Abstract
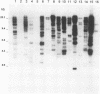
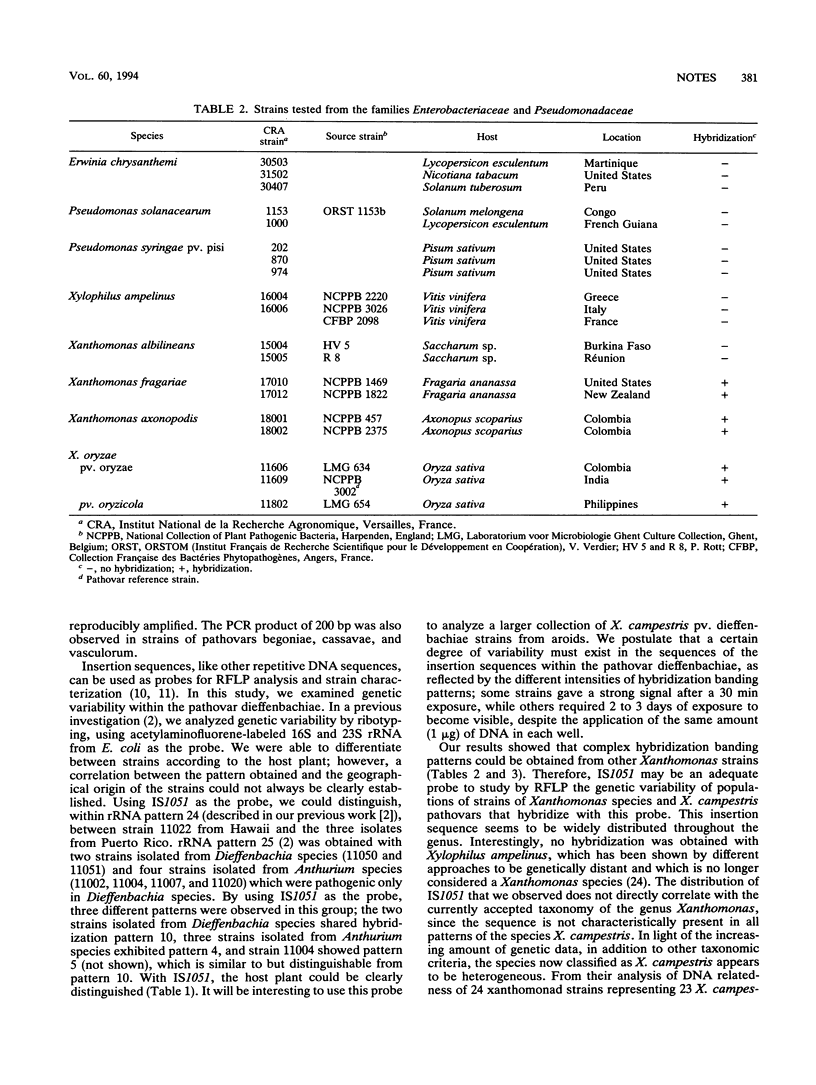
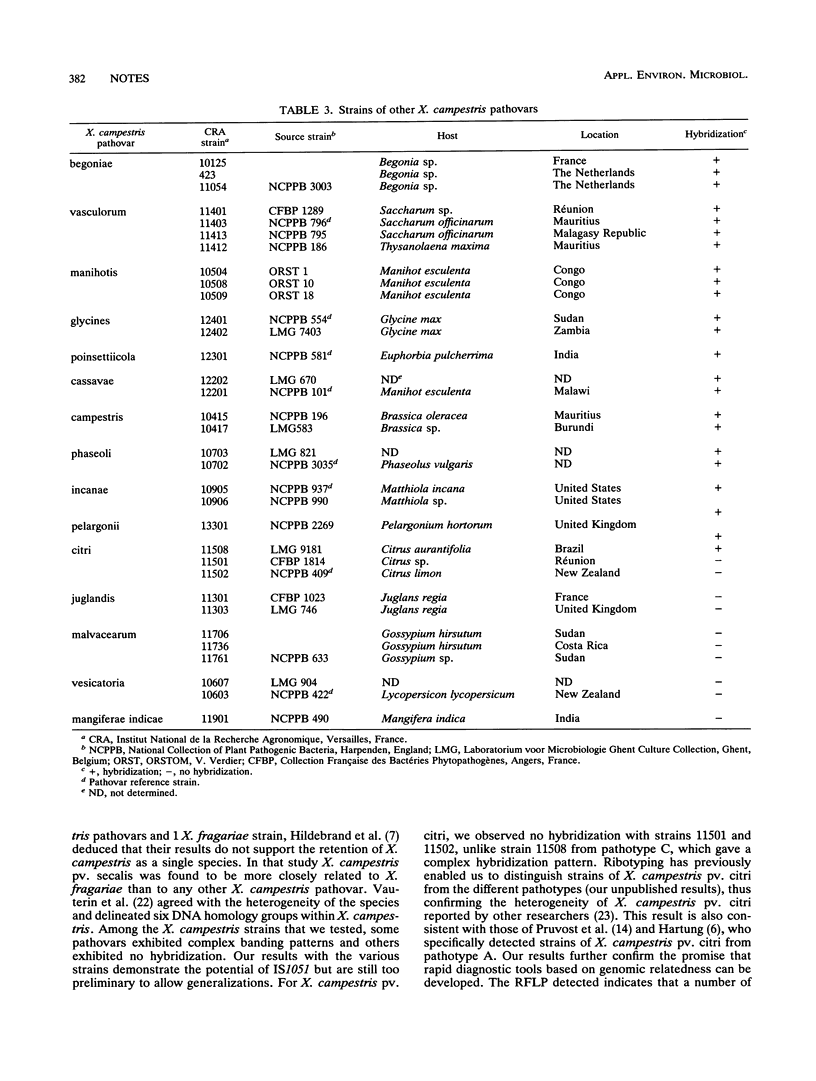
A new insertion sequence was isolated from Xanthomonas campestris pv. dieffenbachiae. Sequence analysis showed that this element is 1,158 bp long and has 15-bp inverted repeat ends containing two mismatches. Comparison of this sequence with sequences in data bases revealed significant homology with Escherichia coli IS5. IS1051, which detected multiple restriction fragment length polymorphisms, was used as a probe to characterize strains from the pathovar dieffenbachiae.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berthier Y., Verdier V., Guesdon J. L., Chevrier D., Denis J. B., Decoux G., Lemattre M. Characterization of Xanthomonas campestris Pathovars by rRNA Gene Restriction Patterns. Appl Environ Microbiol. 1993 Mar;59(3):851–859. doi: 10.1128/aem.59.3.851-859.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler J. A., van Bree M. P. The nucleotide sequence and protein-coding capability of the transposable element IS5. Gene. 1981 Aug;14(3):155–163. doi: 10.1016/0378-1119(81)90111-6. [DOI] [PubMed] [Google Scholar]
- Kearney B., Staskawicz B. J. Characterization of IS476 and its role in bacterial spot disease of tomato and pepper. J Bacteriol. 1990 Jan;172(1):143–148. doi: 10.1128/jb.172.1.143-148.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach J. E., Rhoads M. L., Vera Cruz C. M., White F. F., Mew T. W., Leung H. Assessment of genetic diversity and population structure of Xanthomonas oryzae pv. oryzae with a repetitive DNA element. Appl Environ Microbiol. 1992 Jul;58(7):2188–2195. doi: 10.1128/aem.58.7.2188-2195.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLafferty M. A., Harcus D. R., Hewlett E. L. Nucleotide sequence and characterization of a repetitive DNA element from the genome of Bordetella pertussis with characteristics of an insertion sequence. J Gen Microbiol. 1988 Aug;134(8):2297–2306. doi: 10.1099/00221287-134-8-2297. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thierry D., Cave M. D., Eisenach K. D., Crawford J. T., Bates J. H., Gicquel B., Guesdon J. L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990 Jan 11;18(1):188–188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry D., Vincent V., Clément F., Guesdon J. L. Isolation of specific DNA fragments of Mycobacterium avium and their possible use in diagnosis. J Clin Microbiol. 1993 May;31(5):1048–1054. doi: 10.1128/jcm.31.5.1048-1054.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]