Abstract
The functional specialization or redundancy of the ubiquitous 14-3-3 proteins constitutes a fundamental question in their biology and stems from their highly conserved structure and multiplicity of coexpressed isotypes. We address this question in vivo using mutations in the two Drosophila 14-3-3 genes, leonardo (14-3-3ζ) and D14-3-3ε. We demonstrate that D14-3-3ε is essential for embryonic hatching. Nevertheless, D14-3-3ε null homozygotes survive because they upregulate transcripts encoding the LEOII isoform at the time of hatching, compensating D14-3-3ε loss. This novel homeostatic response explains the reported functional redundancy of the Drosophila 14-3-3 isotypes and survival of D14-3-3ε mutants. The response appears unidirectional, as D14-3-3ε elevation upon LEO loss was not observed and elevation of leo transcripts was stage and tissue specific. In contrast, LEO levels are not changed in the wing disks, resulting in the aberrant wing veins characterizing D14-3-3ε mutants. Nevertheless, conditional overexpression of LEOI, but not of LEOII, in the wing disk can partially rescue the venation deficits. Thus, excess of a particular LEO isoform can functionally compensate for D14-3-3ε loss in a cellular-context-specific manner. These results demonstrate functional differences both among Drosophila 14-3-3 proteins and between the two LEO isoforms in vivo, which likely underlie differential dimer affinities toward 14-3-3 targets.
A fundamental issue concerning members of highly conserved protein families is the extent to which they are functionally redundant or exhibit specialized biological functions. The 14-3-3 proteins compose a highly conserved family of acidic molecules present in all eukaryotes (Aitken 1995; Wang and Shakes 1996; Rosenquist et al. 2000). 14-3-3's share a common structure composed of nine antiparallel α-helices forming a horseshoe shape with a negatively charged interior surface (Fu et al. 2000; Tzivion et al. 2001; Aitken et al. 2002; Bridges and Moorehead 2005; Van Heudsen 2005; Coblitz et al. 2006). Interactions among particular amino acids in the first helix, with ones in helix 2 and helix 3 of another monomer, promote dimerization (Luo et al. 1995; Xiao et al. 1995; Fu et al. 2000; Van Heudsen 2005). Dimerization generates a tandem binding surface, which can simultaneously bind to one or two sites on one target protein or to sites on two different client molecules. The dimers bind clients containing phosphoserine- or phosphothreonine-containing motifs via highly conserved amino acids within the groove (Muslin et al. 1996; Yaffe and Elia 2001; Tzivion and Avruch 2002). 14-3-3 proteins can also bind targets with surfaces outside the conserved phosphopeptide-binding cleft (Benton et al. 2002; Wilker et al. 2005). 14-3-3 binding may allosterically stabilize conformational changes, leading to activation or deactivation of the target or to interaction between two proteins (Yaffe 2002). Furthermore, 14-3-3 binding may mask or expose interaction sites, often leading to changes in the subcellular localization of client proteins (Van Hemert et al. 2001; Aitken et al. 2002; Bridges and Moorehead 2005; Van Heudsen 2005).
An extraordinary feature of this protein family is the high sequence conservation among isotypes, characterized by long stretches of invariant amino acids (Wang and Shakes 1996; Gardino et al. 2006), suggesting functional redundancy. However, despite this extensive sequence identity, multiple 14-3-3 proteins exist in metazoans, indicating at least some functional specificity. Vertebrates contain seven distinct protein isotypes, β, ε, ζ, γ, η, θ, and σ (Aitken et al. 1995). In vertebrate brains where these proteins are highly abundant, there is some specificity in isotype distribution, but generally 14-3-3's are expressed in complex overlapping patterns (Martin et al. 1994; Baxter et al. 2002). In addition, multiple heterodimers are possible in tissues that contain more than one isotype (Jones et al. 1995). It is unclear whether the presence of multiple highly similar proteins with overlapping distribution reflects functional differences among them or represents a mechanism to ensure that ample functionally redundant 14-3-3's are available to mediate the multiple essential cellular functions that require them (Van Heudsen 2005). Thus, the question of 14-3-3 functional specificity in vivo is fundamental in understanding their biology. The highly overlapping isotype distribution in vertebrate models hinders systematic investigation of this question.
To address the issue of functional specificity in vivo, we used Drosophila melanogaster, which offers a simple, but representative, genetically tractable metazoan system. It is simple because it contains only two 14-3-3 genes, an ortholog of the mammalian 14-3-3ζ (88% identity) leonardo and an ortholog of the ε isotype, D14-3-3ε (Skoulakis and Davis 1998). It is representative because the two fly genes belong to the two different 14-3-3 conservation groups (Wang and Shakes 1996; Skoulakis and Davis 1998). leonardo encodes two nearly identical protein isoforms (LEO I and LEO II) via alternative splicing of the primary transcript (Kockel et al. 1997; Philip et al. 2001), with modest tissue specificity (Philip et al. 2001). In contrast, D14-3-3ε encodes a single protein (Chang and Rubin 1997), apparently present in all developmental stages and tissues examined with only slight enrichment in the adult brain (Tien et al. 1999; Philip et al. 2001).
Maternal LEO is required for normal chromosome separation during syncytial mitoses, whereas D14-3-3ε appears required to time them, suggesting distinct functions for the two 14-3-3's in the single-celled syncytial embryo (Su et al. 2001). Maternal LEO is also essential for early Raf-dependent decisions that pattern the embryo (Li et al. 1997). Zygotic leo loss-of-function mutants exhibit functional impairments of their embryonic and adult nervous system (Skoulakis and Davis 1996; Broadie et al. 1997; Philip et al. 2001). D14-3-3ε functions in photoreceptor formation and appears involved in development of the wing (Chang and Rubin 1997), but whether it is important for the function of the nervous system is unknown. LEO and D14-3-3ε appear at least partially redundant for photoreceptor formation (Karim et al. 1996; Chang and Rubin 1997). Furthermore, LEO and D14-3-3ε have been reported to function redundantly in anterior–posterior axis formation of the developing oocyte (Benton et al. 2002) and follicle cell polarity (Benton and St Johnston 2003).
Nevertheless, three reasons motivated a systematic investigation of potential functional specificity of the two Drosophila 14-3-3 isotypes by searching for isotype-specific phenotypes. First, studies to date used a transposon allele of D14-3-3ε (j2B10), which may not be a null allele. In fact, although D14-3-3ε has been reported dispensable for viability (Chang and Rubin 1997), a lethal deficiency uncovering this gene was used to show its involvement in Raf-mediated developmental processes in the embryo (Li et al. 2000). Second, leo mutations are homozygous lethal, suggesting that D14-3-3ε cannot functionally compensate for its loss, although LEO was suggested to at least partially compensate for the lack of D14-3-3ε in embryonic development (Chang and Rubin 1997). Third, the dynamic expression pattern of 14-3-3's during embryonic development and larval and adult nervous systems (Skoulakis and Davis 1996; Tien et al. 1999; Philip et al. 2001) suggested involvement in additional processes other than photoreceptor and oocyte development, which may specifically require one but not the other. Our results demonstrate 14-3-3-isotype-specific functions and a tissue- and temporal-specific transcriptional mechanism to compensate for loss of D14-3-3ε and suggest dynamic temporal and spatial interactions of the two 14-3-3 isotypes.
MATERIALS AND METHODS
Drosophila culture and strains:
Drosophila were cultured in standard wheat–flour–sugar food supplemented with soy flour and CaCl2 at 21°–23°, unless specified otherwise. The D14-3-3εl(3)j2B10 mutant allele, which contains a P-transposon in intron 1 of the gene, has been described previously (Chang and Rubin 1997). Alleles D14-3-3εex5, D14-3-3εex4, and D14-3-3εex24 generated by mobilization of the transposon in D14-3-3εl(3)j2B10 were a kind gift of Henry Chang and G. Rubin. The genetic background of these alleles was normalized using balancer chromosomes in a Cantonized w1118 background for D14-3-3εex5, D14-3-3εex4, and D14-3-3εex24. In contrast, free recombination for six generations following the transposon-borne w+ as a selectable marker was allowed for D14-3-3εl(3)j2B10. Allelism was assessed by complementation tests of alleles normalized over the balancer with D14-3-3εl(3)j2B10 recovered after normalization. The lethal leo12X and leoP1188 alleles have been described previously (Broadie et al. 1997; Philip et al. 2001) and were normalized to the Cantonized w1118 genetic backround using balancer chromosomes.
Complementation tests for viability and wing cross-vein deficits were performed by crossing parents of the appropriate genotypes en masse and scoring the progeny of multiple such crosses per genotype. Viability was measured as the percentage of mutant homozygotes recovered from a cross of balanced parents, relative to the expected number if the homozygotes were fully viable. The expected number of homozygotes, if fully viable, was estimated as one-third of the total progeny recovered because homozygotes for the balancer chromosomes die as embryos. To rescue lethality with heat-shock (HS)-inducible transgenes, crosses were performed and animals were raised to adulthood in programmable cycling incubators (Labline) as described (Philip et al. 2001) or at constant 18° and 23°. Rescue for viability or cross-vein deficits was calculated as the percentage of expected homozygous individuals that increased upon transgene expression over that obtained from the same strain in the absence of transgene [(% viable induced) − (% viable baseline)/(100 − % viable baseline)]. Cross-vein deficit rescue was scored similarly. Each cross was repeated minimally four independent times and the data were pooled.
To determine the lethal phase of null homozygotes, embryos were collected from D14-3-3εl(3)j2B10/TM3SerGFP and D14-3-3εex4/TM3SerGFP flies and manually separated into green fluorescent protein (GFP) fluorescence negative (homozygous mutant) and GFP fluorescence positive. Homozygotes for the balancers were avoided on the basis of their much more intense fluorescence. After hatching, they were monitored in separate food vials until emergence of adult flies at which time their genotype was verified again on the basis of adult visible markers.
The hsleoI, hsleoII, and UAS-mycD14-3-3ε transgenic strains have been described before (Philip et al. 2001; Chen et al. 2003). To generate hsD14-3-3ε, the entire D14-3-3ε cDNA (Chang and Rubin 1997) including the 3′ untranslated region was placed into the P{CaSpeRHS} vector (Bourgouin et al. 1992) and multiple transformant lines on different chromosomes, were obtained. Insertions on the third chromosome were selected and recombined onto the D14-3-3εl(3)j2B10- and D14-3-3εex4-bearing chromosomes with standard crosses. To generate UASleoI and UASleoII, the entire leo open reading frame was inserted in pUAST (Brand and Perrimon 1993) and multiple transformant lines were obtained. Again, insertions on the third chromosome were selected and recombined onto the D14-3-3εl(3)j2B10- and D14-3-3εex4-bearing chromosomes.
Immunohistochemistry:
Embryos were collected on apple juice plates, dechorionated, and fixed in 43.2 mm HEPES, 0.96 mm MgSO4, 0.48 mm EGTA, pH 6.9, 1.6% formaldehyde in 59% heptane, followed by rinses in methanol, 5% EGTA. The embryos were rehydrated to BBT (140 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.4 mm KH2PO4, pH 7.3, 0.1% Tween-20, 1%, bovine serum albumin) and blocked for 1 hr in BBT-250 (BBT, 250 mm NaCl), 10% normal goat serum. Incubation with primary antibodies in 5% normal goat serum BBT-250 was as follows: chicken anti-D14-3-3ε, 1:3000; mAb-22c10, 1:2000 [Developmental Hybridoma Studies Bank (DSHB), University of Iowa, Iowa City, IA]; mAb anti-FASIII, 1:10 (7G10-DSHB); mAb anti-NEUROTACTIN, 1:200 (BP106-DSHB); and rabbit polyclonal anti-MEF2, 1:1000 (Nguyen and Xu 1998). Fluorescent (Molecular Probes, Eugene, OR) and HRP-conjugated (Jackson Immunochemicals) secondary antibodies were used at 1:2000. Homozygous embryos were identified on the basis of their lack of signal against the balancer-chromosome-borne GFP. Embryos homozygous for the balancer were avoided on the basis of their abnormal appearance. Anti-GFP antibodies were a rabbit polyclonal, 1:40 (Santa Cruz), and a mAb 1:2000 (Molecular Probes). Images were captured on a Zeiss Axiovert 200 microscope.
Wing mounting:
Wings were dissected in 95% ethanol and placed in xylene for 10 min, washed twice with ethanol, and mounted in Canada balsam (C-1795, Sigma, St. Louis). Images were captured on a Zeiss Axiovert 35 microscope using a ×20 objective lens.
Western blot analysis:
To obtain extracts from homozygous embryos, GFP fluorescence-negative embryos were hand selected from eggs laid by D14-3-3εl(3)j2B10/TM3SerGFP and D14-3-3εl(3)j2B10/TM3SerGFP parents. Sibling GFP fluorescence-positive heterozygous embryos were selected as controls because they fluoresced and appeared normal. Homozygotes for the balancers were not used and were identified on the basis of their more intense fluorescence and abnormal appearance relative to heterozygotes. The fidelity of the embryonic genotype based on the above criteria was verified on similarly selected embryos by immunohistochemistry. Single flies or an embryo equivalent to three fly heads per lane from control and mutant animals was homogenized in 10 μl of modified radioimmunoprecipitation assay buffer as previously described (Philip et al. 2001). Blots were rabbit anti-LEO, 1:40,000; chicken anti-D14-3-3ε, 1:5000; mAb antitubulin, 1:300 (E7-DSHB); mAb antisyntaxin, 1:500 (8C3-DSHB); and anti-cMyc, 1:200 (9E10-DSHB). Secondary antibodies were used 1:15,000 for anti-rabbit HRP, 1:5000 for anti-chicken HRP, and 1:5000 for anti-mouse HRP and the results were visualized with enhanced chemiluminescence (Pierce, Rockford, IL). The results of at least three independent experiments utilizing different extract preparations were quantified densitometrically and analyzed statistically.
The chicken anti-D14-3-3ε antibody was generated by immunizing hens (Charles River Laboratories) with a his-tagged, bacterially expressed fragment of the D14-3-3ε protein containing the amino-terminal 130 amino acids. IgY was purified from eggs using standard procedures (Charles River Laboratories). Eggs from two different hens yielded antibodies with nearly identical properties, but one of them was used throughout these experiments. The specificity of the anti-D14-3-3ε antibodies was tested against recombinant D14-3-3ε and D14-3-3ζ (LEO) (Skoulakis and Davis 1996) and fly lysates.
Reverse transcription–polymerase chain reaction analysis and quantitative PCR:
Hand-selected embryos and larval wing disk and brain samples were prepared and reverse transcription–polymerase chain reaction (RT–PCR) reactions with leoI, leo II, and D14-3-3ε primers were performed as previously described (Philip et al. 2001). As an internal control, forward and reverse act5C primers were used to quantify the relative amount of RNA in each sample. To identify hsleoI, the leoI forward primer was used with SV40-specific reverse primer and, for hsleoII, the leoII forward primer was used with a hsp70-specific reverse primer. For the quantitative RT–PCR experiments, newly hatched larvae were hand selected on the basis of their lack of GFP fluorescence, and 1 μg of RNA (Philip et al. 2001) was subjected to reverse transcription; the product was diluted 1:100 and 4 μl were used per PCR reaction. Each reverse transcription was sampled four times per PCR run and five independent experiments were performed. leoI, leoII, D14-3-3ε, and act5C primers were used as described above. A calibration curve was constructed for each run and used to fit the values (Pfaffl 2001). Relative quantification was performed using the MJ Opticon Monitor Analysis software (v3.1), with the relative quantification method ΔΔCt (“Guide to Performing Relative Quantification of Gene Expression using Real-Time Quantitative PCR,” Applied Biosystems, Foster City, CA).
Statistical analysis:
Untransformed data from densitometric quantification of protein amounts and the results of cell-counting experiments and complementation tests were analyzed using the JMP3.1 statistical software package (SAS Institute, Cary, NC). Following initial ANOVA, the data were analyzed by Student's t-tests or planned comparisons to a control (Dunnett's test) where appropriate.
RESULTS
Loss of D14-3-3ε compromises viability:
To unequivocally determine whether D14-3-3ε is required for viability, we sought to identify null alleles by characterizing derivatives of transposon mobilization from D14-3-3εl(3)j2B10 (Chang and Rubin 1997). Southern analysis (not shown) demonstrated that D14-3-3εex4 harbors a small deletion removing the first exon and part of the first intron of the gene. Allele D14-3-3εex24 results from a large deletion (>10 kb) extending beyond the D14-3-3ε coding region and likely encompasses at least part of the CG7156 and CG18598 transcription units on either side of the gene (Figure 1A). In contrast, excision of the transposon in D14-3-3εex5 did not result in obvious DNA rearrangements. Furthermore, genomic PCR and high-resolution acrylamide electrophoresis of the DNA flanking the transposon insertion from D14-3-3εex5 homozygotes did not indicate size differences from the w1118 control (not shown). These results, in addition to the full viability of D14-3-3εex5 homozygotes (Table 1) and the lack of the visible phenotypes exhibited by D14-3-3εl(3)j2B10, D14-3-3εex4 homozygotes, suggest that D14-3-3εex5 represents a precise excision allele (Figure 1A). In accord with these results, D14-3-3ε protein was detected in D14-3-3εex5 homozygotes, but it was undetectable in D14-3-3εl(3)j2B10, D14-3-3εex4 homozygotes and heteroallelics with D14-3-3εex24 (Figure 1B). Therefore, by molecular criteria, D14-3-3εex4 and D14-3-3εex24 represent null alleles. Although D14-3-3εl(3)j2B10 lacks detectable protein in these assays, we consider it a strong hypomorph on the basis of the genetic data below.
Figure 1.—
D14-3-3ε mutations and their effects on protein accumulation. (A) The genomic region and mutations of the D14-3-3ε gene. Exons are represented by solid boxes and introns and surrounding nontranscribed regions by lines. The P-element insertion in intron 1 is indicated by the arrow. The deleted DNA in D14-3-3εex4 and D14-3-3εex24 is indicated by the lines flanked by shaded boxes representing regions of uncertainty at the ends of the deficiencies. A perpendicular line indicates the precise excision of the j2B10 transposon in the revertant allele D14-3-3εex5. (B) Mutant homozygotes and heteroallelic combinations yield adult animals lacking D14-3-3ε protein demonstrated by semiquantitative Western blot analysis of whole-animal lysates of the indicated genotypes. The neuronal protein syntaxin (SYX) was used to control for the amount loaded per lane. ex5 stands for D14-3-3εex5, j2B10 for D14-3-3εl(3)j2B10, ex4 for D14-3-3εex4, and ex24 for D14-3-3εex24.
TABLE 1.
Complementation for viability of D14-3-3ε mutants
| Genotype | % viable | n |
|---|---|---|
| D14-3-3εex5/D14-3-3εex5 | 100 | 550 |
| D14-3-3εex5/D14-3-3εl(3)j2B10 | 100 | 413 |
| D14-3-3εex5/D14-3-3εex4 | 100 | 546 |
| D14-3-3εex5/D14-3-3εex24 | 100 | 660 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10 | 75 | 645 |
| D14-3-3εl(3)j2B10/D14-3-3εex4 | 71 | 510 |
| D14-3-3εl(3)j2B10/D14-3-3εex24 | 61 | 510 |
| D14-3-3εex4/D14-3-3εex4 | 42 | 495 |
| D14-3-3εex4/D14-3-3εex24 | 39 | 684 |
| D14-3-3εex24/D14-3-3εex24 | 0 | 650 |
D14-3-3ɛex4 and D14-3-3ɛex24 are novel mutant alleles of D14-3-3ɛ. Viability was calculated as the fraction of adults of each genotype recovered from crosses of balanced individuals over that expected if the mutant homozygotes or heteroallelics were fully viable. n denotes the total number of flies scored per cross.
Although null, homozygotes for the D14-3-3εl(3)j2B10 and D14-3-3εex4 alleles were recovered with lower frequency than expected if fully viable. This observation and the fact that the original D14-3-3εl(3)j2B10 chromosome was associated with a lethal mutation (Chang and Rubin 1997) motivated us to perform complementation tests to determine whether D14-3-3ε is dispensable for viability. To avoid complications, chromosomes bearing D14-3-3ε mutations were introduced into our isogenized w1118 background (see materials and methods). Even in the normalized genetic background, a fraction of the expected D14-3-3εl(3)j2B10 and D14-3-3εex4 homozygotes and heteroallelics were recovered (Table 1). Thus, the reduced viability phenotype is fully recessive, maps exclusively to mutations in the D14-3-3ε gene, and does not appear to be modified by extragenic mutations. Although protein was not detectable in D14-3-3εl(3)j2B10 adult homozygotes, the allele appears to be hypomorphic because of the larger number of D14-3-3εl(3)j2B10 homozygotes and D14-3-3εl(3)j2B10/D14-3-3εex4 heteroallelics recovered compared to D14-3-3εex4 homozygotes. Because homozygotes were never recovered, the D14-3-3εex24 deficiency appears to disrupt neighboring gene(s), as suggested by the molecular data (Figure 1), and was excluded from further analyses.
The reduction in the number of D14-3-3εex4 and D14-3-3εl(3)j2B10 homozygotes was fully rescued by induction of hsD14-3-3ε transgenes (Table 2). We used two independent transgenic lines, the high-expressing hsD14-3-3εH and lower-expressing hsD14-3-3εL (supplemental Figure 1 at http://www.genetics.org/supplemental/) with similar results. Lower, yet significant rescue, especially for D14-3-3εl(3)j2B10 homozygotes, was obtained when the animals were raised at 23°, a consequence of high basal transgene expression (supplemental Figure 1 at http://www.genetics.org/supplemental/). For transgene-carrying mutant animals raised at 18°, the number of homozygotes was similar to that obtained from mutants without the transgene. These results confirm that D14-3-3ε loss results in significantly reduced viability. Given the “leakiness” of the transgenes, to verify that it was indeed elevation of the transgenic protein that rescued the phenotype, we placed UAS-mycD14-3-3ε transgenes into D14-3-3εex4 and D14-3-3εl(3)j2B10 mutant backgrounds. Ubiquitous expression of UAS-mycD14-3-3ε transgenes with the tubPGal4 driver fully rescued the lethality of D14-3-3εex4 (Table 2) and D14-3-3εl(3)j2B10 homozygotes (not shown).
TABLE 2.
Transgenic rescue of D14-3-3ε mutant lethality
| Genotype | Temperature | % viable | % rescue | n |
|---|---|---|---|---|
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10 | 18° | 75 | — | 645 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10 | 23° | 77 | 8 | 526 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10 | HS | 78 | 12 | 522 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10, hsD14-3-3ε | 18° | 74 | — | 462 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10, hsD14-3-3ε | 23° | 81 | 27a | 487 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10, hsD14-3-3ε | HS | 99 | 96.2a | 508 |
| D14-3-3εex4/D14-3-3εex4 | 18° | 42 | — | 382 |
| D14-3-3εex4/D14-3-3εex4 | 23° | 45 | 5 | 411 |
| D14-3-3εex4/D14-3-3εex4 | HS | 45 | 5 | 409 |
| D14-3-3εex4/D14-3-3εex4, hsD14-3-3ε | 18° | 40 | — | 495 |
| D14-3-3εex4/D14-3-3εex4, hsD14-3-3ε | 23° | 44 | 7 | 488 |
| D14-3-3εex4/D14-3-3εex4, hsD14-3-3ε | HS | 92 | 86.7a | 488 |
| D14-3-3εex4, tubPGal4/D14-3-3εex4 | 25° | 39 | — | 245 |
| UASmycD14-3-3εL/+ ; D14-3-3εex4, tubPGal4/D14-3-3εex4 | 25° | 87 | 78.6a | 568 |
| UASmycD14-3-3εH/+ ; D14-3-3εex4/D14-3-3εex4 | 25° | 42 | 5 | 268 |
| UASmycD14-3-3εH/+ ; D14-3-3εex4, tubPGal4/D14-3-3εex4 | 25° | 100 | 100a | 592 |
| elavGal4/+; UASmyc D14-3-3εL/+ ; D14-3-3εex4/D14-3-3εex4 | 25° | 97 | 95a | 812 |
| +/+; UASmyc D14-3-3εH/+ ; D14-3-3εex4/D14-3-3εex4 | 25° | 44 | 8.2 | 327 |
| elavGal4/+; UASmyc D14-3-3εH/+ ; D14-3-3εex4/D14-3-3εex4 | 25° | 100 | 100a | 825 |
Transgenic reversal of D14-3-3ɛ homozygous mutant lethality. All transgenic strains and controls were grown under the three regimes indicated under “Temperature”—constant 18°, constant 23°, and HS conditions of constant 23° with three daily 30-min, 32° heat shocks. All Gal4 driven crosses were performed at constant 25°. Viability was calculated as the fraction of adults of each genotype recovered from crosses of balanced individuals over that expected if the mutant homozygotes were fully viable. “% rescue” was calculated as the percentage increase in mutant homozygotes carrying transgenes over the “baseline” number (denoted by “—”) obtained at 18° for experiments employing HS-inducible transgenes. n denotes the total number of flies scored per cross. Significant rescue was not observed in the absence of the transgenes although a few more homozygotes were obtained under HS conditions.
Significant rescue.
These results indicate that loss of D14-3-3ε results in significantly reduced survival (58% of D14-3-3εex4 homozygotes die). Therefore, the protein is required for complete viability in contrast to previous reports suggesting that the gene is not essential (Chang and Rubin 1997). In contrast, null alleles of the Drosophila 14-3-3ζ gene leonardo are fully lethal when homozygous (Skoulakis and Davis 1996; Broadie et al. 1997).
Morphological characterization of D14-3-3ε homozygous mutant embryos:
We examined the fate of homozygous embryos to determine when D14-3-3ε mutants die. They were identified because they lacked the fluorescence of the balancer-chromosome-borne GFP. Clearly, 100% of null embryos that hatched successfully proceeded to adulthood (Table 3), as their number [72% for D14-3-3ε(3)j2B10 and 40% for D14-3-3εex4] reflected that of the adult homozygotes typically recovered. Therefore, D14-3-3ε does not appear to be required for vital functions in the larval and pupal stages, but it is critical at the time of hatching. In agreement, embryos that failed to hatch remained alive for an additional 6–12 hr as indicated by their occasional peristaltic movements and, if manually removed from the chorion, many survived to adulthood.
TABLE 3.
Lethal phase of D14-3-3ε null homozygotes
| Genotype | % hatching | % eclosed | n |
|---|---|---|---|
| D14-3-3εl(3)j2B10/TM3SerGFP | 100 | 100 | 228 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10 | 72 | 100 | 203 |
| D14-3-3εex4/TM3SerGFP | 100 | 100 | 309 |
| D14-3-3εex4/D14-3-3εex4 | 40 | 100 | 216 |
D14-3-3ε homozygous mutants die as embryos. The genotypes of the embryos were ascertained on the basis of the GFP fluorescence as described in materials and methods and verified upon eclosure of adults. “% hatching” indicates the percentage of embryos yielding larvae, while “% eclosed” denotes the percentage of hatched embryos that became adults. n denotes the total number of embryos assayed per genotype.
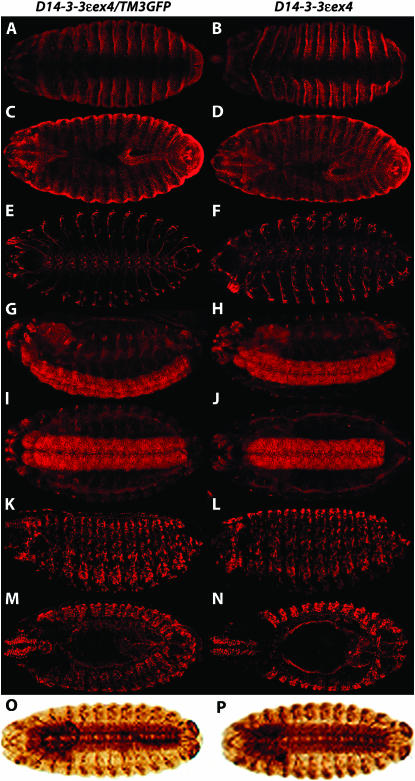
To determine whether null embryos failed to hatch because of developmental defects, we subjected them to immunohistochemical analysis. Most D14-3-3ε mutant embryos appeared smaller in size, but staining with anti-FASIII did not reveal gross morphological changes (Figure 2, A–D). Because we hypothesized that the inability to hatch could be a reflection of neuro-developmental deficits since both D14-3-3ε and LEO are abundant in this tissue (Skoulakis and Davis 1996; Tien et al. 1999), we focused on the nervous system. Staining of D14-3-3εex4 homozygotes with mAb22c10 and anti-NEUROTACTIN did not reveal morphological changes in the central nervous system (CNS) and peripheral nervous system (PNS) (Figure 2, E–J) or in the musculature revealed by anti-MEF staining (Figure 2, K–N). To ascertain that we did not focus on embryos that appeared normal because they would be among ones that hatch successfully, embryos that failed to hatch 24–26 hr post-egg laying (PEL) were collected and stained with anti-FASIII and mAb22c10. These D14-3-3εex4 homozygotes appeared identical to controls as well (Figure 2, O and P). Similar results were obtained with D14-3-3εl(3)j2B10 homozygotes (not shown). Therefore, lack of D14-3-3ε did not result in gross developmental aberrations and homozygotes died as apparently fully formed larvae.
Figure 2.—
Morphology of D14-3-3ε homozygous mutant embryos. Embryos (16–18 hr old) are shown. Anterior is to the left. The genotype ascertained by concurrent staining with anti-GFP (see materials and methods) is shown on top of the two columns. A and B are ventral views, while C and D are dorsal views of embryos stained with anti-FASIII. There were no obvious gross morphological defects. E and F are ventral views of control and mutant embryos, respectively, stained with mAb22c10, and not showing overall deficits in CNS and PNS morphology. This was further demonstrated in homozygotes that failed to hatch (P), compared to heterozygotes 20 hr post-egg laying (O). The CNS and, to a lesser degree, the PNS were also examined with anti-neurotactin. I and J are lateral views, while K and L are ventral views and, in agreement with E and F, do not exhibit obvious structural differences of the CNS (and PNS) in mutant homozygotes compared to their heterozygous siblings. Similar results were obtained with homozygous embryos that failed to hatch (not shown). K and L are lateral views and M and N are dorsal views of embryos stained with anti-MEF-2, which failed to reveal significant changes in the musculature of the mutants.
To determine whether functional deficits of the nervous system, akin to those described for leo nulls (Broadie et al. 1997), result in the observed failure of D14-3-3ε mutant homozygotes to hatch, we targeted transgene expression to the nervous system. UAS-mycD14-3-3ε transgenes driven specifically in the nervous system with the elavGal4 driver (Robinow and White 1988) were strikingly efficient at increasing the number of D14-3-3εex4 homozygotes recovered (Table 2). Thus, lethality of D14-3-3ε mutant homozygotes results from failure of the nervous system to support normal hatching and is consistent with the described preferential distribution of the protein in the CNS and PNS of late embryos (Tien et al. 1999).
Upregulation of LEO in D14-3-3ε mutant embryos:
D14-3-3εex4 or D14-3-3εl(3)j2B10 homozygotes were not recovered if the animals were also made heterozygous for a strong leo mutant allele (Chang and Rubin 1997; E. M. C. Skoulakis, unpublished results). This observation suggested that LEO may compensate for the loss of D14-3-3ε during embryonic development and this may be responsible for the recovery of homozygous mutants. To address this hypothesis, lysates from 18- to 20-hr-old homozygous embryos, hand selected on the basis of their lack of GFP fluorescence, were subjected to semiquantitative Western blot analysis. The representative results in Figure 3A and quantified in Figure 3B show that in homozygous null embryos there was a highly significant increase in the amount of LEO compared to that in sibling heterozygotes or control animals. Because we could not distinguish which of the embryos would hatch and survive, it was not possible to determine whether this increase characterized all embryos, homozygotes destined to die, or only potential survivors. If the latter is the case, then we underestimated the actual level of LEO in survivors because the lysates included embryos destined to die where this elevation may not occur.
Figure 3.—
Elevation of LEO in D14-3-3ε homozygous mutant embryos. (A) A representative blot of embryonic lysates used in acquisition of the data on B. The genotypes of the embryos whose lysates were blotted are indicated on top of the blot: ex5/ex5 for D14-3-3εex5/D14-3-3εex5; j2B10/+ for D14-3-3εl(3)j2B10/D14-3-3εex5; ex4/+ for D14-3-3εex4/D14-3-3εex5; j2B10/j2B10 for D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10; and ex4/ex4 for D14-3-3εex4/D14-3-3εex4. These abbreviations are used in A–D. β-Tub denotes β-tubulin, the protein used to normalize the lanes for the amount loaded. (B) The average ratios (± standard error of the mean or SEM) of the relative levels of LEO/β-Tub and D14-3-3ε/β-Tub is shown from four individual blots similar to the one displayed in A. Ratios are shown relative to those obtained from D14-3-3εex5 homozygotes, which were arbitrarily set to 1. The level of LEO accumulation was significantly higher (**P < 0.001) in D14-3-3εl(3)j2B10 and D14-3-3εex4 homozygotes compared to D14-3-3εex5 controls. (C) A representative blot of embryonic lysates of the indicated genotypes prepared at the particular times PEL. The latest collection was at 24 hr for D14-3-3εex4 homozygotes because of their hatching delay, while the latest time point for control embryos was immediately before hatching at 22 hr. (D) The average ratio of relative levels of LEO/β-Tub ± SEM for D14-3-3εex4 homozygotes compared to D14-3-3εex5 controls estimated from three independent blots similar to the one shown in C. There is a highly significant increase (**P < 0.001) in the amount of LEO in D14-3-3εex4 homozygotes during their 2-hr hatching delay. A smaller increase (*P < 0.05) in D14-3-3εex4 homozygotes was detected at 22-hr PEL in comparison to D14-3-3εex5 controls of the same age. Samples from D14-3-3εex4 homozygotes were not collected at 20-hr PEL and control samples could not be collected at 24-hr PEL because the embryos had hatched to larvae.
To determine when this LEO elevation occurs, we examined lysates from tightly staged embryos. Mutant embryos were individually selected from half-hour egg collections, dechorionated, and visually inspected under the microscope to ascertain stage homogeneity before preparation of lysates. We quantified LEO levels in later stages of embryogenesis relative to maternally provided protein (Li et al. 1997; Philip et al. 2001), which we found relatively invariable over many different experiments (not shown). Thus, compared to 1- to 3-hr control embryos, LEO levels were significantly higher in 22-hr D14-3-3εex4 homozygotes and nearly doubled during their 2-hr hatching delay (Figure 3, C and D). Similar results were obtained with D14-3-3εl(3)j2B10 homozygotes. Therefore, elevation of LEO in D14-3-3ε null embryos occurs late in embryogenesis, particularly during the hatching delay exhibited by mutant homozygotes. These results are consistent with two notions. First, elevation of LEO in D14-3-3ε mutant homozygotes may be the reason a fraction hatches and represents a compensatory mechanism for the loss of D14-3-3ε. Alternatively, LEO elevation occurs in embryos unable to hatch and may represent a stress response that characterizes dying or dead embryos similar to the reported postmortem elevation of certain vertebrate 14-3-3 isotypes (Foundoulakis et al. 2001).
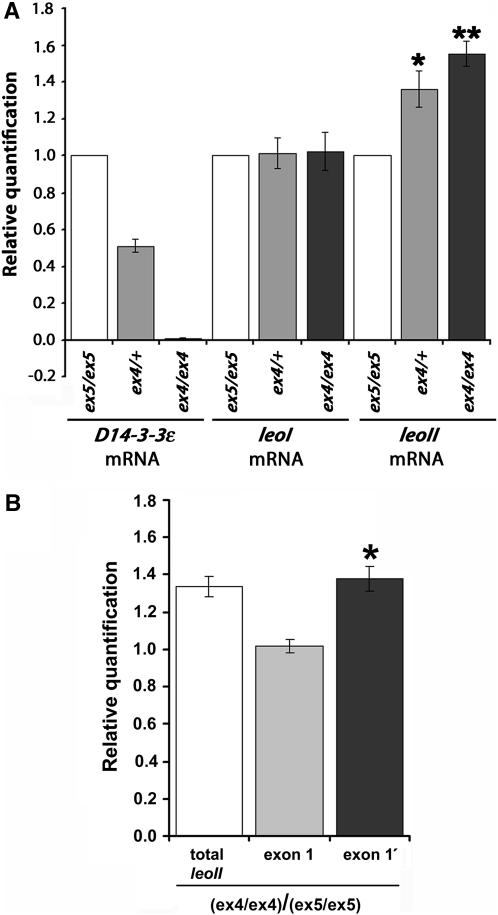
To investigate whether elevation of LEO was a consequence of increased transcription in D14-3-3ε mutants, we estimated the relative levels of the two leo transcripts by quantitative PCR. LEOI and LEOII differ by five amino acids encoded in the alternative, mutually exclusive exons 6 and 6′ (Philip et al. 2001). We used newly hatched homozygous mutant larvae for this quantification because, if detectable, it would indicate that leo elevation occurs in animals that hatch. The 50% reduction in D14-3-3ε transcripts in heterozygotes and the lack of transcripts in homozygous mutants were easily detectable with our experimental conditions (Figure 4A). In these animals, the level of leoI transcripts remained unchanged, but we detected a significant increase in the level of leoII mRNA in heterozygotes and D14-3-3εex4 homozygous mutant larvae. The leo gene contains two alternative 5′ untranslated exons (exon 1 and 1′). Whereas leoI transcripts appear to always utilize exon 1′, leoII transcripts contain either the distal exon 1′ or the proximal exon 1 (Kockel et al. 1997), suggesting differential use of promoters. We investigated whether both alternate exons are utilized in D14-3-3εex4 homozygotes to elevate leoII transcripts. However, the ratio of exon 1-containing transcripts in D14-3-3εex4 homozygotes to those present in control animals remained unchanged (1.015 ± 0.037). In contrast, the number of exon 1′-containing transcripts in the mutants was significantly higher (Figure 4B). This suggests that elevation of leoII transcripts in D14-3-3εex4 homozygotes involves increased utilization of the putative proximal promoter and differential inclusion of exon 1′ in these transcripts. These data strongly suggest that elevation of LEO protein levels are a consequence of upregulation of leoII transcripts. Since this is detected in mutant first instar larvae, the data suggest that elevation of leoII transcripts allows hatching and survival in the fraction of D14-3-3ε homozygotes where it occurs.
Figure 4.—
Elevation of leoII transcripts in D14-3-3εex4 mutant homozygotes. (A) The ratios of D14-3-3ε/act5C, leoI/act5C, and leoII/act5C RT–PCR products in control animals were arbitrarily set to 1 (open bars) and their relative levels in D14-3-3εex4 heterozygotes (shaded bars) and homozygotes (solid bars) were determined. The mean ± SEM of five independent experiments is shown. leoI levels relative to those of act5C were not found significantly different in mutant heterozygotes and homozygotes. In contrast, the relative levels of leoII mRNAs were significantly higher than controls in both mutant heterozygotes (*P < 0.01) and homozygotes (**P < 0.001). (B) The ratios of leoII transcripts in D14-3-3εex4 homozygotes over those of control animals [(ex4/ex4)/(ex5/ex5)] determined in experiments independent from those in A. Elevation of leoII in the mutants is detected because the ratio for “total leoII” is >1. Using primers specific to exon 1 and exon 1′, the levels of transcripts that include exon 1′ in the mutants were found significantly higher (*P < 0.01) than transcripts that include exon 1. The mean ± SEM of four independent experiments is shown.
Conditional overexpression of leo transgenes rescues lethality of D14-3-3ε null homozygotes:
To rigorously test the hypothesis that elevation of LEOII is the reason for hatching and survival of mutant homozygotes, leoI and leoII transgenes were recombined onto the chromosomes bearing the D14-3-3εex4 and D14-3-3εl(3)j2B10 mutations. If endogenous LEOII elevation suffices for successful hatching of D14-3-3ε homozygotes, then further increasing the level of this protein should increase the number of homozygous mutant animals recovered. hsleoI and hsleoII transgenes exhibited leaky expression at 25°, but expression was higher after three daily inductions (HS) and undetectable if flies were kept at 18° (supplemental Figure 1 at http://www.genetics.org/supplemental/). Significantly, two independent insertions of a hsleoII trangene were able to conditionally increase (rescue) the number of D14-3-3εex4 and D14-3-3εl(3)j2B10 homozygotes recovered (Table 4). In contrast, hsleoI transgenes rescued the phenotype only partially, despite the similarity in hsleoI and hsleoII transgene expression (supplemental Figure 1).
TABLE 4.
Rescue of D14-3-3ε lethality with leo transgenes
| Genotype | Temperature | % viable | % rescue | n |
|---|---|---|---|---|
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10 | 18° | 75 | — | 645 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10 | 23° | 77 | 8 | 526 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10 | HS | 78 | 12 | 522 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10, hsleoIL | 18° | 76 | — | 462 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10, hsleoIL | 23° | 84 | 33 | 455 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10, hsleoIL | HS | 88 | 50 | 524 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10, hsleoIIL | 18° | 78 | — | 442 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10, hsleoIIL | 23° | 95 | 77a | 492 |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10, hsleoIIL | HS | 100 | 100a | 425 |
| D14-3-3εex4/D14-3-3εex4 | 18° | 42 | — | 382 |
| D14-3-3εex4/D14-3-3εex4 | 23° | 45 | 5.5 | 411 |
| D14-3-3εex4/D14-3-3εex4 | HS | 45 | 5.5 | 409 |
| D14-3-3εex4/D14-3-3εex4, hsleoIL | 18° | 49 | — | 481 |
| D14-3-3εex4/D14-3-3εex4, hsleoIL | 23° | 56 | 14 | 440 |
| D14-3-3εex4/D14-3-3εex4, hsleoIL | HS | 87 | 74.5a | 493 |
| D14-3-3εex4/D14-3-3εex4, hsleoIH | 18° | 51 | — | 415 |
| D14-3-3εex4/D14-3-3εex4, hsleoIH | 23° | 60 | 18.5 | 433 |
| D14-3-3εex4/D14-3-3εex4, hsleoIH | HS | 85 | 69a | 495 |
| D14-3-3εex4/D14-3-3εex4, hsleoIIL | 18° | 49 | — | 496 |
| D14-3-3εex4/D14-3-3εex4, hsleoIIL | 23° | 69 | 39a | 534 |
| D14-3-3εex4/D14-3-3εex4, hsleoIIL | HS | 100 | 100a | 555 |
| D14-3-3εex4/D14-3-3εex4, hsleoIIH | 18° | 56 | — | 512 |
| D14-3-3εex4/D14-3-3εex4, hsleoIIH | 23° | 91 | 76.5a | 534 |
| D14-3-3εex4/D14-3-3εex4, hsleoIIH | HS | 100 | 100a | 485 |
| D14-3-3εex4/D14-3-3εex4,UASleoI | 25° | 37 | — | 230 |
| elavGal4/+ ; D14-3-3εex4/D14-3-3εex4,UASleoI | 25° | 63 | 41a | 331 |
| D14-3-3εex4/D14-3-3εex4,UASleoII | 25° | 34 | — | 248 |
| elavGal4/+; D14-3-3εex4/D14-3-3εex4,UASleoII | 25° | 100 | 100a | 380 |
leo transgenes rescue the lethality of D14-3-3ε mutant homozygotes. The temperature conditions employed for the crosses are shown under “Temperature.” Under HS conditions, flies were raised under constant 23°, except for three daily 30-min, 32° heat shocks. Viability was calculated as the fraction of adults of each genotype recovered from crosses of balanced individuals over that expected if the mutant homozygotes were fully viable. “% rescue” was calculated as the increase in mutant homozygotes carrying transgenes over the “baseline” number (—). The superscripts H and L for leoI and leoII transgenes denote high- and low-expressing transgenes, respectively. n denotes the total number of flies scored per cross that yielded the mutant homozygotes carrying D14-3-3ε or leo transgenes.
Significant rescue.
Importantly, when driven by the neuronal-specific Gal4 driver elav, UASleoII transgenes rescued fully and UASleoI partially, the lethality of D14-3-3εex4 homozygotes, consistent with the notion that they die because their nervous system is unable to support hatching (Table 4). This is not the result of differences in transgene expression levels, since independent leoI and leoII transgenes inducible by different means yielded similar outcomes. These data suggest that LEOII and, to a lesser degree, LEOI are functionally redundant with D14-3-3ε in the nervous system and confirm that LEOII elevation in a fraction of late D14-3-3ε mutant embryos allows them to hatch. Furthermore, the data indicate that, despite the minor, largely conservative differences (Philip et al. 2001), the two LEO isoforms are not equivalent in compensating for the lack of D14-3-3ε. Expression of leo transgenes in the nervous system is not ectopic, as LEO accumulates abundantly in this tissue (Skoulakis and Davis 1996; Broadie et al. 1997).
The level of D14-3-3ε is not increased in homozygous leo null embryos:
Is the level of one 14-3-3 isotype always elevated when the other is reduced during embryogenesis? Like null D14-3-3ε embryos, homozygotes for the strong hypomorphic transposon insertion allele leoP1188 die as fully formed larvae, while the null leo12X homozygotes exhibit deficits on their dorsal side including incomplete closure (Skoulakis and Davis 1996; Broadie et al. 1997). Semiquantitative Western blot analysis indicated that the level of D14-3-3ε remained relatively unchanged in homozygous leoP1188 and leo12X mutant embryos in comparison to their heterozygous siblings (Figure 5). LEO elevation was again readily detectable in D14-3-3εex4 homozygotes. Therefore, we could not detect reciprocal elevation of D14-3-3ε upon loss of LEO in embryos.
Figure 5.—
D14-3-3ε is not elevated in leo homozygous mutant embryos. The average ratios (±SEM) of LEO/β-TUB and D14-3-3ε/β-TUB are shown from three individual experiments. Ratios are shown relative to those obtained from D14-3-3εex5/D14-3-3εex5 embryos, which were arbitrarily set to 1. The genotypes of the embryos whose lysates were blotted are ex5/ex5 for D14-3-3εex5/D14-3-3εex5; ex4/+ for D14-3-3εex4/D14-3-3εex5; ex4/ex4 for D14-3-3εex4/D14-3-3εex4, whereas full genotypes are shown for leo mutants. Compared to that in D14-3-3εex5 homozygotes, the level of LEO accumulation was significantly higher (**P < 0.001) in D14-3-3εex4 homozygotes and significantly reduced (**P < 0.001) in leo1188 homozygotes. The level of D14-3-3ε was also significantly reduced in late D14-3-3εex4 homozygous embryos.
The level of LEO is unchanged in D14-3-3ε adult heads:
Does LEO remain elevated in adult D14-3-3ε mutant homozygotes? Is D14-3-3ε elevated in adult leo mutant heterozygotes? To address these questions, we determined the relative levels of LEO in isolated heads of D14-3-3εex4 homozygotes and heterozygotes and of D14-3-3ε in leo mutant heterozygotes because both proteins are enriched in adult brains (Skoulakis and Davis 1996; S. F. Acevedo, unpublished observations). Although the 50% reduction of D14-3-3ε in D14-3-3εex4 heterozygotes and leo1188/+; D14-3-3εex4/+ animals was readily detectable, LEO levels in D14-3-3εex4 homozygotes were not significantly different from D14-3-3εex5 (Student's t-tests, P = 0.8678) or from w1118 controls (Figure 6). Similar results were obtained for D14-3-3εl(3)j2B10 homozygotes (supplemental Figure 2 at http://www.genetics.org/supplemental/). Thus, LEO appears to be elevated only in D14-3-3ε mutant embryos around the time of hatching, and therefore it is unlikely that it functionally compensates for D14-3-3ε loss in all tissues. Similarly, D14-3-3ε is not elevated in the heads of leo mutant heterozygotes.
Figure 6.—
LEO is not significantly elevated in the heads of adult D14-3-3ε homozygous mutants, and the level of D14-3-3ε is not changed in the heads of leo mutant heterozygotes. (A) The average ratios (±SEM) of LEO/SYX and D14-3-3ε/SYX is shown from three individual Western blotting experiments, one of which is shown in B. Ratios are shown relative to those obtained from D14-3-3εex5/D14-3-3εex5 adults, which were arbitrarily set to 1. Compared to the levels in D14-3-3εex5/D14-3-3εex5 controls, D14-3-3ε was significantly reduced (*P < 0.01) in D14-3-3εex4/D14-3-3εex5 (ex4/+), leoP1188/+; D14-3-3εex4/+ (leoP1188/+; ex4/+) double heterozygotes and D14-3-3εex4/D14-3-3εex4 (ex4/ex4) homozygotes (**P < 0.001). Similarly, LEO was significantly (*P < 0.01) reduced in leoP1188/+ and leoP1188/+; D14-3-3εex4/+ animals. However, LEO was not significantly elevated in the heads of D14-3-3εex4 homozygotes or D14-3-3ε in the heads of leoP1188 heterozygotes. (B). A representative blot of head lysates from the indicated genotypes quantified in A. The neuronal protein SYNTAXIN (Syx) was utilized to normalize the amount of each lysate loaded.
Loss of D14-3-3ε disrupts wing cross-vein formation:
Adult D14-3-3εex4 and D14-3-3εl(3)j2B10 homozygotes have smaller wings than control flies. Cell counts along the longitudinal veins of homozygous and heteroallelic adults indicated a 10% proportional reduction in length compared to D14-3-3εex5 controls (not shown). This may be a consequence of the overall body-size reduction also observed in null homozygous and heteroallelic embryos (Figure 2), larvae, and adults. In addition, the majority of adult D14-3-3εex4 and D14-3-3εl(3)j2B10 homozygotes exhibited a conspicuous lack of the dorsal part of the posterior cross-vein and, with lesser penetrance, malformation of the anterior cross-vein (Figure 7, A.2–A.5). These defects map to D14-3-3ε, as all mutant animals exhibited the phenotype (Table 5A). Furthermore, posterior and anterior cross-vein deficits can be rescued with hsD14-3-3ε transgenes (Figure 7, A.6 and A.7; Table 5B), demonstrating that cross-vein formation indeed requires D14-3-3ε. We quantified deficits of the posterior cross vein because it exhibited greater penetrance and therefore afforded more sensitivity to rescue experiments and is reported on the “% rescue” column in Table 5, B and C. Significant changes in the fraction of wings that exhibited anterior cross-vein malformations are denoted by footnote a in Table 5.
Figure 7.—
Deficits in cross-vein formation of D14-3-3ε mutants and transgenic rescue by D14-3-3ε and leo transgenes. (A) Posterior cross veins are indicated by arrows, while anterior cross veins are indicated by arrowheads in 1 and 6. Genotypes are 1, D14-3-3εex5/D14-3-3εex5; 2, D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10; 3, D14-3-3εex4/D14-3-3εex4; 4, D14-3-3εex4/D14-3-3εex5; 5, D14-3-3εl(3)j2B10/D14-3-3εex4 heteroallelic, exhibiting anterior cross-vein deficits also; 6, D14-3-3εex4/D14-3-3εex4, hsD14-3-3ε raised under HS conditions; 7, D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10, hsD14-3-3ε raised under HS conditions; 8, D14-3-3εex4/D14-3-3εex4, hsD14-3-3ε raised at 18°; 9, D14-3-3εex4/D14-3-3εex4, hsleoI raised under HS conditions; 10, D14-3-3εex4/D14-3-3εex4, hsleoII raised under HS conditions; 11, A D14-3-3εex4/D14-3-3εex4, hsleoII raised under HS conditions where both anterior and posterior cross veins remained defective. (B) Products of a RT–PCR experiment with RNA from larval wing disks and brains with primers specific for leoI and leoII transcripts (bottom) and amplification of act5C transcripts (top amplicon) as controls for the quality of the transcription. leoII is not expressed in larval wing disks. (C) Products of RT–PCR with transgene-specific primers (Philip et al. 2001), indicating that under HS conditions both leoI and leoII transgenes are expressed in dissected wing disks.
TABLE 5.
Wing cross-vein deficits of D14-3-3ε mutants and transgenic rescue
| Genotype | % anterior malformed | % posterior malformed | % rescue |
|---|---|---|---|
| A. | |||
| D14-3-3εex5/D14-3-3εex5 | 0 | 0 | — |
| D14-3-3εex5/D14-3-3εl(3)j2B10 | 0 | 0 | — |
| D14-3-3εex5/D14-3-3εex4 | 0 | 0 | — |
| D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10 | 25.0 | 75.0 | — |
| D14-3-3εl(3)j2B10/D14-3-3εex4 | 26.7 | 82.4 | — |
| D14-3-3εex4/D14-3-3εex4 | 42.9 | 80.9 | — |
| B. | |||
| D14-3-3εex4/D14-3-3εex4, hsD14-3-3εL (18°) | 40.6 | 83 | — |
| D14-3-3εex4/D14-3-3εex4, hsD14-3-3εL | 9.3a | 14a | 82.3 |
| D14-3-3εex4/D14-3-3εex4, hsD14-3-3εH (18°) | 41.8 | 79.5 | — |
| D14-3-3εex4/D14-3-3εex4, hsD14-3-3εH | 0a | 3.6a | 95.6 |
| C. | |||
| D14-3-3εex4/D14-3-3εex4, hsleoIL (18°) | 43.2 | 82.2 | — |
| D14-3-3εex4/D14-3-3εex4, hsleoIL | 29a | 72 | 11 |
| D14-3-3εex4/D14-3-3εex4, hsleoIH (18°) | 41.3 | 80.9 | — |
| D14-3-3εex4/D14-3-3εex4, hsleoIH | 0a | 60 | 25.8 |
| D14-3-3εex4/D14-3-3εex4, hsleoIIL (18°) | 43.4 | 82.6 | — |
| D14-3-3εex4/D14-3-3εex4, hsleoIIL | 40 | 78.4 | 3.1 |
| D14-3-3εex4/D14-3-3εex4, hsleoIIH (18°) | 41.5 | 81.8 | — |
| D14-3-3εex4/D14-3-3εex4, hsleoIIH | 33 | 76.8 | 5.1 |
The wing cross-vein deficits are rescued by conditional hsD14-3-3ε expression, but not by leo transgenes. (A) Percentage of D14-3-3ε controls and mutant homozygotes that exhibited posterior and anterior cross-vein deficits. Posterior and anterior cross-vein deficits were counted on the same wings and one wing was scored per individual (n > 100). (B) The percentage of D14-3-3εex4/D14-3-3εex4, hsD14-3-3ε exhibiting wing cross-vein deficits raised under conditions of transgene silence (18°) or induction. The “% rescue” indicates the percentage decrease in individuals with deficient wings and is shown only for posterior cross veins. (C). The percentage of D14-3-3εex4/D14-3-3εex4 individuals expressing leoI and leoII transgenes exhibiting wing cross-vein deficits raised under conditions of transgene silence (18°) or induction. The “% rescue” indicates the percentage decrease in individuals with deficient wings and is shown only for posterior cross veins.
Large changes in the percentage of deficient wings (rescue).
Overexpression of leoI partially rescues the wing-venation deficits of D14-3-3ε mutants:
Can LEO compensate for the D14-3-3ε requirement in cross-vein formation as it did for hatching? leoI, but not leoII, is expressed in wing disks (Figure 7B), whereas transcripts for both isoforms could be detected in brains from the same w1118 larvae as reported previously (Philip et al. 2001). To determine whether LEO compensates D14-3-3ε loss in wing cross-vein formation, we examined the wings of D14-3-3εex4 homozygotes rescued from lethality by hsleoI and hsleoII transgenes. Both hsleoI and hsleoII transgenes were expressed in the wing disks after heat shock (Figure 7C). However, although hsleoII is efficient at rescuing lethality, the wings of the same rescued mutant homozygotes retained the posterior cross-vein deficit (Figure 7A.10) and often the anterior cross-vein remained malformed (Figure 7A.11). In contrast, a 25% reduction in posterior cross-vein deficits and a complete rescue of anterior cross-vein malformation was observed in animals rescued from lethality with hsleoI transgenes (Table 5). Similar results were obtained with D14-3-3εl(3)j2B10 homozygotes expressing hsleoI and hsleoII transgenes (not shown). These data indicate that LEOI and D14-3-3ε are partially redundant in processes required for posterior cross-vein formation. Moreover, LEOII is much more inefficient in compensating D14-3-3ε loss in anterior cross-vein formation. Therefore, the two LEO isoforms again are not equivalent in their ability to substitute for the loss of D14-3-3ε.
DISCUSSION
D14-3-3ε is an essential gene:
Our results utilizing null alleles indicate that D14-3-3ε is not dispensable for viability, but its loss is partially compensated by elevation of endogenous leo levels. Consequently, homozygotes survive to adulthood, whose number is higher when the hypomorphic allele D14-3-3εl(3)j2B10 is used. This is the likely reason for the suggestion of previous reports that the gene is not essential (Chang and Rubin 1997; Benton et al. 2002). This interaction is uncovered genetically by the inability to obtain D14-3-3εex4 and D14-3-3εl(3)j2B10 homozygotes when one copy of leo is mutated (i.e., leoP1188/+; D14-3-3εl(3)j2B10/D14-3-3εl(3)j2B10 animals). Embryos homozygous for mutant alleles do not exhibit obvious morphological defects (Figure 2) because maternally provided D14-3-3ε is likely sufficient to fulfill its requirement in syncytial cellular blastoderm and gastrulating animals (Tien et al. 1999; Philip et al. 2001; Su et al. 2001). D14-3-3ε mutant homozygotes die ostensibly because lack of zygotic protein from the nervous system renders them unable to hatch. Similarly, LEO accumulates in embryonic motor neurons innervating the body-wall musculature and its loss in leo mutants is the likely reason for their failure to hatch despite their apparently normal progression through development (Broadie et al. 1997).
14-3-3 homeostasis:
Our results demonstrate that LEOII overaccumulates in late D14-3-3ε null embryos and that this elevation allows a fraction of them to hatch and survive. The conclusion is supported by the striking increase in the number of D14-3-3ε mutant homozygotes that survive upon expression of leoII transgenes in the nervous system (Table 4). Because endogenous LEOII accumulates preferentially in the CNS (Broadie et al. 1997; Philip et al. 2001), our data suggest that its elevation in this tissue leads to successful hatching and survival of D14-3-3ε mutant homozygotes. This “homeostatic” response in D14-3-3ε mutants is specific to late embryogenesis after the maternally supplied D14-3-3ε, which perdures almost until stage 8 (S. F. Acevedo and K. Tsigkari, unpublished results), has decayed. Therefore, the response appears specific to a period when the overall level of either 14-3-3's or D14-3-3ε, specifically, is critically important for survival.
It appears that a mechanism sensing the absence of D14-3-3ε operates in embryos and responds by increasing the level of LEOII. Congruent with this, LEO elevation was not observed in embryos homozygous for the dominant-negative allele D14-3-3εE183K (Chang and Rubin 1997), which compromises D14-3-3ε functionally, but does not change its overall levels in the embryo (supplemental Table 1 at http://www.genetics.org/supplemental/). It is possible that this is the reason that D14-3-3εE183K homozygotes are never recovered. Furthermore, this response appears specific to the loss of D14-3-3ε, because levels of this protein remained normal in homozygous leo mutant embryos and adults (Figures 4 and 5), consistent with their strong lethal phenotype. Clearly, this sensing mechanism responds by increased accumulation of leoII transcripts by preferential utilization of one of two possible promoters and splicing of the primary transcript to include the leoII-specific exon 6′ (Kockel et al. 1997; Philip et al. 2001). How is lack of D14-3-3ε sensed and how could leoII transcription be increased? 14-3-3's have been reported to participate in nuclear/cytoplasmic trafficking of transcription factors (Brunet et al. 2002; Zhao et al. 2004; Berdichevsky and Guarente 2006). Therefore, it is possible that loss of D14-3-3ε enhances transcription from the proximal promoter of the leo gene by not mediating nuclear export of a factor that binds that site. Alternatively, D14-3-3ε may be part of a repressing complex and, upon its loss, transcription from this site is enhanced. In contrast, excessive transgenic elevation in the amount of D14-3-3ε results in recovery of few adults (<10% of expected) homozygous for strong hypomorphic leo mutations (K. Tsigkari and E. M. C. Skoulakis, unpublished results). This suggests that although D14-3-3ε can at least partially compensate for the loss of LEO in high concentrations, an endogenous molecular mechanism to elevate it in leo homozygotes does not appear to exist.
Although leoI transcripts accumulate in the wing disk, LEO does not appear to play a role in wing-vein formation because animals that develop with as low as 10% of normal LEO do not exhibit wing aberrations (Philip et al. 2001). Therefore, the venation deficits are a phenotype specific to D14-3-3ε mutant homozygotes. Interestingly, in congruence with the mechanism proposed above, the leoI transcripts normally expressed in that tissue were not upregulated and leoII transcripts were not ectopically transcribed in D14-3-3ε mutant homozygote wing disks (S. F. Acevedo, unpublished observations). This is because the proposed D14-3-3ε-interacting factor(s) required for exon 1′-containing leoII transcription are likely absent from the wing disk where these transcripts do not normally accumulate. Exon 1-containing leoII transcripts do not appear to require such D14-3-3ε-interacting factor(s), since these transcripts were not upregulated in embryos. Therefore, loss of D14-3-3ε does not alter the tissue specificity of leo transcriptional regulation and specific isoform accumulation.
It is presently unclear whether this compensatory mechanism is operant in other systems where mutant analyses of 14-3-3's have been initiated. Interestingly, single nulls of either 14-3-3-encoding gene in Saccharomyces cerevisiae are viable, while the double mutant is lethal (Roberts et al. 1997) and similar results were obtained for the two Schizosaccharomyces pombe genes (Ford et al. 1994). These observations may reflect similar 14-3-3 “homeostatic” mechanisms in these species. Directed reduction of specific 14-3-3 protein levels during Xenopus laevis development yielded gastrulation and patterning defects for all proteins tested except for 14-3-3ζ (Lau et al. 2006). Unlike Drosophila, Xenopus 14-3-3ζ may not be essential for development, but it is also possible that loss of this isotype is specifically compensated for by elevation of the remaining 14-3-3's. Such mechanisms, if extant in mammals, are likely to hinder genetic analysis of 14-3-3 function, especially in the brain where all family members are expressed (Baxter et al. 2002). Interestingly, mice mutant for 14-3-3ε exhibit severe brain abnormalities and die perinatally, yet a small fraction survive to adulthood appearing smaller, but otherwise normal (Toyo-oka et al. 2003), much like the Drosophila mutants. It is unknown whether 14-3-3ζ or other isotypes are elevated in these animals as predicted by our results.
Functional specificity and redundancy of 14-3-3's:
Functional specificity of 14-3-3 family members may be the result of tissue or temporal-specific gene expression and regulation or of isotype-specific ligand selectivity. Isotypes may have redundant functions within a cell if they are able to interact with the same targets. Even then, affinity differences toward common ligands predicted by their amino-acid sequence and tertiary structure (Gardino et al. 2006) may functionally differentiate coexpressed 14-3-3's.
Although both leoI and leoII transgenes rescued the lethality of D14-3-3ε mutants, they clearly exhibited different efficiency (Table 4). Rescue was invariably higher upon accumulation of LEOII either ubiquitously or specifically in the nervous system. However, rescue required excessive accumulation of LEOII to overcome loss of D14-3-3ε. In fact, the two- to threefold LEO elevation shown in Figure 3B could be as much as a 50–60% underestimate of the level of this protein in D14-3-3ε mutant embryos that hatch. Therefore, a large excess of LEO appears to be necessary to functionally substitute D14-3-3ε in the embryonic nervous system, which may be attained only in a small number of mutant homozygotes. This probably reflects the affinity differences that LEO dimers exhibit toward client proteins normally bound either by D14-3-3ε homodimers or by D14-3-3ε/LEO heterodimers. If so, then even a small amount of D14-3-3ε would increase the number of mutant homozygotes obtained. In agreement with this, more homozygotes were recovered from the transposon allele D14-3-3εl(3)j2B10, which likely contains residual D14-3-3ε (Table 1). Differences in ligand binding between LEOI and LEOII are likely reflected in the large difference with which the two isoforms rescue the lethality of D14-3-3ε mutants. This is the first unequivocal demonstration of functional differences between LEOI and LEOII. These differences must reside in the five unique amino acids of helix 6 that distinguish the two isoforms (Philip et al. 2001). It is unknown whether LEOI, LEOII, or both contribute to the reported redundancy with D14-3-3ε in photoreceptor development and oocyte polarity (Chang and Rubin 1997; Benton et al. 2002; Benton and St Johnston 2003).
Interestingly, the functional redundancy of LEO isoforms with D14-3-3ε is tissue specific. In contrast to the embryonic nervous system, enhanced accumulation of LEOI, and not of LEOII, was able to compensate for anterior cross-vein deficits and partially for the posterior cross vein (Table 5). Again, this suggests that D14-3-3ε ligands in the wing disk necessary for cross-vein formation can be targeted by excess LEOI (and not LEOII). Hence, LEOII can be redundant with D14-3-3ε specifically in the embryonic nervous system where it is presumed to accumulate preferentially, while in the wing disk LEOI, which is normally found in this tissue, is the potential compensating isoform. Similarly, although the two S. cerevisiae 14-3-3 genes are functionally redundant for viability, only one, Bmh1p, is required for efficient forward transport to the endoplasmic reticulum (Michelsen et al. 2006). Thus, redundancy of 14-3-3's largely depends on the specific function and interacting proteins that they engage within a particular tissue or developmental context.
Collectively, our data show a tissue- and temporal-specific upregulation of leo transcription that can account for the apparent functional redundancy between LEO and D14-3-3ε with respect to the lethality of D14-3-3ε mutants and possibly other processes requiring these proteins. In addition, this analysis for the first time demonstrates tissue and temporal functional differences between the two LEO isoforms. Whether these functional differences and the functional redundancy among the Drosophila 14-3-3's will also occur in the adult nervous system where they are all most abundant is currently unknown. A previous study failed to uncover differences between LEOI and LEOII with respect to learning and memory (Philip et al. 2001). Nevertheless, our data strongly support the notion that the existence of multiple 14-3-3 isotypes in metazoans reflects a combination of tissue- and temporal-specific isotype expression, localization, and functional specialization. Importantly, this analysis indicates that understanding the biological roles of 14-3-3's will require identification of proteins engaged by homo- and heterodimers of particular composition in a tissue- and temporal-specific manner.
Acknowledgments
The authors are indebted to H. Chang and G. Rubin for providing mutant stocks and plasmids, to C.-T. Chien for providing an anti-D14-3-3ε antibody used in initial phases of this work, to Charity Moore for help with analysis of the wing phenotype, to Mary McCrady, Courtney Swayze, Alexandros Kanellopoulos, and Maria Anezaki for stock maintenance and husbandry, and to M. Franco-Redrejo for invaluable help with quantitative RT–PCR. We thank Hanh Nguyen for anti-MEF2 antibodies, the Developmental Hybridoma Studies Bank at the University of Iowa (Iowa City, IA) for monoclonal antibodies, and Terry Orr-Weaver and Sumana Datta for valuable discussions and suggestions. This work was supported by a European Commission Marie Curie grant (IRG-003570), by PENED (01EΔ207) from the Greek General Secretariat for Research and Technology, and by the National Science Foundation (grant IBN-0080687).
References
- Aitken, A., 1995. 14–3-3 proteins on the MAP. Trends Biochem. Sci. 20: 95–97. [DOI] [PubMed] [Google Scholar]
- Aitken, A., S. Howell, D. Jones, J. Madrazo and Y. Patel, 1995. 14–3-3 α and δ are the phosphorylated forms of Raf-activating 14–3-3 β and ζ. J. Biol. Chem. 270: 5706–5709. [DOI] [PubMed] [Google Scholar]
- Aitken, A., H. Baxter, T. Dubios, S. Clokie, S. Mackie et al., 2002. 14–3-3 proteins in cell regulation. Biochem. Soc. Trans. 30: 351–360. [DOI] [PubMed] [Google Scholar]
- Baxter, H. C., W.-G. Liu, J. L. Forster, A. Aitken and J. R. Fraser, 2002. Immunolocalisation of 14–3-3 isoforms in normal and scrapie-infected murine brain. Neuroscience 109: 5–14. [DOI] [PubMed] [Google Scholar]
- Benton, R., and D. St Johnston, 2003. Drosophila PAR-1 and 14–3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell 115: 691–704. [DOI] [PubMed] [Google Scholar]
- Benton, R., I. M. Palacios and D. St. Johnston, 2002. Drosophila 14–3-3/PAR-5 is an essential mediator of PAR1 function in axis formation. Dev. Cell 3: 659–671. [DOI] [PubMed] [Google Scholar]
- Berdichevsky, A., and L. Guarente, 2006. A stress response pathway involving sirtuins, forkheads and 14–3-3 proteins. Cell Cycle 5: 2588–2591. [DOI] [PubMed] [Google Scholar]
- Bourgouin, C., S. E. Lundgren and J. B. Thomas, 1992. Apterous is a Drosophila LIM domain gene required for the development of a subset of embryonic muscles. Neuron 9: 549–561. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Bridges, D., and G. B. G. Moorehead, 2005. 14–3-3 proteins: a number of functions for a numbered protein. Sci. STKE 2005: re10. [DOI] [PubMed] [Google Scholar]
- Broadie, K., E. Rushton, E. M. C. Skoulakis and R. L. Davis, 1997. Leonardo, a Drosophila 14–3-3 protein involved in learning, regulates presynaptic function. Neuron 19: 391–402. [DOI] [PubMed] [Google Scholar]
- Brunet, A., F. Kanai, J. Stehn, J. Xu, D. Sarbassova et al., 2002. 14–3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 156: 817–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H. C., and G. M. Rubin, 1997. 14–3-3ε positively regulates Ras mediated signaling in Drosophila. Genes Dev. 11: 1132–1139. [DOI] [PubMed] [Google Scholar]
- Chen, H.-K., P. Fernandez-Funez, S. F. Acevedo, Y. C. Lam, M. D. Kaytor et al., 2003. Interaction of akt-phosphorylated ataxin-1 with 14–3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell 113: 457–468. [DOI] [PubMed] [Google Scholar]
- Coblitz, B., M. Wu, S. Shikano and M. Li, 2006. C-terminal binding: an expanded repertoire and function of 14–3-3 proteins. FEBS Lett. 580: 1531–1535. [DOI] [PubMed] [Google Scholar]
- Ford, J. C., F. al-Khodairy, E. Fotou, K. S. Sheldrick, D. J. Griffiths et al., 1994. 14–3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science 265: 533–535. [DOI] [PubMed] [Google Scholar]
- Foundoulakis, M., R. Hardmeier, H. Hoger and G. Lubec, 2001. Postmortem changes in the level of brain proteins. Exp. Neurol. 167: 86–94. [DOI] [PubMed] [Google Scholar]
- Fu, H., R. R. Subramanian and S. C. Masters, 2000. 14–3-3 proteins: structure, function and regulation. Annu. Rev. Pharmacol. Toxicol. 40: 617–647. [DOI] [PubMed] [Google Scholar]
- Gardino, A. K., S. J. Smerdon and M. B. Yaffe, 2006. Structural determinants of 14–3-3 binding specificities and regulation of subcellular localization of 14–3-3-ligand complexes: a comparison of the X-ray crystal structures of all human 14–3-3 isoforms. Semin. Cancer Biol. 16: 173–182. [DOI] [PubMed] [Google Scholar]
- Jones, D. H. A., H. Martin, J. Madrazo, K. A. Robinson, P. Neilsen et al., 1995. Expression and structural analysis of 14–3-3 proteins. J. Mol. Biol. 245: 375–384. [DOI] [PubMed] [Google Scholar]
- Karim, F. D., H. C. Chang, M. Therrien, D. A. Wassarman, T. Laverty et al., 1996. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics 143: 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockel, L., G. Vorbraggen, H. Jackle, M. Mlodzik and D. Bohnmann, 1997. Requirement for Drosophila 14–3-3 ζ in cell proliferation and Raf-dependent photoreceptor development. Genes Dev. 11: 1140–1147. [DOI] [PubMed] [Google Scholar]
- Lau, J. M., C. Wu and A. J. Muslin, 2006. Differential role of 14–3-3 family members in Xenopus development. Dev. Dyn. 235: 1761–1776. [DOI] [PubMed] [Google Scholar]
- Li, W., E. M. C. Skoulakis, R. L. Davis and N. Perrimon, 1997. The Drosophila 14–3-3 protein Leonardo enhances Torso signaling through D-Raf in a Ras1-dependent manner. Development 124: 4163–4171. [DOI] [PubMed] [Google Scholar]
- Li, W., E. Noll and N. Perrimon, 2000. Identification of autosomal regions involved in Drosophila Raf function. Genetics 156: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Z., X. Zhang, U. Rapp and J. Avruch, 1995. Identification of the 14–3-3ζ domains important for self-association and Raf binding. J. Biol. Chem. 270: 23681–23687. [DOI] [PubMed] [Google Scholar]
- Martin, H., J. Rostas, Y. Patel and A. Aitken, 1994. Subcellular localisation of 14–3-3 isoforms in rat brain using specific antibodies. J. Neurochem. 63: 2259–2265. [DOI] [PubMed] [Google Scholar]
- Michelsen, K., T. Mrowiec, K. E. Duderstadt, S. Frey, D. L. Minor et al., 2006. A multimeric membrane protein reveals 14–3-3 isoform specificity in forward transport in yeast. Traffic 7: 903–916. [DOI] [PubMed] [Google Scholar]
- Muslin, A. J., J. W. Tanner, P. M. Allen and A. S. Shaw, 1996. Interaction of 14–3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84: 889–897. [DOI] [PubMed] [Google Scholar]
- Nguyen, H. T., and X. Xu, 1998. Drosophila mef2 expression during mesoderm development is controlled by a complex array of cis-acting regulatory modules. Dev. Biol. 204: 550–566. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M. W., 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip, N., S. Acevedo and E. M. C. Skoulakis, 2001. Conditional rescue of olfactory learning and memory defects in mutants of the 14–3-3ζ gene leonardo. J. Neurosci. 21: 8417–8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, R. L., H. U. Mosch and G. R. Fink, 1997. 14–3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 89: 1055–1065. [DOI] [PubMed] [Google Scholar]
- Robinow, S., and K. White, 1988. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev. Biol. 126: 294–303. [DOI] [PubMed] [Google Scholar]
- Rosenquist, M., P. Sehnke, R. J. Ferl, M. Sommarin and C. Larsson, 2000. Evolution of the 14–3-3 protein family: Does the large number of isoforms in multicellular organisms reflect functional specificity? J. Mol. Evol. 51: 446–458. [DOI] [PubMed] [Google Scholar]
- Skoulakis, E. M. C., and R. L. Davis, 1996. Olfactory learning deficits in mutants for leonardo, a Drosophila gene encoding a 14–3-3 protein. Neuron 17: 931–944. [DOI] [PubMed] [Google Scholar]
- Skoulakis, E. M. C., and R. L. Davis, 1998. 14–3-3 proteins in neuronal development and function. Mol. Neurobiol. 16: 269–284. [DOI] [PubMed] [Google Scholar]
- Su, T. T., D. H. Parry, B. Donahoe, C.-T. Chien, P. H. O'Farrell et al., 2001. Cell cycle roles for the two 14–3-3 proteins during Drosophila development. J. Cell Sci. 114: 3445–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien, A.-C., H.-Y. Hsei and C.-T. Chien, 1999. Dynamic expression and cellular localization of the Drosophila 14–3-3e during embryonic development. Mech. Dev. 81: 209–212. [DOI] [PubMed] [Google Scholar]
- Toyo-oka, K., A. Shionoya, M. J. Gambello, C. Cardoso, R. Leventer et al., 2003. 14–3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat. Genet. 274: 285. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., and J. Avruch, 2002. 14–3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277: 3061–3064. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., Y. H. Shen and J. Zuh, 2001. 14–3-3 proteins: bringing new definitions to scaffolding. Oncogene 20: 6331–6338. [DOI] [PubMed] [Google Scholar]
- van Hemert, M. J., H. Y. Steensma and G. P. H. van Heudsen, 2001. 14–3-3 proteins: key regulators of cell division, signalling and apoptosis. BioEssays 23: 936–946. [DOI] [PubMed] [Google Scholar]
- van Heudsen, G. P. H., 2005. 14–3-3 proteins: regulators of numerous eukaryotic proteins. IUBMB Life 57: 623–629. [DOI] [PubMed] [Google Scholar]
- Wang, W., and D. Shakes, 1996. Molecular evolution of the 14–3-3 family. Mol. Evol. 43: 384–398. [DOI] [PubMed] [Google Scholar]
- Wilker, E. W., R. A. Grant, S. C. Artim and M. B. Yaffe, 2005. A structural basis for 14–3-3sigma functional specificity. J. Biol. Chem. 280: 18891–18898. [DOI] [PubMed] [Google Scholar]
- Xiao, B., S. Smerdon, D. H. Jones, G. G. Dodson, Y. Soneji et al., 1995. Structure of a 14–3-3 protein and implications for coordination of multiple signalling pathways. Nature 376: 188–191. [DOI] [PubMed] [Google Scholar]
- Yaffe, M. B., 2002. How do 14–3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513: 53–57. [DOI] [PubMed] [Google Scholar]
- Yaffe, M. B., and A. E. H. Elia, 2001. Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol. 13: 131–138. [DOI] [PubMed] [Google Scholar]
- Zhao, X., L. Gan, H. Pan, D. Kan, M. Majeski et al., 2004. Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: characterization of phosphorylation- and 14–3-3-dependent and -independent mechanisms. Biochem. J. 378: 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]