Abstract
The E2A-HLF (hepatic leukemia factor) oncoprotein, generated in pro-B lymphocytes by fusion of the trans-activation domain of E2A to the basic region/leucine zipper (bZIP) domain of HLF, functions as an anti-apoptotic transcription factor in leukemic cell transformation. When introduced into interleukin 3 (IL-3)-dependent mouse pro-B lymphocytes, E2A-HLF prevents apoptosis induced by growth factor deprivation, suggesting that IL-3 mediates cell survival through activation of a transcription factor whose activity can be constitutively replaced by the chimeric oncoprotein. We considered four bZIP transcription factors as candidates for this putative IL-3-regulated factor, each of which binds avidly to the DNA consensus sequence recognized by E2A-HLF and is related to the Caenorhabditis elegans CES-2 (cell death specification protein) neuron-specific mediator of cell death. The expression and binding activity of the Nfil3 protein (also called E4bp4), but not of Hlf, Dbp, or Tef, was found to be regulated by IL-3 in mouse pro-B cell lines (Baf-3 and FL5.12). Northern blot analysis showed that Nfil3/E4bp4 is regulated as a “delayed-early” IL-3-responsive gene, requiring de novo protein synthesis. In the absence of IL-3, enforced expression of the human NFIL3/E4BP4 cDNA promoted the survival but not the growth of IL-3-dependent pro-B cells. Our results implicate NFIL3/E4BP4 (nuclear factor regulated by IL-3/adenovirus E4 promoter binding protein) in a distinct growth factor-regulated signaling pathway that is responsible for the survival of early B-cell progenitors, and whose alteration by E2A-HLF leads to childhood B lineage leukemia.
Keywords: apoptosis, hematopoietic growth factor, signal transduction, leukemia
The elimination of defective T and B lymphocyte progenitors by apoptosis has important implications for the regulation of normal development of the immune system and for malignant transformation in the leukemias and lymphomas (1–3). This process is greatly influenced by cytokines that act on lymphoid and hematopoietic precursors, including interleukin 3 (IL-3), which supports the in vitro proliferation of multipotent stem cells as well as many progenitor cells already committed to particular lineages (4). When IL-3-dependent cell lines are deprived of growth factor, they not only stop proliferating but undergo apoptosis as well, indicating that IL-3 promotes cell survival by suppression of programmed cell death (5–7). Thus, it is important to identify the key molecules through which IL-3-generated survival signals are transduced.
The E2A-HLF (hepatic leukemia factor) fusion protein, which drives the leukemic conversion of pro-B lymphocytes harboring a t(17;19) chromosomal translocation (8, 9), can block apoptosis caused by IL-3 deprivation (10), suggesting that its primary effect is on cell survival rather than cell growth. Because of the close homology between the basic region/leucine zipper (bZIP) DNA binding and dimerization domain of HLF and that of CES-2, a neuron-specific cell-death specification protein in the nematode Caenorhabditis elegans, we postulated that E2A-HLF interferes with an evolutionarily conserved pathway that can have both positive and negative effects on the survival of B-lymphocyte progenitors (10, 11). To explain the ability of E2A-HLF to block apoptosis resulting from IL-3 deprivation, we reasoned that it might constitutively replace the function of a transcription factor whose activation is a normal event within the signaling pathway regulated by IL-3. As initial candidates for this regulatory role, we selected mammalian bZIP transcription factors that are highly related to the C. elegans CES-2 protein and that bind avidly to the same DNA binding sequence. These include members of the mammalian proline- and acidic amino acid-rich (PAR) subfamily of bZIP proteins: HLF (8, 9, 12–14), DBP (albumin gene promoter D-box binding protein) (15, 16), and TEF (thyrotroph embryonic factor) (17). We also analyzed a related bZIP protein called E4BP4, isolated by its ability to recognize the proximal activating transcription factor binding site of the adenovirus E4 promoter (18, 19), and later independently identified as NFIL3, a protein expressed in T cells and capable of binding to a similar sequence motif in the 5′ flanking region of the human IL-3 promoter (20). Here we report that murine Nfil3/E4bp4 is regulated by IL-3 and that enforced expression of its human counterpart in IL-3-deprived cells prevents apoptosis. Thus, this transcription factor normally regulates a pivotal step in a growth-factor responsive anti-apoptotic signaling pathway whose alteration likely contributes to human B-lineage leukemia.
MATERIALS AND METHODS
Cell Culture.
Baf-3 and FL5.12 pro-B lymphocytes were cultured in RPMI 1640 medium containing 10% fetal calf serum, 20 mM Hepes, 55 μM 2-mercaptoethanol, and 10% WEHI-3B conditioned medium as a source of IL-3. In restimulation experiments, recombinant mouse IL-3 (R & D Systems) was added at a concentration of 10 ng/ml. Transfectants were generated in two independent experiments by electroporation using 2 × 107 cells and 80 μg of pMT-CB6+/NFIL3/E4BP4 vector, with the Gene Pulser (Bio-Rad) set at 300 V and 960 μF. Cells were separated into 24 pools immediately after electroporation and selected by incubating cells in the presence of G418 (0.6 mg/ml) for 2 weeks. Nine independent pools of transfected cells were selected for further analysis, and four clones were obtained from one of these pools by limiting dilution. NFIL3/E4BP4 expression was induced by treating cells from each of the nine pools, and the four clones were derived by limiting dilution with 100 μM ZnSO4 for 24 hr prior to growth factor deprivation. Viable cell counts were determined by trypan blue dye exclusion in triplicate assays.
Electrophoretic Mobility Shift Assay.
Extraction of nuclear lysates, radiolabeling of oligonucleotide probes, and electrophoretic mobility shift analysis were performed as described (12). Anti-NFIL3/E4BP4 rabbit sera were prepared from animals injected with recombinant human protein, and 1 μl per lane was used in antibody-perturbed gel-shift analyses.
Immunoblot Analysis.
Cells (7.5 × 105) were lysed in RIPA buffer (150 mM NaCl/1.0% Nonidet P-40/0.5% deoxycholate/0.1% SDS/50 mM Tris, pH 8.0) in the presence of protease inhibitors, and total cellular proteins were separated by SDS/PAGE. After wet electrotransfer onto nitrocellulose membranes, immunoblotting was performed with anti-NFIL3/E4BP4 rabbit serum. Blots were incubated with horseradish-conjugated anti-rabbit Ig secondary antibodies and subjected to autoradiography with enhanced chemiluminescence (Amersham).
Northern Blot Analysis.
Twenty micrograms of total RNA was separated by electrophoresis in 1% agarose gels containing 2.2 M formaldehyde, transferred to nylon membranes, and visualized by autoradiography following hybridization to a murine Nfil3/E4bp4, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) or rat c-fos 32P-labeled cDNA.
RESULTS AND DISCUSSION
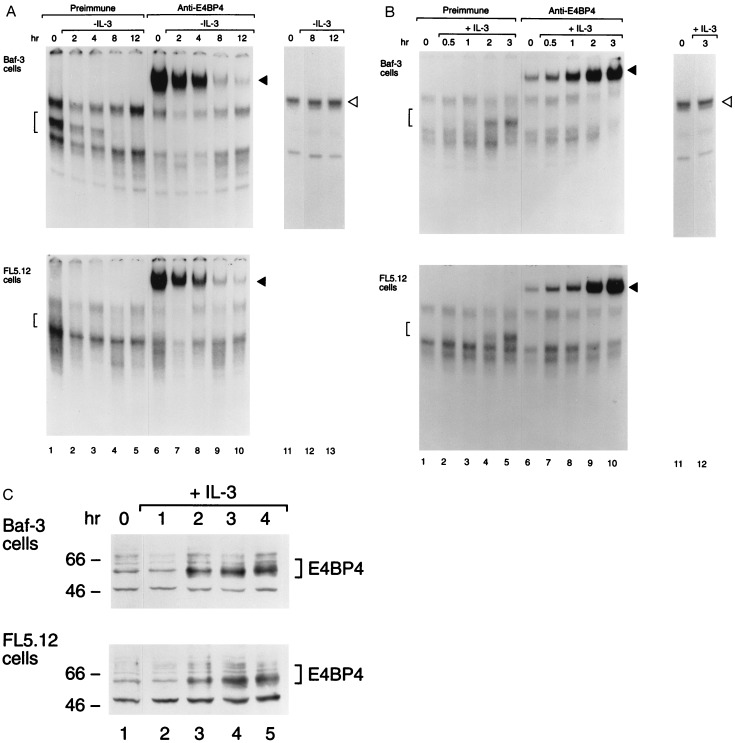
Any one of several mammalian bZIP proteins could serve as a transcription factor that transduces survival signals from the IL-3 receptor and whose function is replaced by the E2A-HLF oncoprotein in leukemic pro-B cells. As a first step in our search for such a growth-factor-regulated anti-apoptotic transcription factor, we employed a sensitive antibody-perturbed electrophoretic mobility shift assay with the HLF consensus sequence (HLF-CS) oligonucleotide probe. Our analysis demonstrated that the binding activity of Nfil3/E4bp4 (Fig. 1), but not that of Hlf, Dbp, or Tef (the mammalian PAR proteins, which were undetectable; data not shown), is regulated by IL-3 in growth factor-dependent murine pro-B lymphocytes [Baf-3 (5) or FL5.12 (21)]. Nfil3/E4bp4 binding activity was rapidly down-regulated after IL-3 deprivation (Fig. 1A), reappearing within 2–3 hr after restoration of the growth factor to the culture medium (Fig. 1B), indicative of its dependence on signals transduced through the IL-3 receptor. By contrast, the binding activity of SP1 (22) remained unchanged (Figs. 1 A and B). Additional controls demonstrated the specificity of native and supershifted Nfil3/E4bp4-DNA complexes, in that they were ablated by an excess of nonradiolabeled HLF-CS oligonucleotide competitor, but unaffected by an oligonucleotide with altered base pairs in the HLF consensus binding sequence (HLF-M4; see ref. 12; data not shown). Levels of the Nfil3/E4bp4 protein paralleled its binding activity (Fig. 1C), suggesting control at the level of synthesis of a relatively short-lived protein.
Figure 1.
Nfil3/E4bp4 binding activity is regulated by IL-3. (A) Nuclear extracts prepared from IL-3-dependent mouse pro-B cells (Baf-3 or FL5.12) were studied by electrophoretic mobility shift assay with a 32P-labeled oligonucleotide probe containing the HLF consensus binding sequence (GTTACGTAAC; lanes 1–10). Lanes 2–5 and 7–10 show results at the indicated intervals after withdrawal of IL-3 from the medium; lanes 1 and 6 depict findings for stable culture in IL-3. Brackets indicate Nfil3/E4bp4-DNA complexes (lanes 1–5), and solid arrowheads indicate the supershifted complex that was evident after incubation with antiserum specific for Nfil3/E4bp4 (lanes 6–10). As a control for extract integrity, the mobility shift assay was also performed with an SP1 transcription factor consensus sequence probe on extracts from cells growing in IL-3 (lane 11) and after its withdrawal (lanes 12 and 13). (B) Same as in A, except that extracts were analyzed from cells deprived of ligand for 8 hr (lanes 1, 6, and 11) and after the indicated intervals following IL-3 addition (lanes 2–5, 7–10, and 12). (C) Western blot analysis showing levels of the ≈56- to 58-kDa Nfil3/E4bp4 protein in Baf-3 and FL5.12 cells deprived of IL-3 for 8 hr (lane 1) and at 1-hr intervals following reinstatement of the cytokines. Nfil3/E4bp4 protein levels were 4.1- and 4.5-fold higher, respectively, in Baf-3 and FL5.12 cells, 4 hr after IL-3 restimulation (lane 5) than in cells deprived of IL-3 for 8 hr (lane 1). Nfil3/E4bp4 levels in cells growing continuously in IL-3 were 2.5- (Baf-3 cells) and 1.8-fold (FL5.12 cells) higher than in IL-3-deprived cells (lane 1).
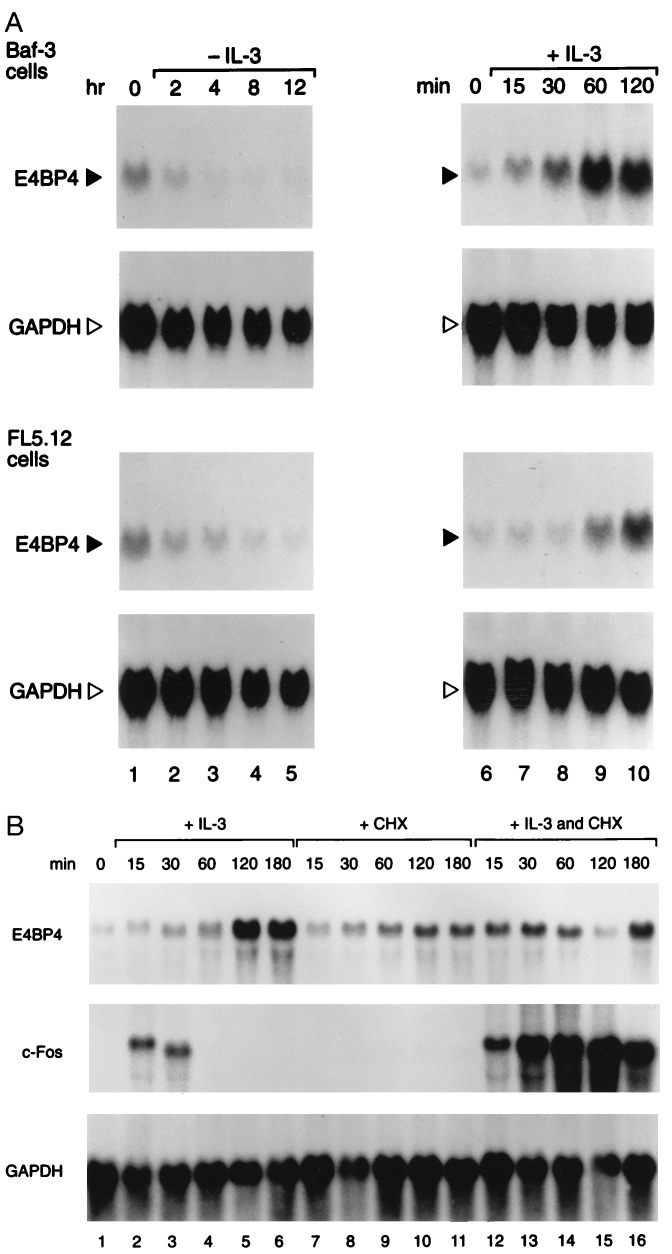
To assess the effects of IL-3 on Nfil3/E4bp4 mRNA levels, we isolated the mouse Nfil3/E4bp4 cDNA, which encodes a protein highly conserved with respect to human E4BP4 (100% identical in the bZIP domain and 84.4% identical overall). Northern blot analysis with this probe demonstrated that Nfil3/E4bp4 mRNA expression depends on the presence of IL-3. A marked decline in the levels of mRNA occurred within 2 hr of IL-3 deprivation, with maximal levels attained 1–2 hr after reintroduction of IL-3 into the medium (Fig. 2A). The increase in Nfil3/E4bp4 mRNA was delayed with respect to that of the “immediate-early” gene c-fos, which reached maximal levels after 15 min of treatment. Moreover, in contrast to c-fos, the induction of Nfil3/E4bp4 by IL-3 was blocked by cycloheximide (Fig. 2B). Thus, Nfil3/E4bp4 is transcriptionally regulated as a “delayed-early” IL-3-responsive gene that is dependent upon de novo protein synthesis.
Figure 2.
Nfil3/E4bp4 expression is regulated by IL-3. (A) Northern blot analysis of total RNA (20 μg per lane) prepared from Baf-3 (Upper) or FL5.12 (Lower) pro-B lymphocytes were grown in IL-3 (lane 1) and then deprived of the ligand for the indicated intervals (lanes 2–5). Cells starved of ligand for 8 hr (lane 7) were restimulated with 10 ng/ml of murine IL-3 and incubated for the indicated times (lanes 8–11). Blots were hybridized with an Nfil3/E4bp4 probe and stripped and rehybridized with the GAPDH control probe. (B) Induction of Nfil3/E4bp4 by IL-3 requires protein synthesis. Asynchronously growing FL5.12 cells were deprived of IL-3 for 16 hr (lane 1) and stimulated with either IL-3 alone for the indicated times (lanes 2–6), or with cycloheximide (CHX, 20 μg/ml) alone (lanes 7–11), or were pretreated with CHX for 15 min and incubated in both IL-3 and CHX for the indicated times (lanes 12–16). Northern blots were sequentially hybridized with probes for Nfil3/E4bp4, c-fos, and GAPDH.
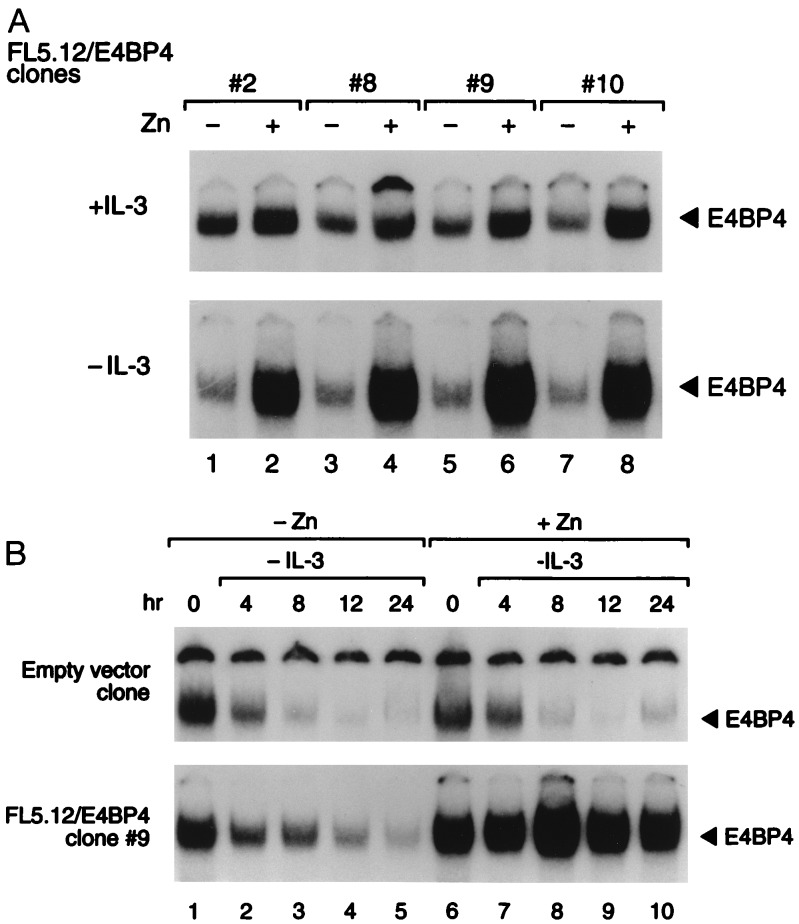
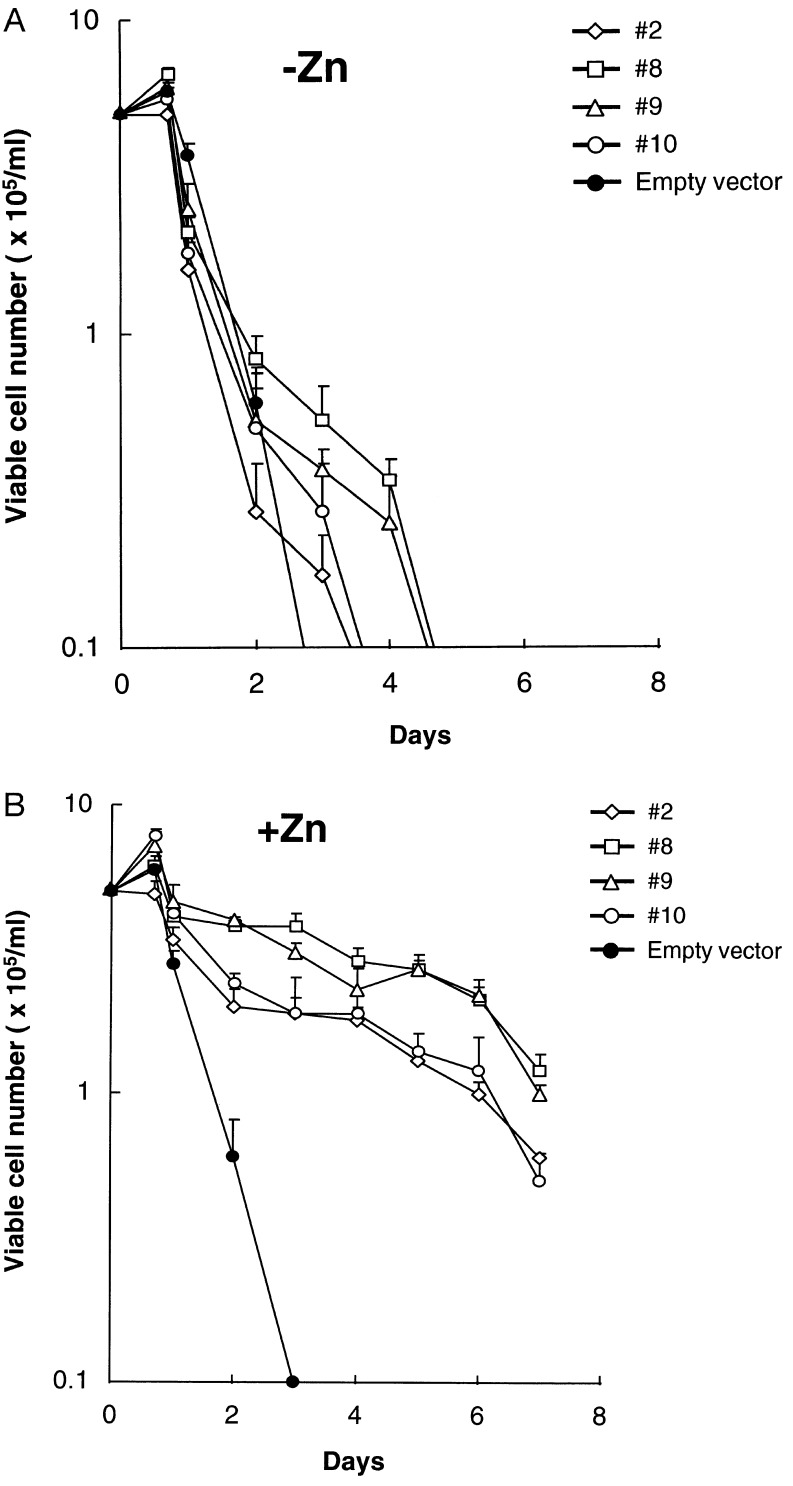
To test the function of NFIL3/E4BP4, we introduced the human NFIL3/E4BP4 cDNA into FL5.12 cells using a zinc-regulated eukaryotic expression vector. In the presence of IL-3, NFIL3/E4BP4 binding activity was only modestly elevated after zinc treatment (Fig. 3A Upper), whereas in cells deprived of IL-3, it was maintained at higher-than-normal levels in the presence of zinc, despite a dramatic decrease in the absence of metal induction. In a kinetic experiment with cells from clone 9 (Fig. 3B), the DNA binding activity of NFIL3/E4BP4 remained elevated in IL-3-deprived cells in the presence of zinc (Lower Right), but it declined rapidly when cells were cultured in medium lacking both zinc and IL-3 (Lower Left). The functional significance of NFIL3/E4BP4 expression was demonstrated in four clones, each of which showed prolonged survival in the presence of zinc after IL-3 deprivation (Fig. 4B). In the absence of zinc, the cells underwent rapid apoptosis (Fig. 4A), as did control cells regardless of zinc concentration.
Figure 3.
Transfected pro-B cell clones conditionally overexpressing NFIL3/E4BP4. (A) Antibody-perturbed electrophoretic mobility shift assay of FL5.12 clones expressing NFIL3/E4BP4 from the zinc-regulated pMT-CB6+ expression plasmid. Solid arrowheads indicate NFIL3/E4BP4–DNA complexes from each of four independent clones grown in the presence (even lanes) or absence (odd lanes) of zinc, in the presence of IL-3 for 24 hr (+IL-3, Upper) or 16 hr after its removal (−IL-3, Lower). (B) A time course of NFIL3/E4BP4 binding activity in FL5.12 cells bearing the empty zinc-regulated vector (Upper) or the vector containing NFIL3/E4BP4 (clone 9; Lower). The mobility assay was performed with extracts from cells cultured in the presence (lanes 6–10) or absence (lanes 1–5) of zinc, and in the presence of IL-3 (lanes 1 and 6), or at the indicated intervals after its removal. Solid arrowheads indicate NFIL3/E4BP4–DNA complexes that supershifted with E4BP4 antiserum.
Figure 4.
NFIL3/E4BP4 suppresses apoptosis of IL-3-deprived murine pro-B cells. (A) Numbers of viable cells at serial times after IL-3 withdrawal in the absence of zinc for FL5.12 clones transfected with zinc-regulatable NFIL3/E4BP4 plasmid or the empty vector (means of triplicate counts, with bars showing standard deviations). (B) Viable cell counts of the same clones after IL-3 withdrawal and culture in the presence of zinc to induce NFIL3/E4BP4 expression. Representative data from seven independent experiments are presented; comparable results were obtained with nine independently derived pools of cells transfected with the NFIL3/E4BP4 plasmid (data not shown).
While dramatic, the effects of enforced NFIL3/E4BP4 expression on survival were less potent than those previously observed for E2A-HLF when it was expressed in Baf-3 cells under the control of the same zinc-inducible vector (10). This appeared to be due to the fact that, after IL-3 had been removed for 48 hr, NFIL3/E4BP4 DNA binding activity in zinc-treated transfectants fell below levels detected in cells growing in medium containing IL-3 (data not shown). Further studies are needed to determine whether protein levels are affected, or whether additional posttranslational modifications, such as phosphorylation (23), are mediated in response to IL-3 and required for full activity of NFIL3/E4BP4 as a mediator of DNA binding and cell survival.
Although NFIL3/E4BP4 has been shown to trans-activate the IL-3 promoter in T cells (20), its effects on the survival of FL5.12 pro-B cells do not appear to be mediated through an autocrine mechanism, as IL-3 mRNA was not detectable in clones overexpressing NFIL3/E4BP4, and conditioned media from these cells did not promote the growth or survival of wild-type cells (data not shown). In addition, murine lymphoblasts with enforced expression of NFIL3/E4BP4 accumulated in G0/G1 phase in the absence of IL-3, and the administration of zinc in the presence of IL-3 did not effect cell growth rate (data not shown). This indicates that NFIL3/E4BP4 promotes survival, but does not transduce proliferative signals mediated by growth factor-induced activation of the IL-3 receptor. No significant changes were observed in levels of the Bcl-2, Bcl-xL, Bax, and Bad proteins (reviewed in refs. 1 and 2) in cells overexpressing NFIL3/E4BP4 (negative data not shown), suggesting that the effects of this transcription factor on survival are not mediated through induction of these regulators of apoptotic signaling. However, we cannot rule out the possibility that the activity of Bcl-2 family members is affected by posttranslational modifications or by alterations in complexes of these proteins. For example, in one recent study, Zha et al. (24) found that phosphorylation of Bad on serine residues in the presence of IL-3 resulted in its binding to the 14–3-3 protein rather than Bcl-xL, plus blocking the pro-apoptotic function of Bad.
Although the precise role of IL-3 in the early stages of B lymphopoiesis is unknown, the growth factor was required to isolate clonal pro-B and pre-B cell lines (5, 21, 25, 26), and the IL-3 gene is activated in some cases of human acute pro-B cell leukemia associated with the t(5;14) chromosomal translocation (27, 28). The apparent leukemogenic effects of IL-3 activation in man strongly suggest that the cytokine mediates cell growth and survival in at least a subset of early B cell progenitors. Here we provide insight into the transcriptional mechanism through which IL-3 influences progenitor cell survival during B lymphopoiesis, demonstrating that NFIL3/E4BP4 is positively regulated by IL-3 and functions downstream of the IL-3 receptor to block the programmed cell death that results from growth factor deprivation.
The oncogenic transcription factor E2A-HLF, which binds the same DNA consensus sequence as NFIL3/E4BP4, also protects pro-B cells from programmed cell death (10), suggesting that both act through a common downstream signaling pathway. Functional studies indicate a positive regulatory role for E2A-HLF in gene expression, mediated through two separate domains (AD1 and AD2) in the E2A fusion element that act as potent trans-activators of gene expression in several contexts (12, 13, 29–31). By contrast, NFIL3/E4BP4 can function as either a trans-repressor or trans-activator of transcription in different cell types and promoter contexts (18–20). The C. elegans cell death specification protein CES-2 is highly related to HLF and NFIL3/E4BP4 in its bZIP domain, and it binds to the same DNA consensus sequence (11). Genetic evidence suggests that the CES-2 protein acts as a transcriptional repressor to induce programmed cell death in the sister cells of serotoninergic neurosecretory motor neurons during C. elegans embryogenesis (11, 32). Accordingly, the consensus binding sequence recognized by HLF, NFIL3/E4BP4, and CES-2 may serve as a versatile regulatory switch in the promoter regions of target genes, either inducing (CES-2) or suppressing (NFIL3/E4BP4, E2A-HLF) apoptosis, depending on site occupancy. We therefore propose that E2A-HLF, due to its constitutive expression in pro-B cells carrying the t(17;19), replaces NFIL3/E4BP4 in a novel IL-3-dependent survival pathway, trans-activating a target gene or genes whose expression is critical for downstream anti-apoptotic signaling (Fig. 5). As these lymphoid progenitors accumulate, they likely acquire additional mutations of growth regulatory genes needed to produce overt human B-lineage acute leukemia.
Figure 5.
Model of NFIL3/E4BP4 function in cell survival pathways and a proposed mechanism of action of the E2A-HLF oncoprotein in the transformation of pro-B lymphocytes. Expression of the NFIL3/E4BP4 gene is tightly regulated by IL-3 through an undefined signal transduction pathway emanating from the ligand-activated IL-3 receptor. The NFIL3/E4BP4 transcription factor binds to a consensus sequence in the promoter of downstream target genes, leading to trans-activation of critical survival signals. Because HLF and NFIL3/E4BP4 can recognize the same DNA binding site, the constitutively expressed E2A-HLF oncoprotein can replace the function of the normal IL-3-regulated transcription factor, accounting for its anti-apoptotic role in leukemogenesis. An additional prediction of the model is that the consensus binding site (TTACGTAA) could serve as a versatile regulatory switch, allowing other, still-to-be-identified mammalian CES-2 homologues to contribute to programmed cell death by acting as trans-repressors of this pathway.
Acknowledgments
We thank R. Ashmun for assistance with flow cytometric analysis; B. Jones, H. Yang, and K. Dittmer for technical assistance; F. Rauscher III for providing the pMT-CB6+ expression vector; J. Gilbert for scientific editing; and T. Curran and C. Sherr for comments on the manuscript. This research was supported by grants from the National Cancer Institute (CA 59571, CA 20180, and Cancer Center Core CA 21765) and the National Institute of Diabetes and Digestive and Kidney Diseases (DK 44158 and DK 43025), and by the American Lebanese Syrian Associated Charities, St. Jude Children’s Research Hospital.
ABBREVIATIONS
- IL-3
interleukin 3
- bZIP
basic region/leucine zipper
- PAR
proline- and acidic amino acid-rich
- NFIL3
nuclear factor regulated by IL-3
- E4BP4
adenovirus E4 promoter binding protein
- HLF
hepatic leukemia factor
- DBP
albumin gene promoter D-box binding protein
- TEF
thyrotroph embryonic factor
- CES-2
cell death specification protein of Caenorhabditis elegans
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
References
- 1.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 2.Yang E, Korsmeyer S J. Blood. 1996;88:386–401. [PubMed] [Google Scholar]
- 3.Strasser A. Curr Opin Immunol. 1995;7:228–234. doi: 10.1016/0952-7915(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 4.Schrader J W. Annu Rev Immunol. 1986;4:205–230. doi: 10.1146/annurev.iy.04.040186.001225. [DOI] [PubMed] [Google Scholar]
- 5.Palacios R, Henson G, Steinmetz M, McKearn J P. Nature (London) 1984;309:126–131. doi: 10.1038/309126a0. [DOI] [PubMed] [Google Scholar]
- 6.Williams G T, Smith C A, Spooncer E, Dexter T M, Taylor D R. Nature (London) 1990;343:76–79. doi: 10.1038/343076a0. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Tarduchy G, Collins M, Lopez-Rivas A. EMBO J. 1990;9:2997–3002. doi: 10.1002/j.1460-2075.1990.tb07492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inaba T, Roberts W M, Shapiro L H, Jolly K W, Raimondi S C, Smith S D, Look A T. Science. 1992;257:531–534. doi: 10.1126/science.1386162. [DOI] [PubMed] [Google Scholar]
- 9.Hunger S P, Ohyashiki K, Toyama K, Cleary M L. Genes Dev. 1992;6:1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- 10.Inaba T, Inukai T, Yoshihara T, Seyschab H, Ashmun R A, Canman C E, Laken S J, Kastan M B, Look A T. Nature (London) 1996;382:541–544. doi: 10.1038/382541a0. [DOI] [PubMed] [Google Scholar]
- 11.Metzstein M M, Hengartner M O, Tsung N, Ellis R E, Horvitz H R. Nature (London) 1996;382:545–547. doi: 10.1038/382545a0. [DOI] [PubMed] [Google Scholar]
- 12.Inaba T, Shapiro L H, Funabiki T, Sinclair A E, Jones B G, Ashmun R A, Look A T. Mol Cell Biol. 1994;14:3403–3413. doi: 10.1128/mcb.14.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunger S P, Brown R, Cleary M L. Mol Cell Biol. 1994;14:5986–5996. doi: 10.1128/mcb.14.9.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falvey E, Fleury-Olela F, Schibler U. EMBO J. 1995;14:4307–4317. doi: 10.1002/j.1460-2075.1995.tb00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller C R, Maire P, Schibler U. Cell. 1990;61:279–291. doi: 10.1016/0092-8674(90)90808-r. [DOI] [PubMed] [Google Scholar]
- 16.Fonjallaz, P., Ossipow, V., Wanner, G. & Schibler, U. (1996) EMBO J. 351–356. [PMC free article] [PubMed]
- 17.Drolet D W, Scully K M, Simmons D M, Wegner M, Chu K T, Swanson L W, Rosenfeld M G. Genes Dev. 1991;5:1739–1753. doi: 10.1101/gad.5.10.1739. [DOI] [PubMed] [Google Scholar]
- 18.Cowell I G, Skinner A, Hurst H C. Mol Cell Biol. 1992;12:3070–3077. doi: 10.1128/mcb.12.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowell I G, Hurst H C. Nucleic Acids Res. 1994;22:59–65. doi: 10.1093/nar/22.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Zhang J, Kornuc M, Kwan K, Frank R, Nimer S D. Mol Cell Biol. 1995;15:6055–6063. doi: 10.1128/mcb.15.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKearn J P, McCubrey J, Fagg B. Proc Natl Acad Sci USA. 1985;82:7414–7418. doi: 10.1073/pnas.82.21.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Courey A J, Tjian R. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 23.Chen W J, Lewis K S, Chandra G, Cogswell J P, Stinnett S W, Kadwell S H, Gray J G. Biochim Biophys Acta. 1995;1264:388–396. doi: 10.1016/0167-4781(95)00182-4. [DOI] [PubMed] [Google Scholar]
- 24.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 25.Rennick D, Jackson J, Moulds C, Lee F, Yang G. J Immunol. 1989;142:161–166. [PubMed] [Google Scholar]
- 26.Palacios R, Steinmetz M. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 27.Grimaldi J C, Meeker T C. Blood. 1989;73:2081–2085. [PubMed] [Google Scholar]
- 28.Meeker T C, Hardy D, Willman C, Hogan T, Abrams J. Blood. 1990;76:285–289. [PubMed] [Google Scholar]
- 29.Henthorn P, Kiledjian M, Kadesch T. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 30.Quong M W, Massari M E, Zwart R, Murre C. Mol Cell Biol. 1993;13:792–800. doi: 10.1128/mcb.13.2.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aronheim A, Shiran R, Rosen A, Walker M D. Proc Natl Acad Sci USA. 1993;90:8063–8067. doi: 10.1073/pnas.90.17.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis R E, Horvitz H R. Development (Cambridge, UK) 1991;112:591–603. doi: 10.1242/dev.112.2.591. [DOI] [PubMed] [Google Scholar]