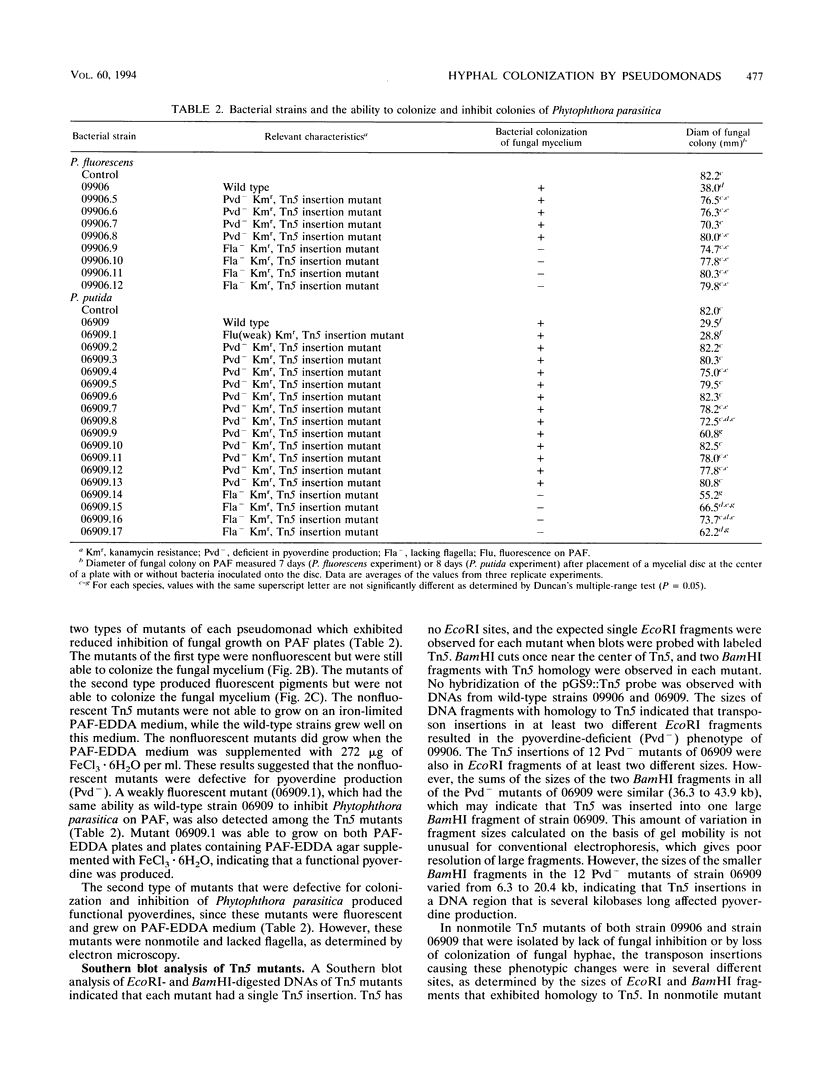
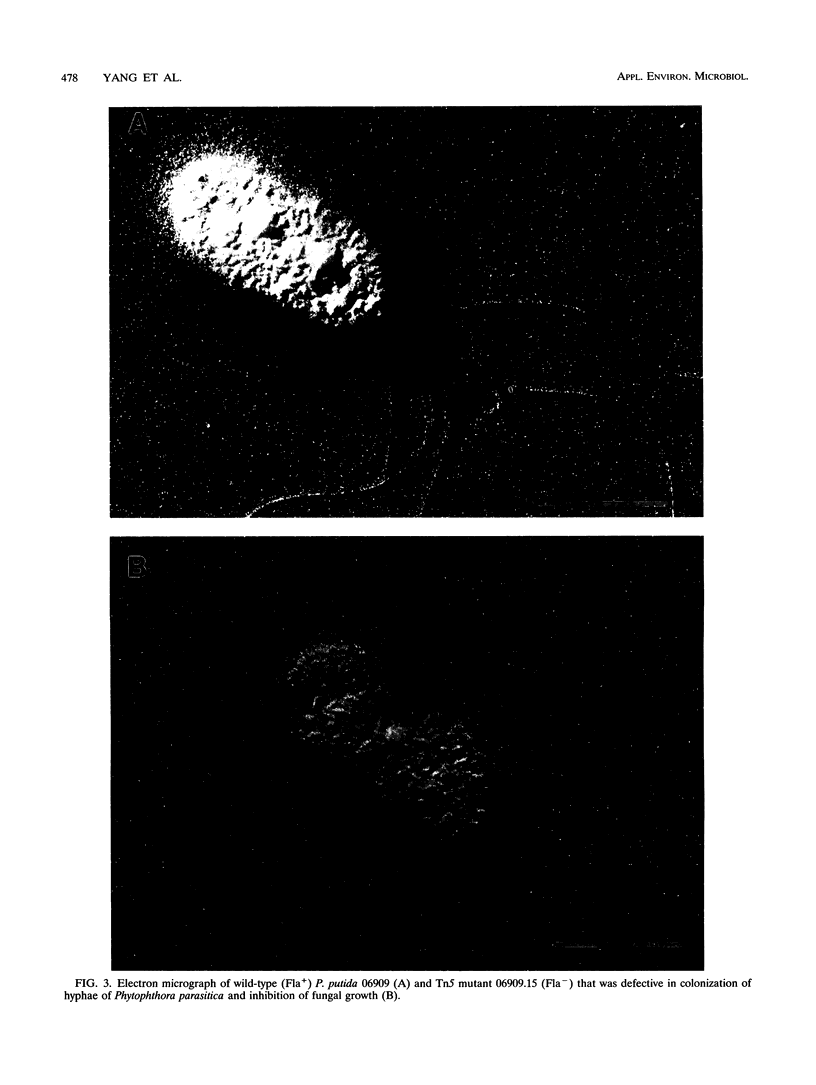
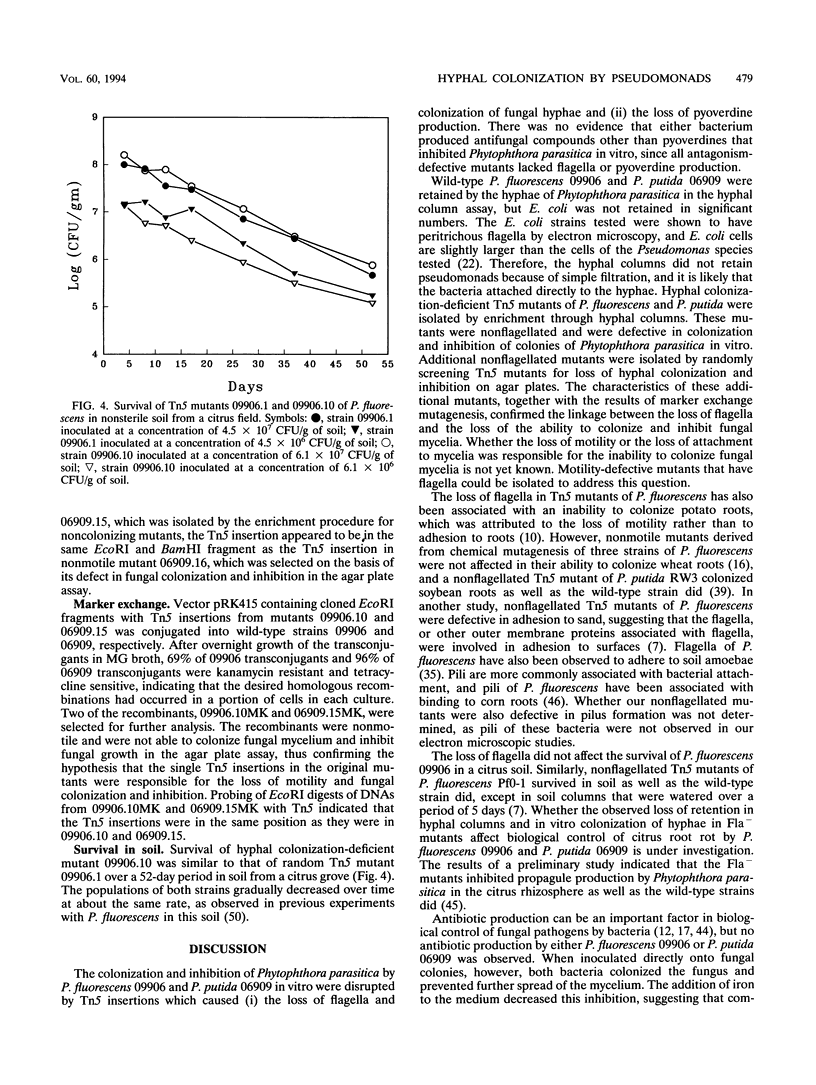
Abstract
In previous studies, Pseudomonas putida 06909 and Pseudomonas fluorescens 09906 suppressed populations of Phytophthora parasitica in the citrus rhizosphere, suggesting that these bacteria may be useful in biological control of citrus root rot. In this study we investigated the mechanisms of antagonism between the bacteria and the fungus. Both bacteria colonized Phytophthora hyphae and inhibited the fungus on agar media. A hyphal column assay was developed to measure the colonization of bacteria on fungal hyphae and to enrich for colonization-deficient mutants. In this way we identified Tn5 mutants of each pseudomonad that were not able to colonize the hyphae and inhibit fungal growth in vitro. Colonization-deficient mutants were nonmotile and lacked flagella. Survival of nonmotile mutants in a citrus soil was similar to survival of a random Tn5 mutant over a 52-day period. Additional screening of random Tn5 mutants of both pseudomonads for loss of fungal inhibition in vitro yielded two distinct types of mutants. Mutants of the first type were deficient in production of pyoverdines and in inhibition of the fungus in vitro, although they still colonized fungal hyphae. Mutants of the second type lacked flagella and were not able to colonize the hyphae or inhibit fungal growth. No role was found for antibiotic production by the two bacteria in the inhibition of the fungus. Our results suggest that both hyphal colonization and pyoverdine production are important in the inhibition of Phytophthora parasitica by P. fluorescens and P. putida in vitro.
Full text
PDF
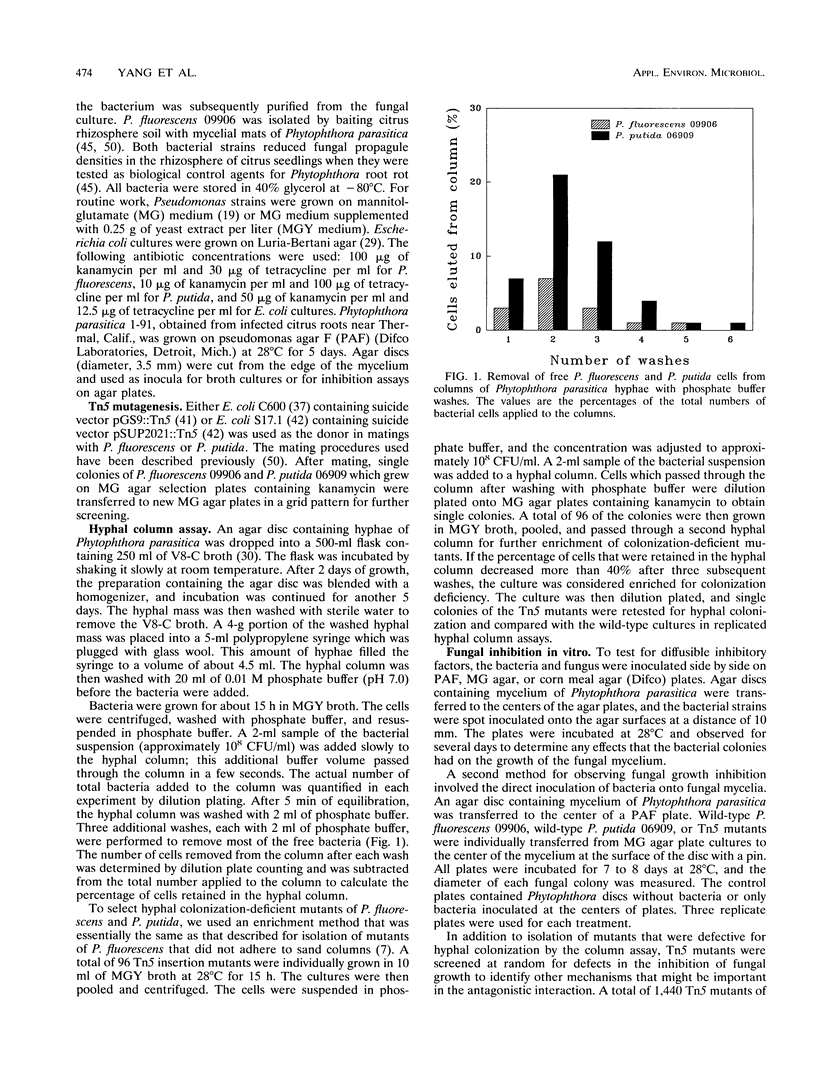

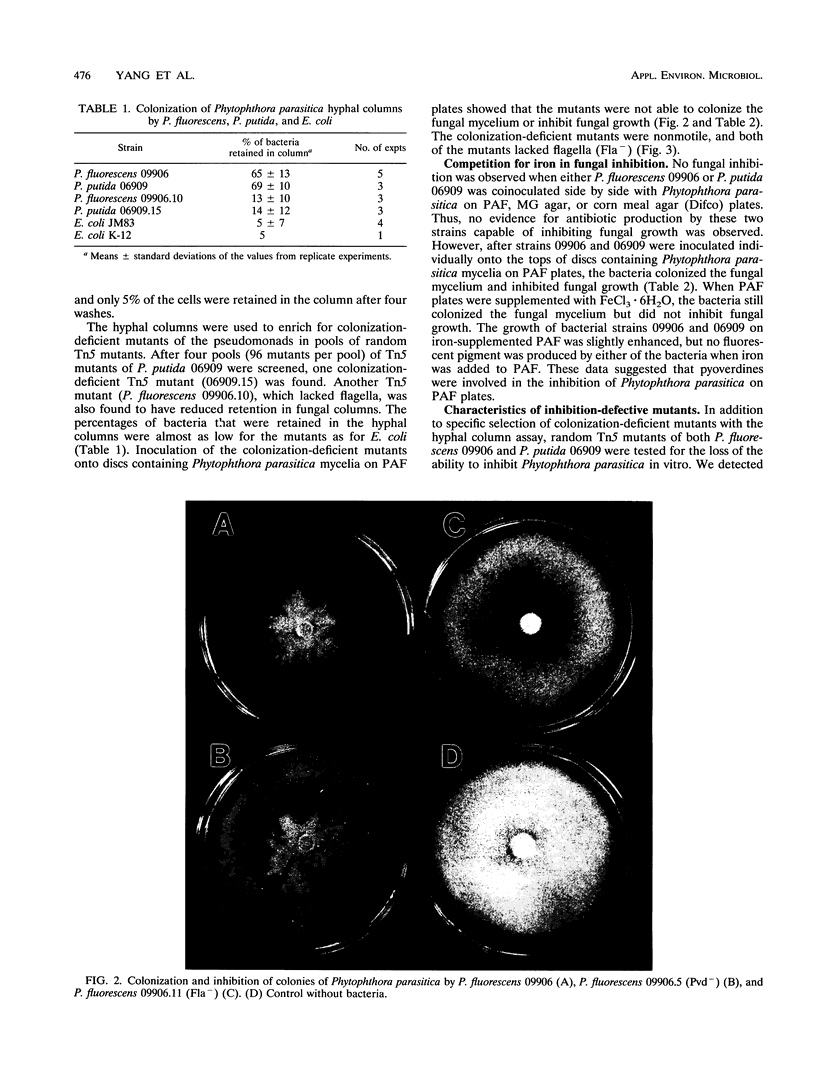


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson A. J., Habibzadegah-Tari P., Tepper C. S. Molecular Studies on the Role of a Root Surface Agglutinin in Adherence and Colonization by Pseudomonas putida. Appl Environ Microbiol. 1988 Feb;54(2):375–380. doi: 10.1128/aem.54.2.375-380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A., Azad H. R., Cha J. S., Lim C. K. Copper resistance gene homologs in pathogenic and saprophytic bacterial species from tomato. Appl Environ Microbiol. 1990 Feb;56(2):431–435. doi: 10.1128/aem.56.2.431-435.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Weger L. A., van der Vlugt C. I., Wijfjes A. H., Bakker P. A., Schippers B., Lugtenberg B. Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J Bacteriol. 1987 Jun;169(6):2769–2773. doi: 10.1128/jb.169.6.2769-2773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deflaun M. F., Tanzer A. S., McAteer A. L., Marshall B., Levy S. B. Development of an Adhesion Assay and Characterization of an Adhesion-Deficient Mutant of Pseudomonas fluorescens. Appl Environ Microbiol. 1990 Jan;56(1):112–119. doi: 10.1128/aem.56.1.112-119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dershaw D. D., Scher H. I. Serial transabdominal sonography of bladder cancer. AJR Am J Roentgenol. 1988 May;150(5):1055–1059. doi: 10.2214/ajr.150.5.1055. [DOI] [PubMed] [Google Scholar]
- Gilbert R. G., Linderman R. G. Increased activity of soil microorganisms near sclerotia of Sclerotium rolfsii in soil. Can J Microbiol. 1971 Apr;17(4):557–562. doi: 10.1139/m71-091. [DOI] [PubMed] [Google Scholar]
- Hamdan H., Weller D. M., Thomashow L. S. Relative importance of fluorescent siderophores and other factors in biological control of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens 2-79 and M4-80R. Appl Environ Microbiol. 1991 Nov;57(11):3270–3277. doi: 10.1128/aem.57.11.3270-3277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S., Kobayashi D., Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988 Oct 15;70(1):191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Ong S. A., Peterson T., Neilands J. B. Agrobactin, a siderophore from Agrobacterium tumefaciens. J Biol Chem. 1979 Mar 25;254(6):1860–1865. [PubMed] [Google Scholar]
- Rogers H. J. Iron-Binding Catechols and Virulence in Escherichia coli. Infect Immun. 1973 Mar;7(3):445–456. doi: 10.1128/iai.7.3.445-456.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj G., Iyer V. N. Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol. 1983 Dec;156(3):1292–1300. doi: 10.1128/jb.156.3.1292-1300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow L. S., Weller D. M. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988 Aug;170(8):3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesper S. J. Production of Pili (Fimbriae) by Pseudomonas fluorescens and Correlation with Attachment to Corn Roots. Appl Environ Microbiol. 1987 Jul;53(7):1397–1405. doi: 10.1128/aem.53.7.1397-1405.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yang C. H., Menge J. A., Cooksey D. A. Role of copper resistance in competitive survival of Pseudomonas fluorescens in soil. Appl Environ Microbiol. 1993 Feb;59(2):580–584. doi: 10.1128/aem.59.2.580-584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]