex McKenzie for helping produce the figures.
Abbreviations
- bFGF
basic fibroblast growth factor
- ECM
extracellular matrix
- GPCR
G-protein coupled receptor
- HGF
hepatocyte growth factor
- HS
heparan sulphate
- HSPGs
heparan sulphate proteoglycans
- KGF
keratin growth factor
- OGS
Oxford GlycoSciences
- SAR
structure–activity relationship
- TGFβ
transforming growth factor β
- VEGF
vascular endothelial growth factor
Conflict of interest
The author states no conflict of interest.
References
- Abboud-Jarrous G, Rangini-Guetta Z, Aingorn H, Atzmon R, Elgavish S, Peretz T, et al. Site-directed mutagenesis, proteolytic cleavage, and activation of human proheparanase. J Biol Chem. 2005;280:13568–13575. doi: 10.1074/jbc.M413370200. [DOI] [PubMed] [Google Scholar]
- Baker E, Crawford J, Sutherland GR, Freeman C, Parish CR, Hulett MD. Human HPA endoglycosidase heparanase Map position 4q21.3. Chromosome Res. 1999;7:319. doi: 10.1023/a:1009235132576. [DOI] [PubMed] [Google Scholar]
- Baraz L, Haupt Y, Elkin M, Peretz T, Vlodavsky I. Tumor suppressor p53 regulates heparanase gene expression. Oncogene. 2006;25:3939–3947. doi: 10.1038/sj.onc.1209425. [DOI] [PubMed] [Google Scholar]
- Beckhove P, Helmke BM, Ziouta Y, Bucur M, Dorner W, Mogler C, et al. Heparanase expression at the invasion front of human head and neck cancers and correlation with poor prognosis. Clin Cancer Res. 2005;11:2899–2906. doi: 10.1158/1078-0432.CCR-04-0664. [DOI] [PubMed] [Google Scholar]
- Belotti D, Paganoni P, Manenti L, Garofalo A, Marchini S, Taraboletti G, et al. Matrix metalloproteinases (MMP9 and MMP2) induce the release of vascular endothelial growth factor (VEGF) by ovarian carcinoma cells: implications for ascites formation. Cancer Res. 2003;63:5224–5229. [PubMed] [Google Scholar]
- Benhamron S, Nechushtan H, Verbovetski I, Krispin A, Abboud-Jarrous G, Zcharia E, et al. Translocation of active heparanase to cell surface regulates degradation of extracellular matrix heparan sulfate upon transmigration of mature monocyte-derived dendritic cells. J Immunol. 2006;176:6417–6424. doi: 10.4049/jimmunol.176.11.6417. [DOI] [PubMed] [Google Scholar]
- Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Braddock PS, Hu DE, Fan TP, Stratford IJ, Harris AL, Bicknell R. A structure–activity analysis of antagonism of the growth factor and angiogenic activity of basic fibroblast growth factor by suramin and related polyanions. Br J Cancer. 1994;69:890–898. doi: 10.1038/bjc.1994.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KJ, Maynes SF, Bezos A, Maguire DJ, Ford MD, Parish CR. A novel in vitro assay for human angiogenesis. Lab Invest. 1996;75:539–555. [PubMed] [Google Scholar]
- Castor CW, Kotlyar A, Edwards BE. Connective tissue activation XXXVIII: heparin/heparanase activity of human platelets resides in a high molecular weight protein, not in connective tissue activating peptide III. J Rheumatol. 2002;29:2337–2344. [PubMed] [Google Scholar]
- Casu B, Guerrini M, Naggi A, Perez M, Torri G, Ribatti D, et al. Short heparin sequences spaced by glycol-split uronate residues are antagonists of fibroblast growth factor 2 and angiogenesis inhibitors. Biochemistry. 2002;41:10519–10528. doi: 10.1021/bi020118n. [DOI] [PubMed] [Google Scholar]
- Chen G, Wang D, Vikramadithyan R, Yagyu H, Saxena U, Pillarisetti S, et al. Inflammatory cytokines and fatty acids regulate endothelial cell heparanase expression. Biochemistry. 2004;43:4971–4977. doi: 10.1021/bi0356552. [DOI] [PubMed] [Google Scholar]
- Cohen E, Atzmon R, Vlodavsky I, Ilan N. Heparanase processing by lysosomal/endosomal protein preparation. FEBS Lett. 2005;579:2334–2338. doi: 10.1016/j.febslet.2005.03.030. [DOI] [PubMed] [Google Scholar]
- Courtney SM, Hay PA, Buck RT, Colville CS, Phillips DJ, Scopes DI, et al. Furanyl-1,3-thiazol-2-yl and benzoxazol-5-yl acetic acid derivatives: novel classes of heparanase inhibitor. Bioorg Med Chem Lett. 2005;15:2295–2299. doi: 10.1016/j.bmcl.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Courtney SM, Hay PA, Buck RT, Colville CS, Porter DW, Scopes DI, et al. 2,3-Dihydro-1,3-dioxo-1H-isoindole-5-carboxylic acid derivatives: a novel class of small molecule heparanase inhibitors. Bioorg Med Chem Lett. 2004;14:3269–3273. doi: 10.1016/j.bmcl.2004.03.086. [DOI] [PubMed] [Google Scholar]
- De Mestre AM, Khachigian LM, Santiago FS, Staykova MA, Hulett MD. Regulation of inducible heparanase gene transcription in activated T cells by early growth response 1. J Biol Chem. 2003;278:50377–50385. doi: 10.1074/jbc.M310154200. [DOI] [PubMed] [Google Scholar]
- De Mestre AM, Rao S, Hornby JR, Soe-Htwe T, Khachigian LM, Hulett MD. Early growth response gene 1 (EGR1) regulates heparanase gene transcription in tumor cells. J Biol Chem. 2005;280:35136–35147. doi: 10.1074/jbc.M503414200. [DOI] [PubMed] [Google Scholar]
- Dempsey LA, Plummer TB, Coombes SL, Platt JL. Heparanase expression in invasive trophoblasts and acute vascular damage. Glycobiology. 2000;10:467–475. doi: 10.1093/glycob/10.5.467. [DOI] [PubMed] [Google Scholar]
- Dong J, Kukula AK, Toyoshima M, Nakajima M. Genomic organization and chromosome localization of the newly identified human heparanase gene. Gene. 2000;253:171–178. doi: 10.1016/s0378-1119(00)00251-1. [DOI] [PubMed] [Google Scholar]
- Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- Edovitsky E, Lerner I, Zcharia E, Peretz T, Vlodavsky I, Elkin M. Role of endothelial heparanase in delayed-type hypersensitivity. Blood. 2006;107:3609–3616. doi: 10.1182/blood-2005-08-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkin M, Cohen I, Zcharia E, Orgel A, Guatta-Rangini Z, Peretz T, et al. Regulation of heparanase gene expression by estrogen in breast cancer. Cancer Res. 2003;63:8821–8826. [PubMed] [Google Scholar]
- Fairbanks MB, Mildner AM, Leone JW, Cavey GS, Mathews WR, Drong RF, et al. Processing of the human heparanase precursor and evidence that the active enzyme is a heterodimer. J Biol Chem. 1999;274:29587–29590. doi: 10.1074/jbc.274.42.29587. [DOI] [PubMed] [Google Scholar]
- Ferro V, Hammond E, Fairweather JK. The development of inhibitors of heparanase, a key enzyme involved in tumour metastasis, angiogenesis and inflammation. Mini Rev Med Chem. 2004;4:693–702. doi: 10.2174/1389557043403729. [DOI] [PubMed] [Google Scholar]
- Firsching A, Nickel P, Mora P, Allolio B. Antiproliferative and angiostatic activity of suramin analogues. Cancer Res. 1995;55:4957–4961. [PubMed] [Google Scholar]
- Fitzgerald M, Hayward MP, Thomas AC, Campbell GR, Campell JH. Matrix metalloproteinase can facilitate the heparanase-induced promotion of phenotype change in vascular smooth muscle cells. Atherosclerosis. 1999;145:97–106. doi: 10.1016/s0021-9150(99)00019-2. [DOI] [PubMed] [Google Scholar]
- Francis DJ, Parish CR, McGarry M, Santiago FS, Lowe HC, Brown KJ, et al. Blockade of vascular smooth muscle cell proliferation and intimal thickening after balloon injury by the sulfated oligosaccharide PI-88: phosphomannopentaose sulfate directly binds FGF-2, blocks cellular signaling, and inhibits proliferation. Circ Res. 2003;92:70–77. doi: 10.1161/01.RES.0000071345.76095.07. [DOI] [PubMed] [Google Scholar]
- Freeman C, Browne AM, Parish CR. Evidence that platelet and tumour heparanases are similar enzymes. Biochem J. 1999;342 Part 2:361–368. [PMC free article] [PubMed] [Google Scholar]
- Freeman C, Parish CR. A rapid quantitative assay for the detection of mammalian heparanase activity. Biochem J. 1997;325 Part 1:229–237. doi: 10.1042/bj3250229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman C, Parish CR. Human platelet heparanase: purification, characterization and catalytic activity. Biochem J. 1998;330 Part 3:1341–1350. doi: 10.1042/bj3301341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann Y, Vlodavsky I, Aingorn H, Aviv A, Peretz T, Pecker I, et al. Expression of heparanase in normal, dysplastic, and neoplastic human colonic mucosa and stroma. Evidence for its role in colonic tumorigenesis. Am J Pathol. 2000;157:1167–1175. doi: 10.1016/S0002-9440(10)64632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingis-Velitski S, Zetser A, Kaplan V, Ben-Zaken O, Cohen E, Levy-Adam F, et al. Heparanase uptake is mediated by cell membrane heparan sulfate proteoglycans. J Biol Chem. 2004;279:44084–44092. doi: 10.1074/jbc.M402131200. [DOI] [PubMed] [Google Scholar]
- Gohji K, Okamoto M, Kitazawa S, Toyoshima M, Dong J, Katsuoka Y, et al. Heparanase protein and gene expression in bladder cancer. J Urol. 2001;166:1286–1290. [PubMed] [Google Scholar]
- Goldshmidt O, Nadav L, Aingorn H, Irit C, Feinstein N, Ilan N, et al. Human heparanase is localized within lysosomes in a stable form. Exp Cell Res. 2002a;281:50–62. doi: 10.1006/excr.2002.5651. [DOI] [PubMed] [Google Scholar]
- Goldshmidt O, Zcharia E, Abramovitch R, Metzger S, Aingorn H, Friedmann Y, et al. Cell surface expression and secretion of heparanase markedly promote tumor angiogenesis and metastasis. Proc Natl Acad Sci USA. 2002b;99:10031–10036. doi: 10.1073/pnas.152070599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmidt O, Zcharia E, Aingorn H, Guatta-Rangini Z, Atzmon R, Michal I, et al. Expression pattern and secretion of human and chicken heparanase are determined by their signal peptide sequence. J Biol Chem. 2001;276:29178–29187. doi: 10.1074/jbc.M102462200. [DOI] [PubMed] [Google Scholar]
- Goldshmidt O, Zcharia E, Cohen M, Aingorn H, Cohen I, Nadav L, et al. Heparanase mediates cell adhesion independent of its enzymatic activity. FASEB J. 2003;17:1015–1025. doi: 10.1096/fj.02-0773com. [DOI] [PubMed] [Google Scholar]
- Hasan J, Shnyder SD, Clamp AR, McGown AT, Bicknell R, Presta M, et al. Heparin octasaccharides inhibit angiogenesis in vivo. Clin Cancer Res. 2005;11:8172–8179. doi: 10.1158/1078-0432.CCR-05-0452. [DOI] [PubMed] [Google Scholar]
- He X, Brenchley PE, Jayson GC, Hampson L, Davies J, Hampson IN. Hypoxia increases heparanase-dependent tumor cell invasion, which can be inhibited by antiheparanase antibodies. Cancer Res. 2004;64:3928–3933. doi: 10.1158/0008-5472.CAN-03-2718. [DOI] [PubMed] [Google Scholar]
- Hoogewerf AJ, Leone JW, Reardon IM, Howe WJ, Asa D, Heinrikson RL, et al. CXC chemokines connective tissue activating peptide-III and neutrophil activating peptide-2 are heparin/heparan sulfate-degrading enzymes. J Biol Chem. 1995;270:3268–3277. doi: 10.1074/jbc.270.7.3268. [DOI] [PubMed] [Google Scholar]
- Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- Hulett MD, Hornby JR, Ohms SJ, Zuegg J, Freeman C, Gready JE, et al. Identification of active-site residues of the pro-metastatic endoglycosidase heparanase. Biochemistry. 2000;39:15659–15667. doi: 10.1021/bi002080p. [DOI] [PubMed] [Google Scholar]
- Ikuta M, Podyma KA, Maruyama K, Enomoto S, Yanagishita M. Expression of heparanase in oral cancer cell lines and oral cancer tissues. Oral Oncol. 2001;37:177–184. doi: 10.1016/s1368-8375(00)00077-4. [DOI] [PubMed] [Google Scholar]
- Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Ishida K, Hirai G, Murakami K, Teruya T, Simizu S, Sodeoka M, et al. Structure-based design of a selective heparanase inhibitor as an antimetastatic agent. Mol Cancer Ther. 2004;3:1069–1077. [PubMed] [Google Scholar]
- Jiang P, Kumar A, Parrillo JE, Dempsey LA, Platt JL, Prinz RA, et al. Cloning and characterization of the human heparanase-1 (HPR1) gene promoter: role of GA-binding protein and Sp1 in regulating HPR1 basal promoter activity. J Biol Chem. 2002;277:8989–8998. doi: 10.1074/jbc.M105682200. [DOI] [PubMed] [Google Scholar]
- Jones AL, Hulett MD, Parish CR. Histidine-rich glycoprotein binds to cell-surface heparan sulfate via its N-terminal domain following Zn2+ chelation. J Biol Chem. 2004;279:30114–30122. doi: 10.1074/jbc.M401996200. [DOI] [PubMed] [Google Scholar]
- Joyce JA, Freeman C, Meyer-Morse N, Parish CR, Hahahan D. A functional heparan sulphate mimetic implicates both heparanase and heparan sulphate in tumor angiogenesis in a mouse model of multistage cancer. Oncogene. 2005;24:4037–4051. doi: 10.1038/sj.onc.1208602. [DOI] [PubMed] [Google Scholar]
- Kahner BN, Shankar H, Murugappan S, Prasad GL, Kunapuli SP. Nucleotide receptor signalling in platelets. J Thromb Haemost. 2006;4:2317–2326. doi: 10.1111/j.1538-7836.2006.02192.x. [DOI] [PubMed] [Google Scholar]
- Karoli T, Liu L, Fairweather JK, Hammond E, Li CP, Cochran S, et al. Synthesis, biological activity, and preliminary pharmacokinetic evaluation of analogues of a phosphosulfomannan angiogenesis inhibitor (PI-88) J Med Chem. 2005;48:8229–8236. doi: 10.1021/jm050618p. [DOI] [PubMed] [Google Scholar]
- Keren Z, LeGrue SJ. Low-molecular-weight membrane component inhibits the metastatic phenotype of B16-F10 melanoma. Clin Exp Metastasis. 1989;7:315–328. doi: 10.1007/BF01753683. [DOI] [PubMed] [Google Scholar]
- Kim AW, Xu X, Hollinger EF, Gattuso P, Godellas CV, Prinz RA. Human heparanase-1 gene expression in pancreatic adenocarcinoma. J Gastrointest Surg. 2002;6:167–172. doi: 10.1016/s1091-255x(01)00087-7. [DOI] [PubMed] [Google Scholar]
- Kizaki K, Okada M, Ito R, Yoshioka K, Hashizume K, Mutoh K, et al. Induction of heparanase gene expression in ventricular myocardium of rats with isoproterenol-induced cardiac hypertrophy. Biol Pharm Bull. 2005;28:2331–2334. doi: 10.1248/bpb.28.2331. [DOI] [PubMed] [Google Scholar]
- Klerk CP, Smorenburg SM, Otten HM, Lensing AW, Prins MH, Piovella F, et al. The effect of low molecular weight heparin on survival in patients with advanced malignancy. J Clin Oncol. 2005;23:2130–2135. doi: 10.1200/JCO.2005.03.134. [DOI] [PubMed] [Google Scholar]
- Koliopanos A, Friess H, Kleeff J, Shi X, Liao Q, Pecker I, et al. Heparanase expression in primary and metastatic pancreatic cancer. Cancer Res. 2001;61:4655–4659. [PubMed] [Google Scholar]
- Kreuger J, Spilmann D, Li JP, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie PH, Hulmes JD, Ludwig DL, Patel S, Navarro EC, Seddon AP, et al. Cloning and functional expression of a human heparanase gene. Biochem Biophys Res Commun. 1999;261:183–187. doi: 10.1006/bbrc.1999.0962. [DOI] [PubMed] [Google Scholar]
- Levidiotis V, Freeman C, Punler M, Martinello P, Creese B, Ferro V, et al. A synthetic heparanase inhibitor reduces proteinuria in passive Heymann nephritis. J Am Soc Nephrol. 2004;15:2882–2892. doi: 10.1097/01.ASN.0000142426.55612.6D. [DOI] [PubMed] [Google Scholar]
- Levidiotis V, Freeman C, Tikellis C, Cooper ME, Power DA. Heparanase inhibition reduces proteinuria in a model of accelerated anti-glomerular basement membrane antibody disease. Nephrology (Carlton) 2005;10:167–173. doi: 10.1111/j.1440-1797.2005.00388.x. [DOI] [PubMed] [Google Scholar]
- Levy-Adam F, Miao HQ, Heinrikson RL, Vlodavsky I, Ilan N. Heterodimer formation is essential for heparanase enzymatic activity. Biochem Biophys Res Commun. 2003;308:885–891. doi: 10.1016/s0006-291x(03)01478-5. [DOI] [PubMed] [Google Scholar]
- Li S, Huang S, Peng SB. Overexpression of G protein-coupled receptor in cancer cells: involvement in tumor progression. Int J Oncol. 2005;27:1329–1339. [PubMed] [Google Scholar]
- Lyon M, Gallagher JT. Bio-specific sequences and domains in heparan sulphate and the regulation of cell growth and adhesion. Matrix Biol. 1998;17:485–493. doi: 10.1016/s0945-053x(98)90096-8. [DOI] [PubMed] [Google Scholar]
- Marchetti D, Li J, Shen R. Astrocytes contribute to the brain-metastatic specificity of melanoma cells by producing heparanase. Cancer Res. 2000;60:4767–4770. [PubMed] [Google Scholar]
- Marchetti D, Liu S, Spohn WC, Carson DD. Heparanase and a synthetic peptide of heparan sulfate-interacting protein recognize common sites on cell surface and extracellular matrix heparan sulfate. J Biol Chem. 1997;272:15891–15897. doi: 10.1074/jbc.272.25.15891. [DOI] [PubMed] [Google Scholar]
- Marchetti D, Nicolson GL. Human heparanase: a molecular determinant of brain metastasis. Adv Enzyme Regul. 2001;41:343–359. doi: 10.1016/s0065-2571(00)00016-9. [DOI] [PubMed] [Google Scholar]
- Marchetti D, Reiland J, Erwin B, Roy M. Inhibition of heparanase activity and heparanase-induced angiogenesis by suramin analogues. Int J Cancer. 2003;104:167–174. doi: 10.1002/ijc.10930. [DOI] [PubMed] [Google Scholar]
- Maxhimer JB, Quiros RM, Stewart R, Dowlatshahi K, Gattuso P, Fan M, et al. Heparanase-1 expression is associated with the metastatic potential of breast cancer. Surgery. 2002;132:326–333. doi: 10.1067/msy.2002.125719. [DOI] [PubMed] [Google Scholar]
- McKenzie E, Young K, Hircock M, Bennett J, Bhaman M, Felix R, et al. Biochemical characterization of the active heterodimer form of human heparanase (Hpa1) protein expressed in insect cells. Biochem J. 2003;373:423–435. doi: 10.1042/BJ20030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry CL, Bullock SL, Swan DC, Backen AC, Lyon M, Beddington RS, et al. The molecular phenotype of heparan sulfate in the Hs2st−/− mutant mouse. J Biol Chem. 2001;276:35429–35434. doi: 10.1074/jbc.M100379200. [DOI] [PubMed] [Google Scholar]
- Mikami S, Ohashi K, Usui Y, Nemoto T, Katsube K, Yanagishita M, et al. Loss of syndecan-1 and increased expression of heparanase in invasive esophageal carcinomas. Jpn J Cancer Res. 2001;92:1062–1073. doi: 10.1111/j.1349-7006.2001.tb01061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KJ, Merry CL, Lyon M, Thompson JE, Roberts IS, Gallagher JT. A new model for the domain structure of heparan sulfate based on the novel specificity of K5 lyase. J Biol Chem. 2004;279:27239–27245. doi: 10.1074/jbc.M401774200. [DOI] [PubMed] [Google Scholar]
- Myler HA, Lipke EA, Rice EE, West JL. Novel heparanase-inhibiting antibody reduces neointima formation. J Biochem (Tokyo) 2006;139:339–345. doi: 10.1093/jb/mvj061. [DOI] [PubMed] [Google Scholar]
- Myler HA, West JL. Heparanase and platelet factor-4 induce smooth muscle cell proliferation and migration via bFGF release from the ECM. J Biochem (Tokyo) 2002;131:913–922. doi: 10.1093/oxfordjournals.jbchem.a003182. [DOI] [PubMed] [Google Scholar]
- Nadav L, Eldor A, Yacoby-Zeevi O, Zamir E, Pecker I, Ilan N, et al. Activation, processing and trafficking of extracellular heparanase by primary human fibroblasts. J Cell Sci. 2002;115:2179–2187. doi: 10.1242/jcs.115.10.2179. [DOI] [PubMed] [Google Scholar]
- Naggi A, Casu B, Perez M, Torri G, Cassinelli G, Penco S, et al. Modulation of the heparanase-inhibiting activity of heparin through selective desulfation, graded N-acetylation, and glycol splitting. J Biol Chem. 2005;280:12103–12113. doi: 10.1074/jbc.M414217200. [DOI] [PubMed] [Google Scholar]
- Naomoto Y, Gunduz M, Takaoka M, Okawa T, Gunduz E, Nobuhisa T, et al. Heparanase promotes angiogenesis through Cox-2 and HIF1α. Med Hypotheses. 2007;68:162–165. doi: 10.1016/j.mehy.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Niers TM, Klerk CP, Dinisio M, Van Noorden CJ, Buller HR, Reitsma PH, et al. Mechanisms of heparin induced anti-cancer activity in experimental cancer models. Crit Rev Oncol Hematol. 2006;60:1–88. doi: 10.1016/j.critrevonc.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Ogishima T, Shiina H, Breault JE, Tabatabai L, Bassett WW, Enokida H, et al. Increased heparanase expression is caused by promoter hypomethylation and up-regulation of transcriptional factor early growth response-1 in human prostate cancer. Clin Cancer Res. 2005a;11:1028–1036. [PubMed] [Google Scholar]
- Ogishima T, Shiina H, Breault JE, Terashima M, Honda S, Enokida H, et al. Promoter CpG hypomethylation and transcription factor EGR1 hyperactivate heparanase expression in bladder cancer. Oncogene. 2005b;24:6765–6772. doi: 10.1038/sj.onc.1208811. [DOI] [PubMed] [Google Scholar]
- Ohtawa Y, Naomoto Y, Shirakawa Y, Takaoka M, Murata T, Sonoda R, et al. The close relationship between heparanase and cyclooxygenase-2 expressions in signet-ring cell carcinoma of the stomach. Hum Pathol. 2006;37:1145–1152. doi: 10.1016/j.humpath.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Oosta GM, Favreau LV, Beeler DL, Rosenberg RD. Purification and properties of human platelet heparitinase. J Biol Chem. 1982;257:11249–11255. [PubMed] [Google Scholar]
- Pan W, Miao HQ, Xu YJ, Navarro EC, Tonra JR, Corcoran E, et al. 1-[4-(1H-benzoimidazol-2-yl)-phenyl]-3-[4-(1H-benzoimidazol-2-yl)-phenyl]- urea derivatives as small molecule heparanase inhibitors. Bioorg Med Chem Lett. 2006;16:409–412. doi: 10.1016/j.bmcl.2005.09.069. [DOI] [PubMed] [Google Scholar]
- Parish CR, Freeman C, Brown KJ, Francis DJ, Cowden WB. Identification of sulfated oligosaccharide-based inhibitors of tumor growth and metastasis using novel in vitro assays for angiogenesis and heparanase activity. Cancer Res. 1999;59:3433–3441. [PubMed] [Google Scholar]
- Parish CR, Freeman C, Hulett MD. Heparanase a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–M108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- Pesenti E, Sola F, Mongelli N, Grandi M, Spreafico F. Suramin prevents neovascularisation and tumour growth through blocking of basic fibroblast growth factor activity. Br J Cancer. 1992;66:367–372. doi: 10.1038/bjc.1992.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechter M, Lider O, Cahalon L, Baharav E, Dekel M, Seigel D, et al. A cellulose-binding domain-fused recombinant human T cell connective tissue-activating peptide-III manifests heparanase activity. Biochem Biophys Res Commun. 1999;255:657–662. doi: 10.1006/bbrc.1999.0181. [DOI] [PubMed] [Google Scholar]
- Reiland J, Kempf D, Roy M, Denkins Y, Marchetti D. FGF2 binding, signalling and angiogenesis are modulated by heparanase in metastatic melanoma cells. Neoplasia. 2006;8:596–606. doi: 10.1593/neo.06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickles FR. If heparanase is the answer, what is the question. J Thromb Haemost. 2006;4:557–559. doi: 10.1111/j.1538-7836.2006.01828.x. [DOI] [PubMed] [Google Scholar]
- Rohloff J, Zinke J, Schoppmeyer K, Tannapfel A, Witzigmann H, Mossner J, et al. Heparanase expression is a prognostic indicator for postoperative survival in pancreatic adenocarcinoma. Br J Cancer. 2002;86:1270–1275. doi: 10.1038/sj.bjc.6600232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson RD, Yang Y, Suva LJ, Kelly T. Heparan sulfate proteoglycans and heparanase – partners in osteolytic tumor growth and metastasis. Matrix Biol. 2004;23:341–352. doi: 10.1016/j.matbio.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Sasaki N, Higashi N, Taka T, Nakajima M, Irimura T. Cell surface localization of heparanase on macrophages regulates degradation of extracellular matrix heparan sulfate. J Immunol. 2004;172:3830–3835. doi: 10.4049/jimmunol.172.6.3830. [DOI] [PubMed] [Google Scholar]
- Sato T, Yamaguchi A, Goi T, Hirono Y, Takeuchi K, Katayama K, et al. Heparanase expression in human colorectal cancer and its relationship to tumor angiogenesis, hematogenous metastasis, and prognosis. J Surg Oncol. 2004;87:174–181. doi: 10.1002/jso.20097. [DOI] [PubMed] [Google Scholar]
- Schubert SY, Ilan N, Shushy M, Ben-Izhak O, Vlodavsky I, Goldshmidt O. Human heparanase nuclear localization and enzymatic activity. Lab Invest. 2004;84:535–544. doi: 10.1038/labinvest.3700084. [DOI] [PubMed] [Google Scholar]
- Shafat I, Vlodavsky I, Ilan N. Characterization of mechanisms involved in secretion of active heparanase. J Biol Chem. 2006a;281:23804–23811. doi: 10.1074/jbc.M602762200. [DOI] [PubMed] [Google Scholar]
- Shafat I, Zcharia E, Nisman B, Nadir Y, Nakhoul F, Vlodavsky I, et al. An ELISA method for the detection and quantification of human heparanase. Biochem Biophys Res Commun. 2006b;341:958–963. doi: 10.1016/j.bbrc.2006.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozawa H, Takahashi M, Takatsu T, Kinoshita T, Tanzawa K, Hosoya T, et al. Trachyspic acid, a new metabolite produced by Talaromyces trachyspermus, that inhibits tumor cell heparanase: taxonomy of the producing strain, fermentation, isolation, structural elucidation, and biological activity. J Antibiot (Tokyo) 1995;48:357–362. doi: 10.7164/antibiotics.48.357. [DOI] [PubMed] [Google Scholar]
- Shteper PJ, Zcharia E, Ashhab Y, Peretz T, Vlodavsky I, Ben-Yehuda D. Role of promoter methylation in regulation of the mammalian heparanase gene. Oncogene. 2003;22:7737–7749. doi: 10.1038/sj.onc.1207056. [DOI] [PubMed] [Google Scholar]
- Simizu S, Ishida K, Wierzba MK, Osada H. Secretion of heparanase protein is regulated by glycosylation in human tumor cell lines. J Biol Chem. 2004a;279:2697–2703. doi: 10.1074/jbc.M300541200. [DOI] [PubMed] [Google Scholar]
- Simizu S, Ishida K, Osada H. Heparanase as a molecular target of cancer chemotherapy. Cancer Sci. 2004b;95:553–558. doi: 10.1111/j.1349-7006.2004.tb02485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotnikov I, Hershkoviz R, Grabovsky V, Ilan N, Cahalon L, Vlodavsky I, et al. Enzymatically quiescent heparanase augments T cell interactions with VCAM-1 and extracellular matrix components under versatile dynamic contexts. J Immunol. 2004;172:5185–5193. doi: 10.4049/jimmunol.172.9.5185. [DOI] [PubMed] [Google Scholar]
- Stein CA, LaRocca RV, Thomas R, McAtee, Myers CE. Suramin: an anticancer drug with a unique mechanism of action. J Clin Oncol. 1989;7:499–508. doi: 10.1200/JCO.1989.7.4.499. [DOI] [PubMed] [Google Scholar]
- Takaoka M, Naomoto Y, Ohkawa T, Uetsuka H, Shirakawa Y, Uno F, et al. Heparanase expression correlates with invasion and poor prognosis in gastric cancers. Lab Invest. 2003;83:613–622. doi: 10.1097/01.lab.0000067482.84946.bd. [DOI] [PubMed] [Google Scholar]
- Tang W, Nakamura Y, Tsujimoto M, Sato M, Wang X, Kurozumi K, et al. Heparanase: a key enzyme in invasion and metastasis of gastric carcinoma. Mod Pathol. 2002;15:593–598. doi: 10.1038/modpathol.3880571. [DOI] [PubMed] [Google Scholar]
- Tas F, Duranyildiz D, Oguz H, Camlica H, Yasasever V, Topuz E. Circulating serum levels of angiogenic factors and vascular endothelial growth factor receptors 1 and 2 in melanoma patients. Melanoma Res. 2006;16:405–411. doi: 10.1097/01.cmr.0000222598.27438.82. [DOI] [PubMed] [Google Scholar]
- Temkin V, Aingorn H, Puxeddu I, Goldshmidt O, Zcharia E, Gleich GJ, et al. Eosinophil major basic protein: first identified natural heparanase-inhibiting protein. J Allergy Clin Immunol. 2004;113:703–709. doi: 10.1016/j.jaci.2003.11.038. [DOI] [PubMed] [Google Scholar]
- Toyoshima M, Nakajima M. Human heparanase. Purification, characterization, cloning, and expression. J Biol Chem. 1999;274:24153–24160. doi: 10.1074/jbc.274.34.24153. [DOI] [PubMed] [Google Scholar]
- Uno F, Fujiwara T, Takata Y, Ohtani S, katsuda K, Takaoka M, et al. Antisense-mediated suppression of human heparanase gene expression inhibits pleural dissemination of human cancer cells. Cancer Res. 2001;61:7855–7860. [PubMed] [Google Scholar]
- Vaday GG, Lider O. Extracellular matrix moieties, cytokines, and enzymes: dynamic effects on immune cell behavior and inflammation. J Leukoc Biol. 2000;67:149–159. doi: 10.1002/jlb.67.2.149. [DOI] [PubMed] [Google Scholar]
- Van Den Born J, Salmivirta K, Henttinen T, Ostman N, Ishimaru T, Miyaura S, et al. Novel heparan sulfate structures revealed by monoclonal antibodies. J Biol Chem. 2005;280:20516–20523. doi: 10.1074/jbc.M502065200. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Eldor A, Haimovitz-Friedman A, Matzner Y, Ishai-Michaeli R, Lider O, et al. Expression of heparanase by platelets and circulating cells of the immune system: possible involvement in diapedesis and extravasation. Invasion Metastasis. 1992;12:112–127. [PubMed] [Google Scholar]
- Vlodavsky I, Goldshmidt O, Zcharia E, Atzmon R, Rangini-Guatta Z, Elkin M, et al. Mammalian heparanase: involvement in cancer metastasis, normal development and angiogenesis. Semin Cancer Biol. 2002;12:121–129. doi: 10.1006/scbi.2001.0420. [DOI] [PubMed] [Google Scholar]
- Vreys V, Delande N, Zhang Z, Coomans C, Roebroek A, Durr J, et al. Cellular uptake of mammalian heparanase precursor involves low density lipoprotein receptor-related proteins, mannose 6-phosphate receptors, and heparan sulfate proteoglycans. J Biol Chem. 2005;280:33141–33148. doi: 10.1074/jbc.M503007200. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhang Z, Yu B. Total synthesis of CRM646-A and -B, two fungal glucuronides with potent heparinase inhibition activities. J Org Chem. 2005;70:8884–8889. doi: 10.1021/jo051384k. [DOI] [PubMed] [Google Scholar]
- Xu X, Quiros RM, Maxhimer JB, Jiang P, Marcinek R, Ain KB, et al. Inverse correlation between heparan sulfate composition and heparanase-1 gene expression in thyroid papillary carcinomas: a potential role in tumor metastasis. Clin Cancer Res. 2003;9:5968–5979. [PubMed] [Google Scholar]
- Xu YJ, Miao HQ, Pan W, Navarro EC, Tonra JR, Mitelman S, et al. N-(4-{[4-(1H-Benzoimidazol-2-yl)-arylamino]-methyl}-phenyl)-benzamide derivatives as small molecule heparanase inhibitors. Bioorg Med Chem Lett. 2006;16:404–408. doi: 10.1016/j.bmcl.2005.09.070. [DOI] [PubMed] [Google Scholar]
- Zetser A, Levy-Adam F, Kaplan V, Gingis-Velitski S, Bashenko Y, Schubert S, et al. Processing and activation of latent heparanase occurs in lysosomes. J Cell Sci. 2004;117:2249–2258. doi: 10.1242/jcs.01068. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Bates M, Madura JD. Structure modeling, ligand binding, and binding affinity calculation (LR-MM-PBSA) of human heparanase for inhibition and drug design. Proteins. 2006;65:580–592. doi: 10.1002/prot.21065. [DOI] [PubMed] [Google Scholar]