Abstract
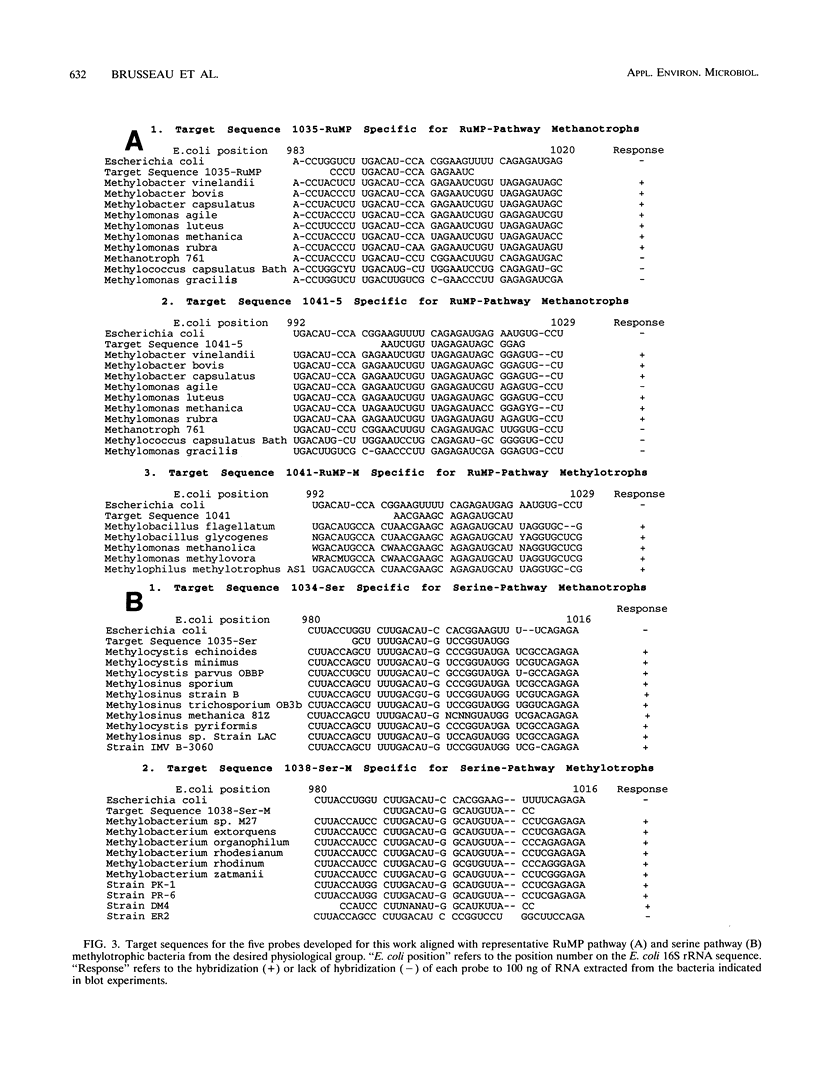
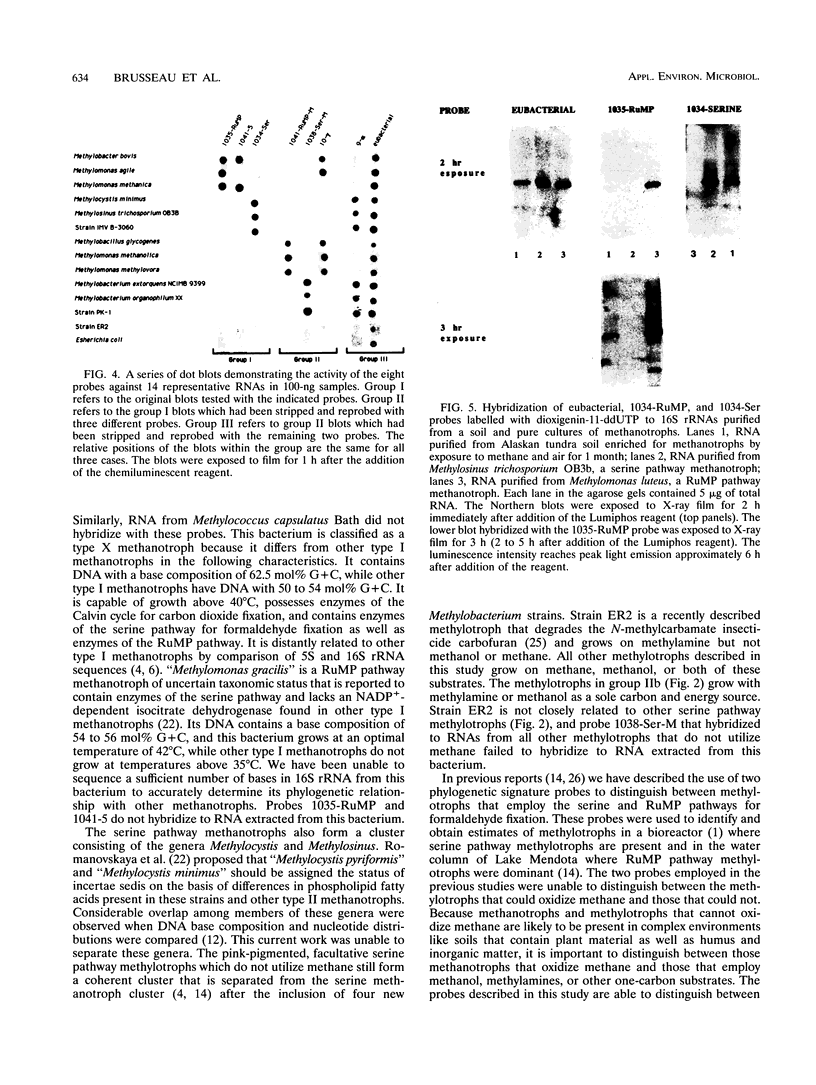
Fifteen small-subunit rRNAs from methylotrophic bacteria have been sequenced. Comparisons of these sequences with 22 previously published sequences further defined the phylogenetic relationships among these bacteria and illustrated the agreement between phylogeny and physiological characteristics of the bacteria. Phylogenetic trees were constructed with 16S rRNA sequences from methylotrophic bacteria and representative organisms from subdivisions within the class Proteobacteria on the basis of sequence similarities by using a weighted least-mean-square difference method. The methylotrophs have been separated into coherent clusters in which bacteria shared physiological characteristics. The clusters distinguished bacteria which used either the ribulose monophosphate or serine pathway for carbon assimilation. In addition, methanotrophs and methylotrophs which do not utilize methane were found to form distinct clusters within these groups. Five new deoxyoligonucleotide probes were designed, synthesized, labelled with digoxigenin-11-ddUTP, and tested for the ability to hybridize to RNA extracted from the bacteria represented in the unique clusters and for the ability to detect RNAs purified from soils enriched for methanotrophs by exposure to a methane-air atmosphere for one month. The 16S rRNA purified from soil hybridized to the probe which was complementary to sequences present in 16S rRNA from serine pathway methanotrophs and hybridized to a lesser extent with a probe complementary to sequences in 16S rRNAs of ribulose monophosphate pathway methanotrophs. The nonradioactive detection system used performed reliably at amounts of RNA from pure cultures as small as 10 ng.
Full text
PDF
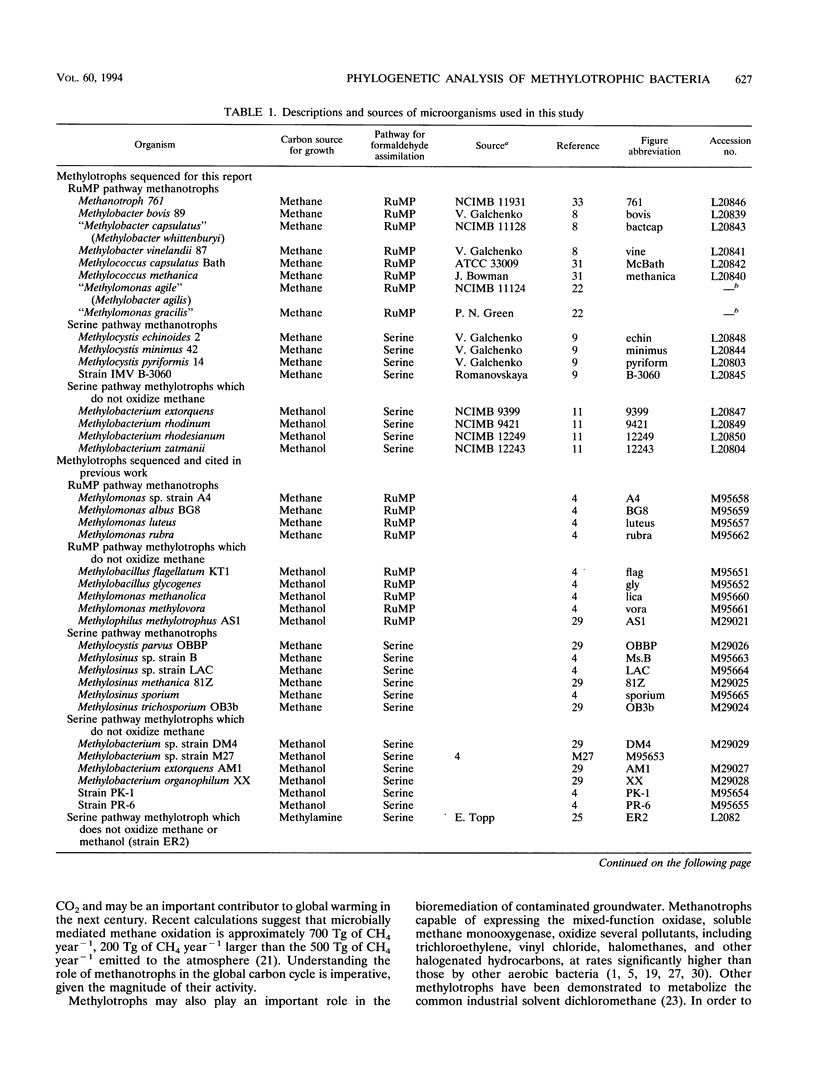


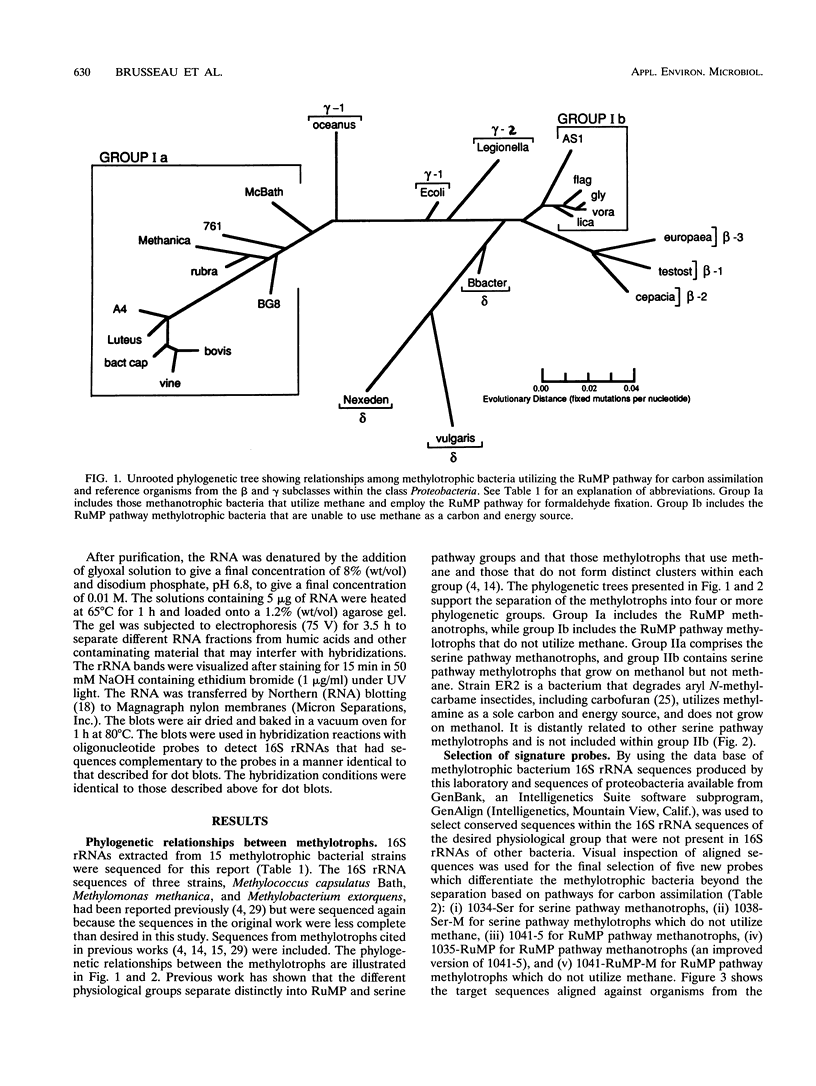
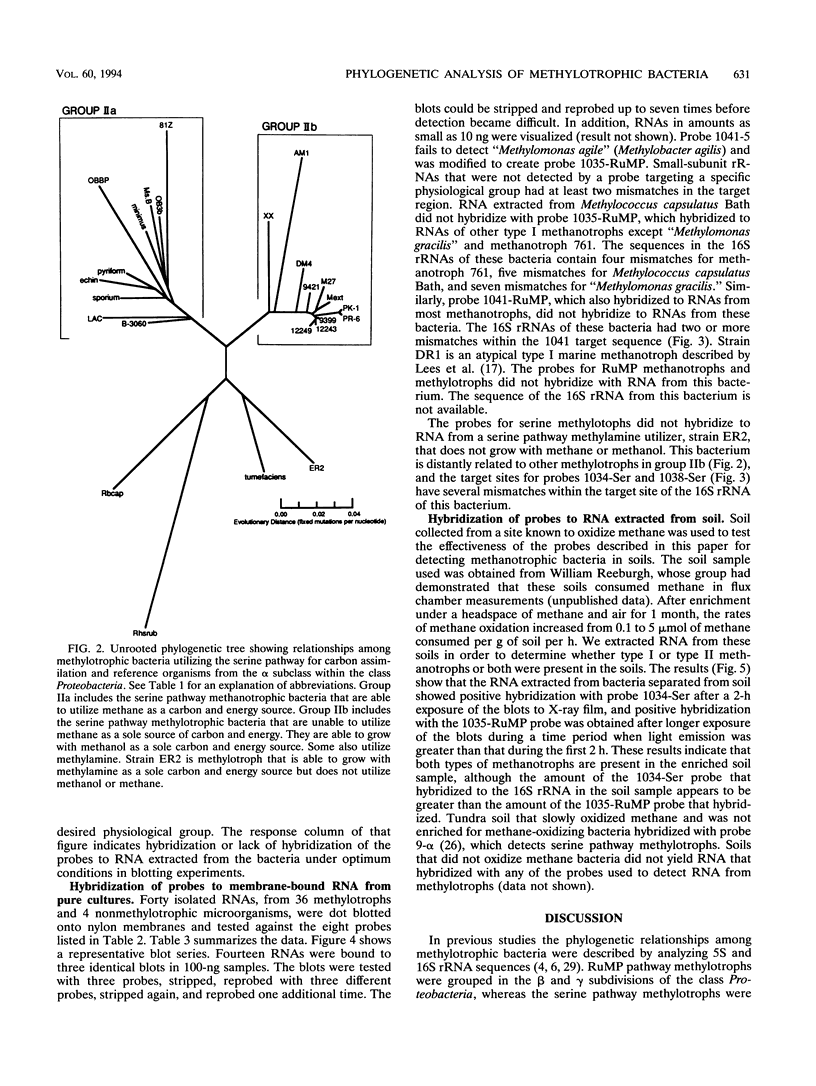



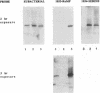
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alvarez-Cohen L., McCarty P. L., Boulygina E., Hanson R. S., Brusseau G. A., Tsien H. C. Characterization of a methane-utilizing bacterium from a bacterial consortium that rapidly degrades trichloroethylene and chloroform. Appl Environ Microbiol. 1992 Jun;58(6):1886–1893. doi: 10.1128/aem.58.6.1886-1893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratina B. J., Brusseau G. A., Hanson R. S. Use of 16S rRNA analysis to investigate phylogeny of methylotrophic bacteria. Int J Syst Bacteriol. 1992 Oct;42(4):645–648. doi: 10.1099/00207713-42-4-645. [DOI] [PubMed] [Google Scholar]
- Brusseau G. A., Tsien H. C., Hanson R. S., Wackett L. P. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation. 1990;1(1):19–29. doi: 10.1007/BF00117048. [DOI] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guckert J. B., Ringelberg D. B., White D. C., Hanson R. S., Bratina B. J. Membrane fatty acids as phenotypic markers in the polyphasic taxonomy of methylotrophs within the Proteobacteria. J Gen Microbiol. 1991 Nov;137(11):2631–2641. doi: 10.1099/00221287-137-11-2631. [DOI] [PubMed] [Google Scholar]
- Oldenhuis R., Vink R. L., Janssen D. B., Witholt B. Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Environ Microbiol. 1989 Nov;55(11):2819–2826. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J. Phylogenetic analysis using ribosomal RNA. Methods Enzymol. 1988;164:793–812. doi: 10.1016/s0076-6879(88)64084-5. [DOI] [PubMed] [Google Scholar]
- Scholtz R., Wackett L. P., Egli C., Cook A. M., Leisinger T. Dichloromethane dehalogenase with improved catalytic activity isolated from a fast-growing dichloromethane-utilizing bacterium. J Bacteriol. 1988 Dec;170(12):5698–5704. doi: 10.1128/jb.170.12.5698-5704.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp E., Hanson R. S., Ringelberg D. B., White D. C., Wheatcroft R. Isolation and characterization of an N-methylcarbamate insecticide-degrading methylotrophic bacterium. Appl Environ Microbiol. 1993 Oct;59(10):3339–3349. doi: 10.1128/aem.59.10.3339-3349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Bratina B. J., Tsuji K., Hanson R. S. Use of oligodeoxynucleotide signature probes for identification of physiological groups of methylotrophic bacteria. Appl Environ Microbiol. 1990 Sep;56(9):2858–2865. doi: 10.1128/aem.56.9.2858-2865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Brusseau G. A., Hanson R. S., Waclett L. P. Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1989 Dec;55(12):3155–3161. doi: 10.1128/aem.55.12.3155-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K., Tsien H. C., Hanson R. S., DePalma S. R., Scholtz R., LaRoche S. 16S ribosomal RNA sequence analysis for determination of phylogenetic relationship among methylotrophs. J Gen Microbiol. 1990 Jan;136(1):1–10. doi: 10.1099/00221287-136-1-1. [DOI] [PubMed] [Google Scholar]
- Wackett L. P., Brusseau G. A., Householder S. R., Hanson R. S. Survey of microbial oxygenases: trichloroethylene degradation by propane-oxidizing bacteria. Appl Environ Microbiol. 1989 Nov;55(11):2960–2964. doi: 10.1128/aem.55.11.2960-2964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittenbury R., Phillips K. C., Wilkinson J. F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970 May;61(2):205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. J., Hanson R. S. Variants of the Obligate Methanotroph Isolate 761M Capable of Growth on Glucose in the Absence of Methane. Appl Environ Microbiol. 1984 Oct;48(4):807–812. doi: 10.1128/aem.48.4.807-812.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]