Abstract
Siz1 and Siz2/Nfi1 are the two Siz/PIAS SUMO E3 ligases in Saccharomyces cerevisiae. Here we show that siz1Δ siz2Δ mutants fail to grow in the absence of the homologous recombination pathway or the Fen1 ortholog RAD27. Remarkably, the growth defects of mutants such as siz1Δ siz2Δ rad52Δ are suppressed by mutations in TOP1, suggesting that these growth defects are caused by topoisomerase I activity. Other mutants that affect SUMO conjugation, including a ulp1 mutant and the nuclear pore mutants nup60Δ and nup133Δ, show similar top1-suppressible synthetic defects with DNA repair mutants, suggesting that these phenotypes also result from reduced SUMO conjugation. siz1Δ siz2Δ mutants also display TOP1-independent genome instability phenotypes, including increased mitotic recombination and elongated telomeres. We also show that SUMO conjugation, TOP1, and RAD27 have overlapping roles in telomere maintenance. Top1 is sumoylated, but Top1 does not appear to be the SUMO substrate involved in the synthetic growth defects. However, sumoylation of certain substrates, including Top1 itself and Tri1 (YMR233W), is enhanced in the absence of Top1 activity. Sumoylation is also required for growth of top1Δ cells. These results suggest that the SUMO pathway has a complex effect on genome stability that involves several mechanistically distinct processes.
POST-TRANSLATIONAL attachment of the ubiquitin-related protein SUMO (small ubiquitin-related modifier) to other proteins is involved in many important biological processes including maintenance of genome integrity, transcriptional regulation, and signal transduction (Gill 2004; Johnson 2004; Müller et al. 2004; Hay 2005). Sumoylation is essential for viability of most eukaryotic cells, including Saccharomyces cerevisiae, but the essential function(s) is unknown.
SUMO conjugation is carried out by a three-step pathway that involves a SUMO-activating enzyme (E1) called Uba2-Aos1, a SUMO-conjugating enzyme (E2) called Ubc9, and one of several SUMO ligases (E3's). SUMO E3's in yeast include the two Siz/PIAS (protein inhibitor of activated STAT) proteins Siz1 and Siz2/Nfi1, Mms21, and the meiotic E3 Zip3/Cst9 (Johnson and Gupta 2001; Takahashi et al. 2001; Zhao and Blobel 2005; Cheng et al. 2006). SUMO is often attached to the side chain of the Lys residue in the consensus motif ΨKXE, where Ψ is a hydrophobic residue. In yeast, two SUMO-specific proteases Ulp1 and Ulp2 remove SUMO from modified proteins. Ulp1 also produces mature SUMO from the SUMO precursor and therefore is required for both sumoylation and desumoylation of proteins. UBA2, AOS1, UBC9, ULP1, and SMT3, the gene encoding SUMO, are all essential genes, while siz1Δ, siz2Δ, ulp2Δ, zip3Δ, and mutants that eliminate the sumoylation activity of MMS21 are viable (Ouspenski et al. 1999; Johnson 2004; Zhao and Blobel 2005).
Siz1 and Siz2 belong to the conserved family of PIAS proteins, which share several conserved domains, including the SP-RING (Siz/PIAS-RING), a zinc binding domain that is required for the SUMO E3 ligase activity, and the SAP (SAF-A/B, Acinus, PIAS) domain, which is required for sumoylation of many, but not all, substrates (Sachdev et al. 2001; Takahashi et al. 2001; Tan et al. 2002; Okubo et al. 2004; Reindle et al. 2006). Siz1 and Siz2 are required for most SUMO conjugation in yeast, as a siz1Δ siz2Δ mutant shows <10% of the wild-type (wt) levels of SUMO conjugation (Johnson and Gupta 2001). SIZ1 and SIZ2 each have unique functions, but also show functional overlap. The siz1Δ siz2Δ double mutant accumulates up to 40-fold higher levels of the endogenous yeast plasmid, the 2-μm circle, than do wt cells, and this 2-μm accumulation causes growth defects and heterogeneity between different lineages (Chen et al. 2005). siz1Δ siz2Δ mutants also have a defect in minichromosome maintenance that results from deficient SUMO attachment to Top2, which can be sumoylated by either Siz protein (Takahashi et al. 2005). Sumoylation of many other proteins can also be stimulated by either Siz1 or Siz2 (Reindle et al. 2006).
Here we describe another phenotype of the siz1Δ siz2Δ mutant: it shows synthetic growth defects with mutants in the homologous recombination (HR) pathway. In mitotic cells the main function of HR is to repair double-strand breaks (DSBs) and to restart collapsed replication forks (Paques and Haber 1999; Krogh and Symington 2004). This process is carried out by products of the RAD52 epistasis group. These genes fall into several subgroups. In S. cerevisiae, most DSB repair is carried out by the gene conversion pathway, which is performed by the Rad51 subgroup: Rad51, Rad52, Rad54, Rad55, and Rad57. DSBs can also be repaired by break-induced replication (BIR). BIR can take place either by a Rad51-dependent mechanism or by a Rad51-independent mechanism that requires Rad52, Rad59, Rdh54/Tid1, and the Mre11-Rad50-Xrs2 complex (MRX). A distinct mechanism of DSB repair, nonhomologous end joining (NHEJ), is carried out by the MRX complex along with Ku70 (YKU70), Ku80 (YKU80), Dnl4, and Lif1.
The genetic interaction we observed between siz1Δ siz2Δ mutants and HR genes also involved DNA topoisomerase I (Top1). Top1 participates in DNA replication, transcription, and chromosome condensation by relaxing both positively and negatively supercoiled DNA (Champoux 2001; Li and Liu 2001; Wang 2002). Top1 acts by transiently cleaving the phosphodiester backbone of one strand, generating an intermediate where the active site tyrosine of Top1 is covalently linked to the 3′ phosphate of the nicked strand. The DNA is relaxed by rotating the Top1-DNA complex around the intact DNA, followed by the religation of the cleaved strand. In S. cerevisiae, Top1 is encoded by a single gene TOP1, which is not essential for viability. The Top1-DNA intermediate is potentially toxic to the cell since it involves a single-strand break. The anticancer drug camptothecin (CPT) stabilizes the Top1-DNA intermediate, resulting in formation of DSBs when the intermediate is encountered by a replication fork (Li and Liu 2001). Many yeast genes are involved in repairing CPT-induced damage, including RAD52 (Pouliot et al. 2001; Vance and Wilson 2002; Deng et al. 2005).
Several connections have been made between Top1 and the SUMO pathway. In mammalian cells, Top1 is sumoylated upon treatment of cells with CPT (Mao et al. 2000; Horie et al. 2002; Mo et al. 2002; Christensen et al. 2004), but the functional consequences of this are not clear. Horie et al. also found that a catalytically inactive version of Top1 is more heavily sumoylated than wt Top1. Yeast Top1 is also sumoylated, but the function is also unknown (Reindle et al. 2006). A ubc9 mutant that is sensitive to CPT has been identified, but this mutant is also sensitive to many other DNA damaging agents, suggesting that it has a primary defect in DNA repair rather having a Top1-specific defect (Jacquiau et al. 2005).
In this work, we identified Top1 as the cause of the loss of viability in yeast mutants that are defective for both SUMO conjugation and DNA repair. We also demonstrated that SUMO conjugation, TOP1, and RAD27 have interrelated effects on telomere maintenance. The genetic interactions among SUMO pathway mutants, DNA repair mutants, and top1Δ were complex and suggested that the observed phenotypes reflect defects in several mechanistically distinct processes.
MATERIALS AND METHODS
Media and genetic techniques:
Standard techniques were used (Ausubel et al. 2000). Rich yeast medium containing 2% glucose (YPD) and synthetic yeast media were prepared as previously described (Sherman et al. 1986). SGE is a synthetic medium containing 2% glycerol and 2% ethanol. 5-fluooroorotic acid (5-FOA) plates were prepared in synthetic medium and contained 1 g/liter 5-FOA.
Plasmid and yeast-strain constructions:
S. cerevisiae strains used are listed in supplemental Table 1 at http://www.genetics.org/supplemental/. Strains were either constructed in cir° strains or were cured of 2 μm as described (Tsalik and Gartenberg 1998). C-terminally tagged proteins were constructed by a PCR-based approach (Johnson and Blobel 1999). Briefly, a PCR product containing ∼500 bp of the 3′ end of the desired open reading frame, followed by the tag, a STOP codon, a marker, and then the 3′-flanking region of the gene, was made by assembly PCR. This results in an insertion at the 3′ end of the gene being tagged and should not affect the function of neighboring genes. The sequence of the hemagglutinin (HA)-His8 tag on Top1 and mutant derivatives was GYPYDVPDYAAFLHHHHHHHH. The sequence of the His8-HA tag on Tri1 and Uaf30 was GHHHHHHHHGYPYDVPDYAAFL. The K65R, K91, 92R, K600, 601R, and Y727F versions of TOP1 were constructed by similar PCR-based approaches. The hphMX4 marker conferring hygromycin resistance was derived from pAG32 (Goldstein and Mccusker 1999). kanMX-marked gene deletions were made by transforming JD52-derived strains with PCR products containing deletion alleles from the yeast knockout collection including ∼500 bp of flanking sequence on each side. Oligonucleotide sequences are available on request. TOP2-SNM strains were constructed in our strain background using plasmid pML251 (Bachant et al. 2002), generously provided by S. Elledge. Plasmids were a pRS316-based plasmid containing RAD52, a pRS416-based plasmid expressing Siz1-HA from its own promoter (Reindle et al. 2006), and three similar pRS413-based plasmids expressing wt, ΔSAP, or C400A versions of Siz-HA. Construction details available on request.
Selection for suppressor mutations of siz1Δ siz2Δ rad52Δ growth defect:
A siz1Δ siz2Δ rad52Δ strain containing RAD52 on a URA3-marked plasmid was transformed with NotI-digested DNA from a yeast library containing Tn5∷LEU2 insertions (Burns et al. 1994). These integrated into the chromosomal DNA, generating a collection of mutants containing marked insertions. LEU2 colonies were replica plated to a 5-FOA plate, to select for suppressor mutants that were able to grow without the RAD52-containing plasmid. The DNA containing the insertion in these mutants was isolated by vectorette PCR (http://genomics.princeton.edu/botstein/protocols/vectorette.html) and sequenced.
Yeast growth and plating assays:
To measure doubling times, yeast cultures were grown in YPD at 30° to an OD600 of ∼0.1. OD600 readings were taken every ∼1–2 hr until they reached ∼0.8. For most of the strains, three to four independent cultures were examined to obtain the average generation time. For plating assays to assess drug sensitivity, saturated-overnight cultures were subjected to serial 10-fold dilutions, and aliquots were spotted onto YPD plates containing designated chemicals. Plates were photographed after incubation at 30° for 2 days. Sensitivity to methyl methanesulfonate (MMS, 0.01%), hydroxyurea (HU, 0.1 m), CPT (50 μg/ml in 1% DMSO), and UV irradiation (150 J/m2) were assessed.
Loss of heterozygosity assay:
Diploid yeast strains were heterozygous for a version of chromosome (Chr) VII containing hphMX4 between ERP6 and ERG26 on the left side of the centromere, ade3Δ∷URA3 at the ADE3 locus on the right arm 409 kb from the centromere, and mal13Δ∷kanMX near the right telomere 162 kb from ade3Δ. All strains were constructed from the same haploid MATα siz1Δ∷LEU2 siz2Δ∷TRP1 top1Δ∷HIS3 strain containing this marked chromosome by mating to appropriate MATa strains with an unmarked Chr VII. Thus, they are homozygous at the loci indicated in Table 2 and are heterozygous at the other loci. Cultures from six single colonies for each strain were grown to saturation in SGE −ura −his −trp −leu. This selects for heterozygosity at the ADE3/ade3Δ∷URA3 locus, because ADE3 is required for histidine prototrophy. Aliquots were plated directly on SD-complete 5-FOA plates, to select for loss of URA3, or diluted and plated on SGE-complete plates, to determine the total number of colony-forming units. The loss rate of the URA3 marker was calculated as (no. of colonies on the 5-FOA plate)/(no. of colony-forming units in the same amount of culture). To determine whether kanMX and hphMX4 were also lost, colonies on 5-FOA plates were replica plated to YPD plates containing G418 or hygromycin.
TABLE 2.
Mitotic recombination frequencies in siz mutants
| Genotype | URA3-loss frequency (× 10−4) | HygR KanS (%)a | HygR KanR (%)a | HygS KanS (%)a |
|---|---|---|---|---|
| WT | 1.9 ± 0.7b | 92.1 | 4.7 | 2.4 |
| siz1Δ | 2.0 ± 0.6 | 87.1 | 10.4 | 0.7 |
| siz2Δ | 1.9 ± 0.8 | 93.0 | 7.0 | —c |
| siz1Δ siz2Δ | 8.5 ± 1.7 | 95.6 | 4.0 | 0.4 |
| top1Δ | 0.3 ± 0.2 | 91.8 | 8.2 | — |
| siz1Δ siz2Δ top1Δ | 1.8 ± 0.8 | 97.4 | 2.6 | — |
% of FOAR colonies with the given phenotype.
± SD.
—, none detected.
Telomere length analysis:
Southern blot hybridization was performed as described (Ausubel et al. 2000). Lanes were loaded with 10 μg of yeast DNA prepared using glass beads and phenol/chloroform (Hoffman and Winston 1987) and digested with XhoI, which cuts in the subtelomeric Y′ element found at over half of yeast telomeres. Agarose gels (1.5%) were run in 0.5× TBE at 2.2 V/cm for ∼30 hr. The probe contained Y′ sequences telomere-proximal to the XhoI site and was labeled using the DIG High Prime DNA labeling kit (Roche; Applied Science). Signal was detected according to the manufacturer's instructions.
Antibodies and immunoblot analyses:
Yeast whole-cell lysates were prepared by lysis in NaOH (Yaffe and Schatz 1984). HA- and His8-tagged proteins were purified from yeast by Ni-nitrilotriacetic acid (NTA) affinity chromatography in the presence of 6 m guanidine-HCl as described (Wohlschlegel et al. 2004; Chen et al. 2005) and subjected to immunoblotting followed by chemiluminescent detection (Johnson and Blobel 1999). Antibodies were an affinity-purified rabbit polyclonal antibody (Ab) against Smt3 (SUMO) (Johnson and Blobel 1999) and the 16B12 monoclonal Ab against the HA epitope (Covance Research Products, Emeryville, CA). For quantification of immunoblot signals, secondary antibodies coupled to fluorescent dyes IRDye 800 (Rockland Immunochemicals, Gilbertsville, PA) and Alexa Fluor 680 (Molecular Probes, Eugene, OR) were used with an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE) (Figure 6, B and C) or the film from the chemiluminescent blot was scanned and analyzed using a Kodak Image Station and 1D software (Kodak Digital Science, Rochester, NY) (Figure 6A).
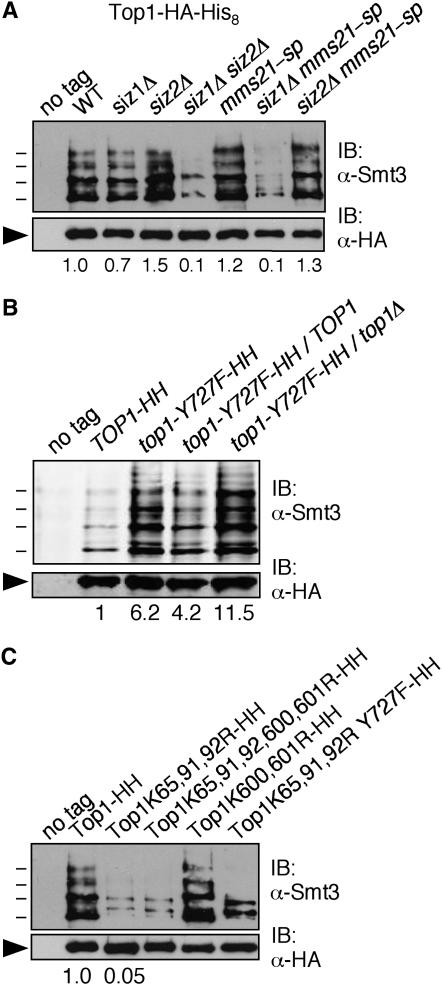
Figure 6.—
Top1 is modified by SUMO. (A) Sumoylation of Top1 in SUMO E3 ligase mutants. Proteins from indicated strains containing Top1-HA-His8 were purified by Ni-NTA affinity chromatography and analyzed by SDS–PAGE and immunoblotting with Abs against Smt3 (top) and HA (bottom). Arrowhead indicates unmodified Top1, and lines indicate SUMO-modified Top1. HH, HA and His8 tags. Numbers under the lanes indicate the ratio of the total signal from the sumoylated species to the signal from the unmodified species, normalized to wt Top1-HA-His8. (B) Top1 sumoylation is induced by deficient Top1 activity. Strains containing Top1-HA-His8 or active site mutant Top1-Y727F-HA-His8 were analyzed as in A. Diploid strains contained untagged TOP1 (lane 4) or top1Δ (lane 5) at the other locus. (C) Sumoylation of Top1 containing mutations in sumoylation consensus motifs. Indicated versions of Top1-HA-His8 were analyzed as in A.
RESULTS
siz1Δ siz2Δ strains require homologous recombination for viability:
Before we realized that the G2/M-arrest phenotype of the siz1Δ siz2Δ [cir+] mutant (Johnson and Gupta 2001) was caused by accumulation of 2 μm (Chen et al. 2005), we hypothesized that this phenotype might reflect a role for SIZ genes in genome stability. Therefore, we tested the siz1Δ siz2Δ strain for synthetic defects with various DNA repair mutants and found that the siz1Δ siz2Δ rad52Δ strain was barely viable, while rad52Δ, siz1Δ siz2Δ, siz1Δ rad52Δ, and siz2Δ rad52Δ mutants all grew well (Figure 1; Table 1; not shown). This is true in either the presence or the absence of 2 μm, indicating that this effect is independent of 2-μm amplification. To avoid effects from 2-μm amplification, all strains used in this work lack 2 μm; i.e., they are cir°. Because the HR pathway, of which RAD52 is the central member, functions in repair of DSBs and collapsed replication forks, this result suggested that siz1Δ siz2Δ contains such lesions.
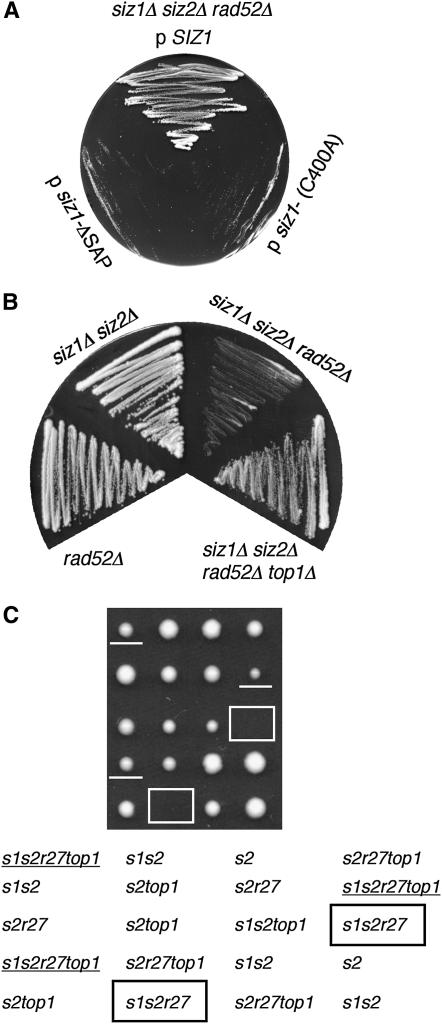
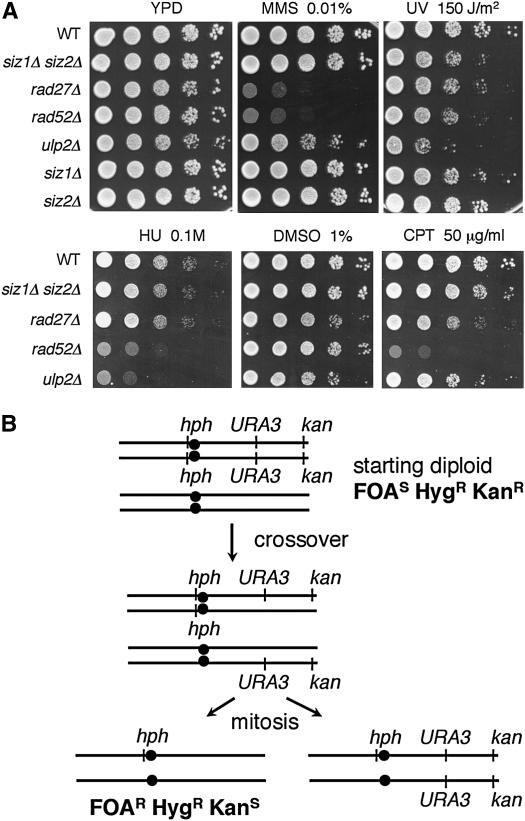
Figure 1.—
siz1Δ siz2Δ shows synthetic growth defects with rad52Δ and rad27Δ that are suppressed by top1Δ. (A) Domains in SIZ1 required to complement siz1Δ siz2Δ rad52Δ growth defect. HIS3-marked plasmids expressing SIZ1, SIZ1-ΔSAP, or SIZ1-C400A (in SP-RING) were introduced into siz1Δ siz2Δ rad52Δ containing SIZ1 on a URA3-marked plasmid. Transformants were streaked onto a 5-FOA plate to select against the URA3-marked plasmid and grown for 3 days at 30°. (B) Suppression of siz1Δ siz2Δ rad52Δ growth defect by top1Δ. Strains of the indicated genotypes were grown at 30° on a YPD plate for 2 days. (C) Suppression of siz1Δ siz2Δ rad27Δ synthetic lethality by top1Δ. Tetrads from a siz1Δ/SIZ1 siz2Δ/siz2Δ rad27Δ/RAD27 top1Δ/TOP1 diploid were dissected and incubated for 3 days at 30°. siz1Δ siz2Δ rad27Δ colonies are boxed and siz1Δ siz2Δ rad27Δ top1Δ are underlined. Mutant alleles present in each segregant are diagrammed at the bottom. s1, siz1Δ; s2, siz2Δ; r27, rad27Δ; top1, top1Δ. A siz2Δ/ siz2Δ strain was used to reduce the number of mutant combinations in the tetrads, but lethality with rad27Δ depended on absence of both SIZ1 and SIZ2.
TABLE 1.
Generation times of SIZ1, SIZ2, TOP1, and RAD52 pathway mutants
| Genotype | WT | top1Δ | siz1Δ siz2Δ | siz1Δ siz2Δ top1Δ |
|---|---|---|---|---|
| WT | 1.66 ± 0.05a | 2.04 ± 0.02 | 1.82 ± 0.01 | 2.2 ± 0.2 |
| rad52Δ | 2.17 ± 0.06 | 2.54 ± 0.04 | 7.7 ± 1.5b | 3.2 ± 0.1 |
| rad51Δ | 1.85 ± 0.05 | 4.5 ± 0.4 | 2.56 ± 0.07 | |
| rad54Δ | 4.32 ± 0.09 | 2.36 ± 0.05 | ||
| rad55Δ | 4.3 ± 0.4 | 2.38 ± 0.05 | ||
| rad57Δ | 3.7 ± 0.3 | 2.52 ± 0.06 | ||
| rad59Δ | 1.91 | 2.04 | ||
| rad50Δ | 3.7 ± 0.2 | 3.5 ± 0.1 | ||
| mre11Δ | 2.2 ± 0.1 | 3.8 ± 0.2 | 3.3 ± 0.2 | |
| xrs2Δ | 3.7 ± 0.3 | 3.5 ± 0.1 |
Doubling times are ± SD. Measurements with no error noted are single experiments.
The doubling time of siz1Δ siz2Δ rad52Δ varies from culture to culture due to the slow growth rate and spontaneous emergence of suppressor mutations.
To investigate which aspects of Siz activity are required for this role, we tested whether plasmids expressing mutant forms of SIZ1 complement the viability of this triple mutant (Figure 1A). Plasmids expressing SIZ1, siz1-ΔSAP, or siz1-C400A, which contains a point mutation in the SP-RING, were introduced into siz1Δ siz2Δ rad52Δ by a plasmid shuffle. All versions of Siz1 were present at comparable levels (Reindle et al. 2006; not shown). Only wild-type SIZ1 complemented the growth defect of siz1Δ siz2Δ rad52Δ. Since the SP-RING is required for the sumoylation activity of Siz1, this result suggested that Siz-dependent sumoylation, rather than a SUMO-independent function of Siz proteins, is important for preventing DNA damage.
Growth defects are suppressed by deleting TOP1:
When the slowly growing isolates of siz1Δ siz2Δ rad52Δ were grown for several days, rapidly growing colonies always emerged. These colonies contained suppressor mutations (not shown). To identify genes containing suppressor mutations, we carried out a screen for suppressors of siz1Δ siz2Δ rad52Δ using a Tn5∷LEU2 insertion library as the mutagen (see materials and methods). The two suppressor strains that were isolated both contained insertions near the 5′ end of the coding region of the TOP1 gene. These would be expected to be null mutations. To confirm that eliminating TOP1 could suppress the siz1Δ siz2Δ rad52Δ growth defect, we analyzed tetrads from a siz1Δ/SIZ1 siz2Δ/SIZ2 rad52Δ/RAD52 top1Δ/TOP1 heterozygous diploid. As expected, siz1Δ siz2Δ rad52Δ grew very poorly (doubling time 7.7 hr; Figure 1B; Table 1). In contrast, siz1Δ siz2Δ rad52Δ top1Δ isolates immediately grew rapidly and uniformly (doubling time 3.2 hr). An active site mutation in TOP1 also suppressed the siz1Δ siz2Δ rad52Δ growth defect: the siz1Δ siz2Δ rad52Δ top1-Y727F strain had a doubling time of 3.9 hr. Thus, absence of Top1 catalytic activity suppresses the growth defect of the siz1Δ siz2Δ rad52Δ mutant. The simplest explanation for this result would be that in the absence of Siz-dependent SUMO conjugation, Top1 directly causes DNA damage that requires the RAD52 pathway for repair, although other explanations are possible (see discussion).
RAD52 pathway genes are required for growth of siz1Δ siz2Δ:
We next tested other mutants of the RAD52 epistasis group for genetic interactions with siz1Δ siz2Δ and top1Δ. Table 1 shows that, except for RAD59, mutations in all RAD52 pathway genes (RAD50, RAD51, RAD54, RAD55, RAD57, MRE11, and XRS2) resulted in synthetic growth defects with siz1Δ siz2Δ. A cir+ version of rdh54Δ/tid1Δ siz1Δ siz2Δ did not have a substantial growth defect. None of these triple mutants grew as poorly as the rad52Δ triple mutant, consistent with the role of RAD52 in both Rad51-dependent and MRX-dependent branches of the pathway. Notably, RAD51 subgroup mutants had more severe synthetic defects than the MRX subgroup. Another difference between the two subgroups was that the growth defects of siz1Δ siz2Δ mutants lacking RAD51 subgroup genes were strongly suppressed by top1Δ, while MRX subgroup mutants were suppressed weakly, if at all. These results suggest that the TOP1-related DNA damage in siz1Δ siz2Δ mutants is repaired primarily by the RAD51 branch of the pathway. These results also suggest that siz1Δ siz2Δ cells contain TOP1-independent DNA damage that is primarily repaired by the MRX branch.
RAD27 is required for viability of siz1Δ siz2Δ:
We also tested for synthetic growth defects between siz1Δ siz2Δ and several other genes involved in DNA repair-related functions. None of rad1Δ, tdp1Δ, rad6Δ, rad9Δ, yku70Δ, srs2Δ, slx1Δ, mgs1Δ, or a rad1Δ tdp1Δ double mutant showed substantial additional defects when combined with siz1Δ siz2Δ (not shown). Deleting RAD6 did completely eliminate growth of the siz1Δ siz2Δ rad52Δ mutant, indicating that the marginal viability of this strain requires the DNA damage tolerance pathway (not shown).
However, we found that a siz1Δ siz2Δ rad27Δ mutant was completely inviable (Figure 1C). These segregants germinated, but stopped growing after four to five cell divisions. This synthetic lethality was also suppressed by deletion of TOP1. Rad27 is the S. cerevisiae homolog of the FEN1 5′-flap-exo/endonuclease (Liu et al. 2004). It plays important roles in Okazaki fragment processing as well as long-patch base-excision repair. rad27Δ is synthetically lethal with mutations in all RAD52 pathway genes (Debrauwere et al. 2001). However, the lethality of the rad27Δ rad52Δ mutant was not suppressed by deletion of TOP1 (not shown). Thus, the rad27Δ rad52Δ synthetic lethality is mechanistically distinct from the siz1Δ siz2Δ rad52Δ and siz1Δ siz2Δ rad27Δ defects.
Genetic interactions between RAD52, RAD27, TOP1, and other SUMO pathway genes:
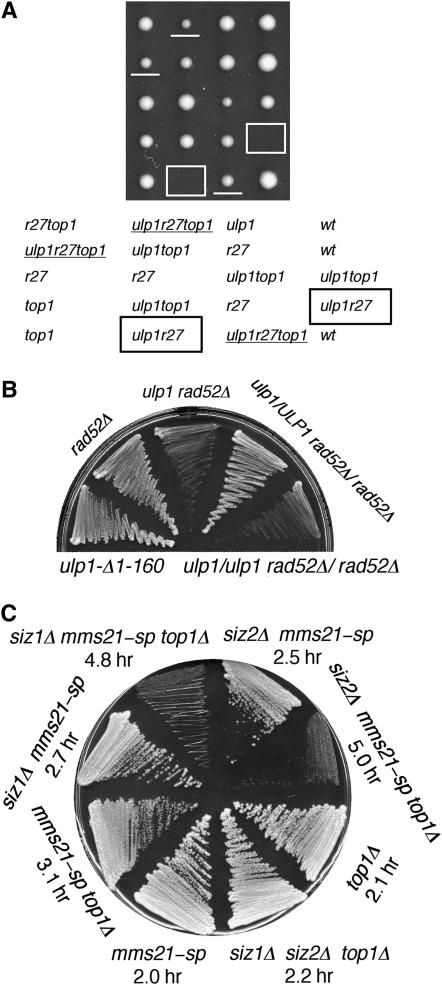
A mutant in the essential SUMO-specific protease ULP1 has been shown previously to show synthetic lethality with rad52Δ and rad27Δ (Soustelle et al. 2004). We found that a less severely affected allele of ulp1, lacking the N-terminal 160 amino acids of Ulp1, shows virtually identical growth defects to siz1Δ siz2Δ when combined with rad52Δ and rad27Δ: the rad52Δ ulp1-Δ1-160 mutant grew very slowly while the rad27Δ ulp1-Δ1-160 mutant was completely inviable (not shown; Figure 2A). Importantly, both mutants' growth defects were rescued by deleting TOP1. The similarity between these phenotypes suggests that the ulp1-Δ1-160 phenotype, like the siz1Δ siz2Δ phenotype, results from deficient SUMO attachment to one or more substrates, rather than from elevated SUMO attachment, as has been proposed (Soustelle et al. 2004; see discussion).
Figure 2.—
Genetic interactions between top1Δ and other SUMO pathway mutants. (A) top1Δ suppresses synthetic lethality between rad27Δ and ulp1-Δ1-160. Tetrads from a ulp1-Δ1-160/ULP1 rad27Δ/RAD27 top1Δ/TOP1 diploid were dissected and incubated for 3 days at 30°. ulp1-Δ1-160 rad27Δ colonies are boxed and ulp1-Δ1-160 rad27Δ top1Δ are underlined. Designations are as in Figure 1C. ulp1, ulp1-Δ1-160. (B) ulp1-Δ1-160 dominance test. Strains of the indicated genotypes bearing a URA3-marked plasmid containing RAD52 were streaked onto a 5-FOA plate to select against the plasmid and grown for 3 days at 30°. (C) Synthetic growth defects between mms21-sp and top1Δ. Strains of the indicated genotypes were grown at 30° on YPD plate for 2 days. Doubling time for each mutant is indicated.
ulp1 mutants containing deletions in their N-terminal regulatory domain, such as ulp1-Δ1-160, are defective in localizing to the nuclear pore complex (NPC) but retain catalytic activity (Li and Hochstrasser 2003; Panse et al. 2003). Two models could explain the defects of these mutants. One is that Ulp1 must be localized correctly to the NPC to carry out its activity, while the other is that the mislocalized mutant Ulp1 catalyzes unregulated desumoylation. The first model predicts that ulp1-Δ1-160 phenotypes should be recessive, while the second predicts that they would be dominant. We found that the synthetic growth defect of ulp1-Δ1-160 with rad52Δ was recessive, as a ulp1-Δ1-160/ULP1 rad52Δ/rad52Δ diploid grew well, while a ulp1-Δ1-160/ulp1-Δ1-160 rad52Δ/rad52Δ diploid grew poorly (Figure 2B). Thus, this phenotype results from insufficient Ulp1 activity rather than from uncontrolled desumoylation.
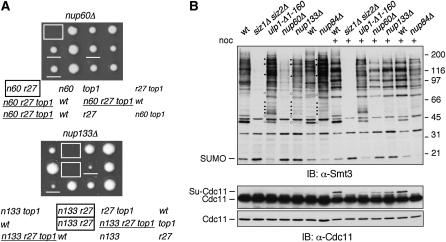
Correct localization of Ulp1 to the NPC requires the nucleoporin Nup60, in addition to several other proteins (Panse et al. 2003; Zhao et al. 2004). Other investigators have shown that the nup60Δ mutant shows synthetic lethality with both rad52Δ and rad27Δ, as do several other NPC mutants that have not previously been connected to the SUMO pathway, including nup133Δ, nup120Δ, and nup84Δ (Loeillet et al. 2005; Pan et al. 2006). We tested whether the synthetic lethality of nup60Δ and nup133Δ with rad27Δ would be suppressed by top1Δ and found that it was (Figure 3A). Furthermore, the nup60Δ, nup133Δ, and nup84Δ mutants all perturbed the pattern of bulk SUMO conjugates (Figure 3B). Thus, it is likely that the synthetic lethality of these NPC mutants with rad52Δ and rad27Δ is an indirect consequence of their effects on SUMO conjugation (see discussion).
Figure 3.—
Nuclear pore mutants show SUMO-related phenotypes. (A) top1Δ suppresses the synthetic lethality between nup60Δ or nup133Δ and rad27Δ. Tetrads from a nup60Δ/NUP60 rad27Δ/RAD27 top1Δ/TOP1 diploid (top) or a nup133Δ/NUP133 rad27Δ/RAD27 top1Δ/TOP1 diploid (bottom) were dissected and incubated for 3 days at 30°. nup rad27Δ colonies are boxed and nup rad27Δ top1Δ are underlined. Designations are as in Figure 1C. n60, nup60Δ; n133, nup133Δ. (B) nup mutants have sumoylation defects. Whole cell lysates from log phase or nocodazole-treated (noc) cultures from strains of the indicated genotypes were analyzed by SDS–PAGE and immunoblotting with an Ab against Smt3 (yeast SUMO) or against the septin Cdc11. Bands corresponding to free SUMO, Cdc11, and sumoylated Cdc11 (Su-Cdc11) are indicated. Solid circles indicate bands that are increased in ulp1, nup133Δ, and nup84Δ lanes. Open circles indicate bands that are decreased. nup84Δ and the wt control to its left are in a different strain background from the other strains.
Unlike most SUMO pathway mutants, mms21-sp, a mutant version of MMS21 that is unable to stimulate sumoylation (Andrews et al. 2005; Reindle et al. 2006), did not show a synthetic growth defect with rad52Δ (not shown). However, mms21-sp did show synthetic growth defects with top1Δ (Figure 2C). This effect was even stronger in siz1Δ mms21-sp or siz2Δ mms21-sp mutants, indicating that a SUMO-dependent activity that is normally carried out by MMS21 is important for growth in the absence of TOP1 activity, but that SIZ1 and SIZ2 can also carry out this function. Thus, SIZ1 and SIZ2 participate both in preventing TOP1-dependent growth defects when TOP1 is present and in compensating for absence of TOP1.
siz1Δ siz2Δ mutants have minor effects on sensitivity to DNA damaging agents:
Our results could be explained by a role for SUMO conjugation either in preventing TOP1-dependent DNA damage or in repairing naturally occurring levels of TOP1-dependent damage. To test for roles of SIZ genes in DNA repair, we examined sensitivity to different DNA damaging agents in cir° versions of siz1Δ siz2Δ. siz1Δ siz2Δ was not sensitive to the alkylating agent MMS or to hydroxyurea, but was slightly sensitive to UV irradiation (Figure 4A). The siz1Δ siz2Δ mutant was also slightly sensitive to the Top1-trapping drug CPT. A concentration of CPT that reduced the plating efficiency of a rad52Δ mutant by several orders of magnitude had little effect on the plating efficiency of a siz1Δ siz2Δ mutant, but did reduce its colony size (Figure 4A). This slight sensitivity to CPT would be consistent either with a role for Siz proteins in repairing Top1-dependent DNA damage or with the possibility that absence of Siz1 and Siz2 causes Top1-dependent damage, thereby increasing the load of Top1-dependent DNA damage in CPT-treated cells.
Figure 4.—
Phenotypes of siz1Δ siz2Δ mutants. (A) Sensitivity of SUMO pathway mutants to DNA damaging agents. Stationary phase cultures with the indicated genotypes were serially diluted (10-fold) and spotted onto YPD containing the indicated concentration of MMS, HU, DMSO, CPT, or subjected to UV irradiation, and incubated for 2–3 days at 30°. One set of dilutions is shown at the top and another set is shown at the bottom. (B) Diagram of loss of heterozygosity assay. Diploid strains were heterozygous for a version of Chr VII marked with the hphMX4 gene conferring hygromycin resistance (HygR), near the centromere, with URA3 conferring sensitivity to 5-FOA (FOAS) on the right arm, and with kanMX conferring G418 resistance (KanR) near the right telomere. Cultures were plated on 5-FOA to select for isolates that have lost the URA3 gene and are thus FOAR. Illustrated is how a reciprocal crossover could give rise to a FOAR HygR KanS isolate. BIR could also generate FOAR HygR KanS colonies. Chromosome loss would give FOAR HygS KanS colonies, while a mutation in URA3 would give FOAR HygR KanR colonies.
siz1Δ siz2Δ mutants display increased mitotic recombination:
The model that siz1Δ siz2Δ contains DNA damage that is repaired by the RAD52 pathway predicts that this mutant might show increased mitotic recombination. We first assayed for recombination between direct repeats with an intervening marker (Petukhova et al. 1999), but these experiments were inconclusive, with at most an approximately twofold increase in gene conversions or pop-outs in the siz1Δ siz2Δ strain (not shown). Next we tested for mitotic recombination by looking for loss of heterozygosity (LOH) in diploid strains with one triply marked chromosome (Figure 4B). By selecting for loss of the middle marker (URA3) and testing whether the flanking markers (at the centromere and near the telomere) were also lost, it was possible to distinguish between recombination events (such as reciprocal exchange or BIR), chromosome loss, and mutations in URA3. This experiment showed an approximately fivefold increase in the frequency of recombination events in the siz1Δ siz2Δ mutant relative to wt or either single mutant (Table 2). However, this effect was not dependent on Top1, as the siz1Δ siz2Δ top1Δ mutant also showed an approximately fivefold increase over top1Δ [although the overall levels of 5-FOA resistant (ura3) colonies in both top1Δ mutants were lower than those in the corresponding TOP1 strains]. We obtained similar results examining LOH at a different locus (URA3 inserted at the MLP2 locus on Chr IX), indicating that this effect is not specific to Chr VII. This result provides additional evidence for TOP1-independent genome instability in siz1Δ siz2Δ.
Sumoylation and Top1 affect telomere length:
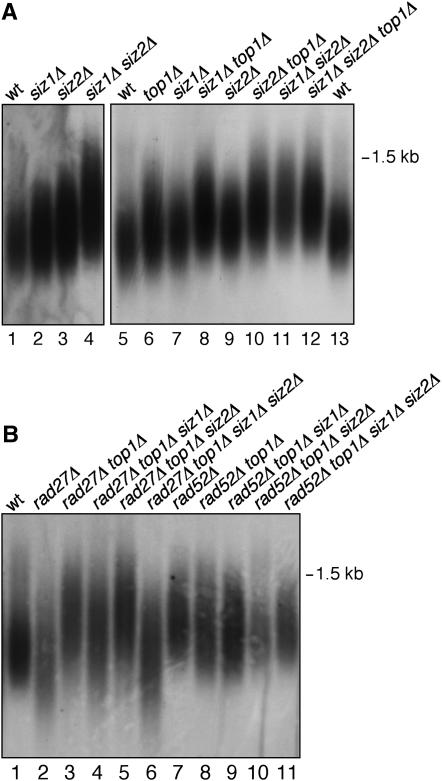
It has been shown that siz2Δ has slightly elongated telomeres, while a S. pombe mutant lacking the Siz/PIAS homolog has dramatically elongated telomeres (Askree et al. 2004; Xhemalce et al. 2004, 2007). We next asked whether the synthetic phenotypes of siz1Δ siz2Δ mutants and suppression by top1Δ were related to telomere length. We detected a slight increase in telomere length in siz2Δ, and possibly in siz1Δ as well, that was clearly exacerbated in the siz1Δ siz2Δ double mutant (Figure 5A). This suggests that SIZ1 and SIZ2 have an overlapping function in telomere maintenance. Next, we tested whether this effect depended on TOP1. The top1Δ mutant alone had very slightly elongated telomeres, but there was a notable increase in telomere length in both top1Δ siz1Δ and top1Δ siz2Δ compared to any of the single mutants (Figure 5A). siz1Δ siz2Δ and siz1Δ siz2Δ top1Δ telomeres were similar to each other in length. Thus, the telomere elongation in siz1Δ siz2Δ is not dependent on TOP1, since telomere elongation is not suppressed by top1Δ. Instead TOP1 and Siz-dependent sumoylation appear to play related, partially overlapping roles in preventing telomere elongation.
Figure 5.—
SIZ genes, TOP1, and RAD27 have overlapping functions in telomere length control. (A and B) Genomic DNA from strains of the indicated genotypes was analyzed by restriction digestion with XhoI, agarose gel electrophoresis, and Southern blotting using a probe against the Y′ subtelomeric element. The 1500-bp marker is indicated. The region containing the 1000-bp marker is not shown.
Next we tested whether these effects on telomere length involved RAD52 or RAD27, which have both been linked to telomere maintenance (Parenteau and Wellinger 1999; Bhattacharyya and Lustig 2006; Figure 5B). rad27Δ showed heterogeneity in telomere length, as has been observed previously (Parenteau and Wellinger 1999). Further deletion of TOP1 resulted in elongation of these telomeres. Remarkably, this telomere elongation in rad27Δ top1Δ depended on the SUMO pathway: deleting both SIZ1 and SIZ2 from this strain restored a pattern similar to RAD27 alone, while deleting SIZ1 had a partial effect. ulp1-Δ1-160, nup60Δ, and nup133Δ mutants had similar effects: ulp1/nup top1Δ rad27Δ mutants had short telomeres, similar to rad27Δ alone (not shown). These results suggest that RAD27 and the SUMO pathway have overlapping functions in telomere maintenance: top1Δ siz1Δ siz2Δ and top1Δ rad27Δ mutants both have elongated telomeres, while the quadruple mutant does not, indicating that either RAD27 or SIZ genes must be present for telomere elongation to occur in the top1Δ mutant. Thus, our results point to both negative and positive effects of sumoylation on telomere length: in otherwise wild-type strains, Siz-dependent sumoylation prevents telomere elongation, whereas in top1Δ rad27Δ mutants, Siz-dependent sumoylation promotes telomere elongation.
The rad52Δ strain had slightly longer telomeres than wt, but there was little additional change when TOP1 and SIZ genes were also deleted. This result is difficult to interpret. One possibility is that RAD52, TOP1, and SIZ genes all act in the same pathway related to telomere length control.
Top1 is sumoylated:
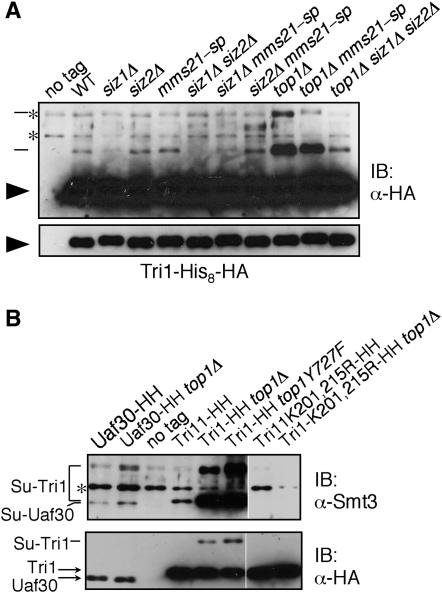
The genetic interactions we observed suggested that reduced Siz-dependent SUMO attachment to an unknown substrate(s) results in Top1-dependent loss of viability. One candidate substrate would be Top1 itself, which could require sumoylation to complete one of its activities properly. Yeast Top1 is sumoylated, with at least four SUMO-modified Top1 species detectable on a His8- and HA-tagged version of Top1 (Reindle et al. 2006; Figure 6). This tagged version of Top1 is at least partially functional, since the tagged strain did not have a growth defect and since siz1Δ siz2Δ rad52Δ TOP1-His8-HA was dead, demonstrating that tagged TOP1 complements the suppression phenotype. Most, but not all, SUMO attachment to Top1 depends on SIZ1 and SIZ2 (Reindle et al. 2006). Here we tested whether MMS21 also plays a role in Top1 sumoylation and found that while similar levels of sumoylation took place in wt and in siz1Δ, siz2Δ, mms21-sp, and siz2Δ mms21-sp mutants, sumoylation was decreased ∼10-fold in siz1Δ siz2Δ and siz1Δ mms21-sp double mutants (Figure 6A). Thus, MMS21 also participates in Top1 sumoylation.
An active site mutant of mammalian topoisomerase I is sumoylated more heavily than the wt protein (Horie et al. 2002). We found that this was also true in yeast, as Top1-Y727F, which lacks the active site tyrosine, was sumoylated approximately sixfold more heavily than wt Top1 (Figure 6B). Interestingly, Top1-Y727F-HA-His8 expressed in a heterozygous diploid with top1Δ at the other locus was sumoylated more heavily than Top1-Y727F-HA-His8 from a heterozygous diploid with wt untagged TOP1 at the other locus (Figure 6B). This result suggests that the level of SUMO attachment to Top1 may be affected by the global level of TOP1 activity in the cell, not just the activity of the Top1 polypeptide that is being sumoylated.
If Top1 itself is the substrate whose sumoylation prevents Top1-dependent DNA damage, then a mutant version of TOP1 lacking the SUMO attachment sites should show synthetic growth defects with rad52Δ and rad27Δ, similarly to the siz1Δ siz2Δ mutant. To test this, we identified SUMO attachment sites in yeast Top1. Mammalian topoisomerase I contains three major SUMO attachment sites in its N-terminal noncatalytic domain (Rallabhandi et al. 2002). This domain is not conserved, but yeast Top1 also contains two consensus sumoylation sites in its N-terminal domain, IKTE including Lys65 and IKKE containing Lys91 and 92. These are the major SUMO attachment sites in yeast Top1, as Lys to Arg mutations at these three Lys residues reduced Top1 sumoylation by ∼95% (Figure 6C). However, there was significant residual sumoylation in this mutant. Yeast Top1 contains one other sumoylation consensus site at LKKE including Lys600 and 601. Mutating these Lys residues had no detectable effect on sumoylation of either wt Top1 or Top1 containing mutations at the other three lysines (Figure 6C). Thus, the residual sumoylation of the Top1-K65,91,92R mutant takes place at nonconsensus sites. The 769-residue Top1 protein contains 103 other Lys residues, complicating the task of identifying additional sites. We found that the rad52Δ TOP1-K65,91,92R double mutant grew indistinguishably from a rad52Δ single mutant (doubling time 2.2 hr) and unlike the rad52Δ siz1Δ siz2Δ strain (7.7 hr). A rad27Δ TOP1-K65,91,92R mutant also did not have a notable growth defect beyond that of rad27Δ. The most likely interpretation of these results is that Top1 itself is not the relevant SUMO target in preventing TOP1-dependent DNA damage. However, it is still possible that Top1 is the relevant substrate, but that the residual sumoylation of Top1-K65,91,92R is sufficient to carry out this function. The TOP1-K65,91,92R mutant does retain topoisomerase activity, as it was functional in topoisomerase assays in vitro, and siz1Δ siz2Δ rad52Δ TOP1-K65,91,92R was dead, indicating that it complements the suppression phenotype (not shown).
SUMO modification of Top1-interacting proteins:
Another possible explanation for the absence of a phenotype in the TOP1-K65,91,92R rad52Δ and rad27Δ mutants is that sumoylation of Top1 may be redundant with sumoylation of other associated proteins. SUMO is often attached to multiple subunits within protein complexes (Wohlschlegel et al. 2004), and sumoylation of different proteins within the same complex may be functionally redundant. The Tri1 (YMR233W) protein has been reported to interact with Top1 and was identified as a SUMO substrate by a proteomic study (Uetz et al. 2000; Hannich et al. 2005). We confirmed that Tri1 is sumoylated and found that its sumoylation depended primarily on SIZ1 but was decreased to a greater extent in siz1Δ siz2Δ (Figure 7A). Remarkably, sumoylation of Tri1 was strongly stimulated by deletion of TOP1 (Figure 7, A and B). This top1Δ-induced sumoylation was primarily Siz-dependent, but Tri1 sumoylation was also increased in siz1Δ siz2Δ top1Δ relative to siz1Δ siz2Δ. Sumoylation of Tri1 was increased to a similar level in a strain containing inactive Top1-Y727F, indicating that it is the absence of Top1 activity, rather than the absence of Top1 protein, that results in the increase in Tri1 sumoylation (Figure 7B). S. cerevisiae contains a gene that is 41% identical to TRI1, called UAF30. Uaf30 was sumoylated to a low level, but its sumoylation was not strongly increased by top1Δ (Figure 7B). SUMO attachment to Pol30, Prp45, Gcn5, Abf1, Rsc2, Top2, and Cdc3 also was not affected by deleting TOP1 (not shown). Furthermore, no change in global sumoylation was observed upon deletion of TOP1 in either wt or siz1Δ siz2Δ cells (not shown). This suggests that reduced Top1 activity stimulates SUMO attachment to a limited subset of SUMO substrates that includes at least Top1 itself and Tri1.
Figure 7.—
Sumoylation of Tri1 is induced by absence of Top1 activity. (A) E3 and TOP1 dependence of Tri1 sumoylation. Indicated strains containing Tri1-His8-HA were analyzed as in Figure 6 and detected by immunoblotting with an Ab against HA. The bottom section is a lighter exposure of the blot shown in the top. Arrowheads indicate unmodified Tri1, and lines indicate SUMO-modified Tri1. Asterisks indicate bands that cross-react with the Ab. HH, HA and His8 tags. (B) Strains of the indicated genotypes expressing versions of Tri1-His8-HA or Uaf30-His8-HA, as indicated, were analyzed as in Figure 6 and detected by immunoblotting with Abs against Smt3 (top) or HA (bottom). Identities of bands are indicated. An asterisk indicates a band that cross-reacts with the Ab.
Tri1 contains two sumoylation consensus sequences near its C terminus at Lys201 and Lys215, and mutating the Lys in these sequences to Arg eliminated SUMO attachment to Tri1 (Figure 7B). This mutant had no obvious phenotypes either alone or when combined with top1Δ, rad52Δ, rad27Δ, TOP1-K65,91,92R, or with the double mutants rad52Δ TOP1-K65,91,92R, or rad27Δ TOP1-K65,91,92R (not shown). The doubling time of TRI1-K201,215R rad52Δ was 2.1 hr and of TRI1-K201,215R TOP1-K65,91,92R rad52Δ was 2.3 hr, neither of which was significantly different from rad52Δ alone. Thus, neither Top1 nor Tri1—nor Top1 and Tri1 acting redundantly—is the substrate that must be sumoylated to prevent TOP1-related DNA damage.
We also tested mutants in other yeast SUMO substrates for synthetic interactions with rad52Δ and rad27Δ. These experiments showed that it is unlikely that deficient SUMO attachment to Pol30 or Top2 or deficient SUMO chain formation is involved in the Top1-dependent synthetic growth defects. Rad52 is also unlikely to be the relevant substrate, although Rad52 is sumoylated. (Sacher et al. 2006). The fact that SIZ1 and SIZ2 are required for viability of rad52Δ indicates that the relevant substrate is being sumoylated in the rad52Δ mutant, which lacks Rad52 protein. Thus, the SUMO substrate associated with the Top1-dependent synthetic growth defects remains to be identified.
DISCUSSION
We describe several new discoveries that provide insight into the role of the SUMO pathway in maintaining genome integrity. Most importantly, we have shown that inactivating topoisomerase I suppresses the synthetic growth defects that arise from simultaneously inhibiting sumoylation and inactivating certain DNA repair pathways. This discovery provides a tool that allowed us both to identify other mutants that affect the SUMO pathway, such as the nuclear pore mutant nup133Δ, and to define the mechanistic relationships between various phenotypes of known SUMO pathway mutants. This second aspect is particularly important given the fact that the yeast SUMO pathway has hundreds of substrates that presumably act through many distinct mechanisms, yet there is a single SUMO-specific protease that processes the SUMO precursor protein (Ulp1), a single E1 (Uba2·Aos1) and E2 (Ubc9) for SUMO, and only three known E3's that function during vegetative growth (Siz1, Siz2, and Mms21). Thus, mutating UBC9 or ULP1 may cause hundreds of distinct defects (Figure 8). For example, a recent article shows that a conditional mutant in ubc9 displays synthetic lethality with mutants including rad51Δ and srs2Δ and also describes a role for MMS21-dependent sumoylation in the response of replication forks to DNA damage (Branzei et al. 2006). Our work suggests that these phenotypes reflect at least three distinct phenomena. One is the MMS21-dependent response to DNA damage. Second is the synthetic lethality of ubc9 with rad51Δ and other HR mutants. This is almost certainly the same top1Δ-suppressible phenomenon we have studied, which results from defective Siz-dependent sumoylation. [Deficient MMS21-dependent sumoylation does not result in synthetic lethality with rad52Δ (not shown).] Third is the synthetic lethality between ubc9 and srs2Δ, which we did not observe in srs2Δ siz1Δ siz2Δ. This either reflects a strain difference or is the result of a process that is defective in ubc9 but not in siz1Δ siz2Δ. In either case, it is a distinct effect.
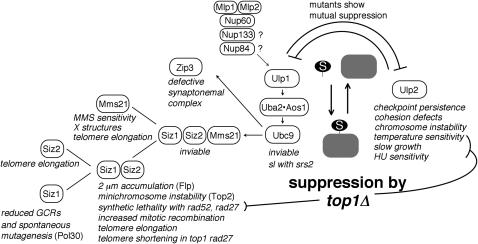
Figure 8.—
Hierarchy of genome stability phenotypes in SUMO pathway mutants. Ulp1, Uba2·Aos1, and Ubc9 are required for all SUMO (S, solid oval) conjugation to all substrates (shaded area) in S. cerevisiae. Mutants in these genes can display all phenotypes associated with downstream mutants, while E3 mutants show subsets of phenotypes. Ulp1 and Ulp2 have similar enzyme activities, but genetically, Ulp1 appears to act in SUMO conjugation (see text), while Ulp2 acts in deconjugation. ulp2Δ has a unique set of phenotypes (Li and Hochstrasser 2000; Schwienhorst et al. 2000; Strunnikov et al. 2001; Bachant et al. 2002). Nup60, Mlp1, and Mlp2 are required for correct localization of Ulp1, and therefore these mutants display a subset of ulp1 phenotypes (Zhao et al. 2004). nup133Δ and nup84Δ also affect SUMO conjugation (this work) and may also act through Ulp1 localization. The SUMO E3's Siz1, Siz2, and Mms21 have both unique and overlapping functions during vegetative growth, while Zip3 acts during meiosis (Stelter and Ulrich 2003; Askree et al. 2004; Chen et al. 2005; Takahashi et al. 2005; Zhao and Blobel 2005; Branzei et al. 2006; Cheng et al. 2006; Motegi et al. 2006; this work). Several phenotypes of ulp2Δ, as well as the synthetic lethality between siz1Δ siz2Δ and rad52Δ or rad27Δ are suppressed by mutations in TOP1 (Jacquiau et al. 2005; this work). Phenotypes associated with mutants in a particular gene or set of genes are listed in italics. Proteins in parentheses are SUMO substrates associated with the phenotype. Arrows point toward downstream components of the pathway. Lines indicate hierarchical relationships. Proteins illustrated vertically each have unique roles in a particular function, while proteins illustrated side by side have redundant roles in the indicated function. sl, synthetic lethal; GCR, gross chromosomal rearrangement.
We identified three additional phenotypes of deficient Siz-dependent SUMO conjugation that are distinct from those described above and probably from each other. First is the increased mitotic recombination in siz1Δ siz2Δ, as detected by LOH in diploid cells (Table 2). Second is the telomere elongation that takes place upon deletion of SIZ genes from wt or from top1Δ cells (Figure 5). Third is the decrease in telomere length that occurs upon deletion of SIZ genes in rad27Δ top1Δ cells. There are two reasons for thinking that these telomere-related effects are distinct. One is that in the first case, Siz activity promotes shorter telomeres, while in the second case it generates longer telomeres. Furthermore, the siz1Δ and siz2Δ single mutants have different effects on these phenomena. The siz2Δ mutant has slightly longer telomeres than siz1Δ, suggesting that SIZ2 plays a greater role in controlling telomere length in wt cells. In contrast, SIZ1 has a greater effect in rad27Δ top1Δ: telomere length is reduced more dramatically upon deleting SIZ1 from this mutant than upon deleting SIZ2 (Figure 5).
We also showed that several previously described SUMO-related phenotypes are distinct from the TOP1-dependent loss of viability in sumoylation- and DNA repair-deficient mutants. An unsumoylatable TOP2 mutant, top2-SNM, has defects in centromere cohesion and minichromosome maintenance (Bachant et al. 2002; Takahashi et al. 2005). However, since top2-SNM does not show synthetic defects with rad52Δ or rad27Δ, these phenotypes are not related to the Top1-dependent synthetic growth defects. Likewise, a mutant version of SUMO that cannot form SUMO chains suppresses many of the phenotypes of a ulp2Δ mutant (Bylebyl et al. 2003), but deficient SUMO chain formation is also not the cause of the synthetic growth defects. Sumoylation of PCNA at K164 depends solely on SIZ1 (Hoege et al. 2002) and so is clearly not responsible for the synthetic defects, but sumoylation at K127 could have been responsible. However, this is not the case. Cumulatively, this set of results suggests that the SUMO pathway has multiple related, yet mechanistically distinct, roles in maintaining the integrity of the genome.
Our current results also emphasize another trend in the phenotypes of SUMO pathway mutants that is often not discussed clearly in the literature: the vast majority of the phenotypes of ulp1 mutants result from reductions in SUMO conjugation, rather than from accumulation of sumoylated substrates. This is not immediately obvious because ulp1 mutants accumulate some sumoylated species and have reductions in others (Li and Hochstrasser 1999). However, in all cases where it has been examined, ulp1 phenotypes are identical to those of mutants that are defective in attaching SUMO. One or more of the SUMO attachment pathway mutants ubc9, uba2, or siz1Δ siz2Δ display all of the following phenotypes of ulp1 mutants: accumulation of the 2 μm circle; synthetic lethality with rad52Δ, rad27Δ, and srs2Δ; suppression of the phenotypes of ulp2Δ; and defects in cell cycle progression, nuclear transport, and trafficking of ribosomal subunits (Li and Hochstrasser 2000; Schwienhorst et al. 2000; Stade et al. 2002; Soustelle et al. 2004; Chen et al. 2005; Dobson et al. 2005; Branzei et al. 2006; Panse et al. 2006). Since these other proteins participate only in SUMO attachment, while Ulp1 plays roles both in generating free SUMO and in removing SUMO from specific conjugates, it is reasonable to conclude that these phenotypes have the same cause in all mutants: deficient SUMO conjugation.
In contrast, the defects associated with ulp2Δ appear to be caused primarily by abnormal accumulation of SUMO conjugates. The reason for believing this is that the major phenotypes of ulp2Δ mutants are suppressed by mutations that decrease SUMO conjugation, including uba2, ubc9, siz1Δ siz2Δ, and ulp1, as well as the unsumoylatable versions of TOP2 and SUMO (Li and Hochstrasser 2000; Schwienhorst et al. 2000; Bachant et al. 2002; Bylebyl et al. 2003). Thus, given our results, it is surprising that deleting TOP1 also suppresses many of these same phenotypes of ulp2Δ (Jacquiau et al. 2005). This means that top1Δ suppresses phenotypes associated both with increased sumoylation (in ulp2Δ) and with decreased sumoylation (in siz1Δ siz2Δ rad52Δ). It is not obvious how this works. One possibility is that top1Δ suppression affects entirely different phenomena in these two cases. Another possibility is that undersumoylation and oversumoylation of the same Top1-related protein have different deleterious effects that are both suppressed by deleting TOP1.
The simplest model to explain how inactivating Top1 would suppress DNA repair-related phenotypes is that Top1 could directly cause DNA damage in these mutants. It is easy to imagine how aberrant Top1 activity could cause DNA damage, since single-strand breaks (SSBs) form during the catalytic mechanism of Top1. However, we have not detected evidence of this. In a ulp1 catalytic domain mutant with a severe growth defect, Soustelle et al. (2004) detected SSBs during DNA replication, but we have not detected SSBs in the rapidly growing siz1Δ siz2Δ mutant. Aberrant Top1 activity can result from the presence of certain DNA abnormalities, such as abasic sites, that delay religation (Champoux 2001; Wang 2002). We did not detect differing levels of abasic sites between wt and siz1Δ siz2Δ DNA (not shown). We also did not observe higher levels of either Top1 protein or Top1 activity in vitro in lysates from siz1Δ siz2Δ cells (not shown). Furthermore, the mitotic recombination and telomere elongation phenotypes in the siz1Δ siz2Δ mutant were not suppressed by top1Δ, indicating that these phenotypes are not caused by Top1 activity. These results may mean that we have not yet detected the defect that is associated with TOP1-dependent cell death in these mutants.
It is also possible that the role of TOP1 in synthetic lethality is indirect. The relevant SIZ-dependent process may be carried out by an entirely different mechanism in the absence of TOP1. Alternatively, inactivating TOP1 may allow cell viability even though the relevant DNA-related defect is still present. For example, SIZ genes and RAD27 have overlapping functions in telomere maintenance, such that telomeres are shorter in siz1Δ siz2Δ rad27Δ top1Δ than in either siz1Δ siz2Δ top1Δ or rad27Δ top1Δ. It is possible that in the presence of TOP1 the telomere shortening in siz1Δ siz2Δ rad27Δ is even more dramatic and results in inviability. We also cannot exclude the possibility that the suppression of siz1Δ siz2Δ mutants' synthetic growth defects by top1Δ results from the increased sumoylation of certain proteins in this mutant.
Another new finding in this work is that the increased sumoylation of defective Top1—either CPT poisoned or with an active site mutation—that has been observed by others appears to be a global effect resulting from reduced Top1 activity in the cell. Previous investigators assumed that the specific CPT-bound or inactivated topo I molecule is targeted for sumoylation (Mao et al. 2000; Horie et al. 2002; Mo et al. 2002; Christensen et al. 2004). In contrast, our results show that there is a general upregulation of SUMO attachment to certain proteins, including Top1 and Tri1, when TOP1 activity is reduced or eliminated.
Finally, we have also shown that, like nup60Δ, mutants in the nucleoporin genes NUP133 and NUP84 have defects in SUMO conjugation and that nup133Δ, at least, shares some of the phenotypes of SUMO pathway mutants. Nup60 is involved in NPC localization of Ulp1 (Zhao et al. 2004), and this is a likely mechanism for the effect of the other NPC mutants as well, although this remains to be proven. There were substantial differences in the patterns of SUMO conjugates among the nup and ulp1 mutants. nup133Δ and nup84Δ showed some increased and some decreased species, somewhat like ulp1-Δ1-160, while nup60Δ showed a greater overall reduction in SUMO conjugation (Figure 3B). This is likely explained by the fact that tethering of Ulp1 to the NPC is mediated by two different karyopherins that bind to different parts of the Ulp1 N-terminal domain (Panse et al. 2003). The ulp1-Δ1-160 mutant lacks only one of these and retains partial nuclear envelope localization (Li and Hochstrasser 2003). nup60Δ shows greater reductions in Ulp1 nuclear envelope localization (Zhao et al. 2004) and consequently may carry out greater unregulated desumoylation. The NPCs in nup133Δ form a single cluster, and Ulp1 has been shown to colocalize with this cluster (Schwienhorst et al. 2000). This indicates that Ulp1 still localizes to the NPC in this mutant, although the NPCs themselves are mislocalized. Interestingly, these nup mutants, as well as mutants in MLP1 and MLP2, which are also involved in localization of Ulp1 to the NPC, have a variety of other phenotypes including defects in subtelomeric silencing, in repair of double-strand breaks near the telomere, and in tethering of telomeres to the nuclear envelope (Galy et al. 2000; Hediger et al. 2002; Therizols et al. 2006). Some of these may also be secondary effects of their defects in SUMO metabolism and may be mechanistically related to the phenotypes of siz1Δ siz2Δ that we have characterized. Determining what the relevant SUMO substrates are and distinguishing phenotypes involving sumoylation of one protein from those involving sumoylation of others will be a challenge for the future.
Acknowledgments
We are grateful to M. A. Bjornsti for helpful suggestions and J. Dohmen for comments on the manuscript. We also thank S. Elledge and H. Klein for strains and E. Alnemri's lab for use of their LiCor Odyssey imaging system. This work was supported in part by GM62268 from the National Institutes of Health and in part by a reapplication enhancement award from Thomas Jefferson University. H.R.S. was supported by National Research Service Award T32-DK07705. C.A. was a participant in Thomas Jefferson University's Summer Undergraduate Research Opportunities Program.
References
- Andrews, E. A., J. Palecek, J. Sergeant, E. Taylor, A. R. Lehmann et al., 2005. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 25: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree, S. H., T. Yehuda, S. Smolikov, R. Gurevich, J. Hawk et al., 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 101: 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith et al., 2000. Current Protocols in Molecular Biology. Wiley-Interscience, New York.
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner and S. J. Elledge, 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9: 1169–1182. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, M. K., and A. J. Lustig, 2006. Telomere dynamics in genome stability. Trends Biochem. Sci. 31: 114–122. [DOI] [PubMed] [Google Scholar]
- Branzei, D., J. Sollier, G. Liberi, X. Zhao, D. Maeda et al., 2006. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522. [DOI] [PubMed] [Google Scholar]
- Burns, N., B. Grimwade, P. B. Ross-Macdonald, E. Y. Choi, K. Finberg et al., 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8: 1087–1105. [DOI] [PubMed] [Google Scholar]
- Bylebyl, G. R., I. Belichenko and E. S. Johnson, 2003. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278: 44113–44120. [DOI] [PubMed] [Google Scholar]
- Champoux, J. J., 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70: 369–413. [DOI] [PubMed] [Google Scholar]
- Chen, X. L., A. Reindle and E. S. Johnson, 2005. Misregulation of 2 μm circle copy number in a SUMO pathway mutant. Mol. Cell. Biol. 25: 4311–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C. H., Y. H. Lo, S. S. Liang, S. C. Ti, F. M. Lin et al., 2006. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20: 2067–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, M. O., R. M. Krokowski, H. U. Barthelmes, R. Hock, F. Boege et al., 2004. Distinct effects of topoisomerase I and RNA polymerase I inhibitors suggest a dual mechanism of nucleolar/nucleoplasmic partitioning of topoisomerase I. J. Biol. Chem. 279: 21873–21882. [DOI] [PubMed] [Google Scholar]
- Debrauwere, H., S. Loeillet, W. Lin, J. Lopes and A. Nicolas, 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98: 8263–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, C., J. A. Brown, D. You and J. M. Brown, 2005. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson, M. J., A. J. Pickett, S. Velmurugan, J. B. Pinder, L. A. Barrett et al., 2005. The 2 μm plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease. Mol. Cell. Biol. 25: 4299–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy, V., J. C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou et al., 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403: 108–112. [DOI] [PubMed] [Google Scholar]
- Gill, G., 2004. SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes Dev. 18: 2046–2059. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Hannich, J. T., A. Lewis, M. B. Kroetz, S. J. Li, H. Heide et al., 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280: 4102–4110. [DOI] [PubMed] [Google Scholar]
- Hay, R. T., 2005. SUMO: a history of modification. Mol. Cell 18: 1–12. [DOI] [PubMed] [Google Scholar]
- Hediger, F., K. Dubrana and S. M. Gasser, 2002. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J. Struct. Biol. 140: 79–91. [DOI] [PubMed] [Google Scholar]
- Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis and S. Jentsch, 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. [DOI] [PubMed] [Google Scholar]
- Hoffman, C. S., and F. Winston, 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272. [DOI] [PubMed] [Google Scholar]
- Horie, K., A. Tomida, Y. Sugimoto, T. Yasugi, H. Yoshikawa et al., 2002. SUMO-1 conjugation to intact DNA topoisomerase I amplifies cleavable complex formation induced by camptothecin. Oncogene 21: 7913–7922. [DOI] [PubMed] [Google Scholar]
- Jacquiau, H. R., R. C. van Waardenburg, R. J. Reid, M. H. Woo, H. Guo et al., 2005. Defects in SUMO (small ubiquitin-related modifier) conjugation and deconjugation alter cell sensitivity to DNA topoisomerase I-induced DNA damage. J. Biol. Chem. 280: 23566–23575. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73: 355–382. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., and G. Blobel, 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. S., and A. A. Gupta, 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744. [DOI] [PubMed] [Google Scholar]
- Krogh, B. O., and L. S. Symington, 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38: 233–271. [DOI] [PubMed] [Google Scholar]
- Li, S. J., and M. Hochstrasser, 1999. A new protease required for cell-cycle progression in yeast. Nature 398: 246–251. [DOI] [PubMed] [Google Scholar]
- Li, S. J., and M. Hochstrasser, 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20: 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. J., and M. Hochstrasser, 2003. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 160: 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. K., and L. F. Liu, 2001. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 41: 53–77. [DOI] [PubMed] [Google Scholar]
- Liu, Y., H. I. Kao and R. A. Bambara, 2004. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 73: 589–615. [DOI] [PubMed] [Google Scholar]
- Loeillet, S., B. Palancade, M. Cartron, A. Thierry, G. F. Richard et al., 2005. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair 4: 459–468. [DOI] [PubMed] [Google Scholar]
- Mao, Y., M. Sun, S. D. Desai and L. F. Liu, 2000. SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proc. Natl. Acad. Sci. USA 97: 4046–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, Y. Y., Y. Yu, Z. Shen and W. T. Beck, 2002. Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J. Biol. Chem. 277: 2958–2964. [DOI] [PubMed] [Google Scholar]
- Motegi, A., K. Kuntz, A. Majeed, S. Smith and K. Myung, 2006. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, S., A. Ledl and D. Schmidt, 2004. SUMO: a regulator of gene expression and genome integrity. Oncogene 23: 1998–2008. [DOI] [PubMed] [Google Scholar]
- Okubo, S., F. Hara, Y. Tsuchida, S. Shimotakahara, S. Suzuki et al., 2004. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J. Biol. Chem. 279: 31455–31461. [DOI] [PubMed] [Google Scholar]
- Ouspenski, II, S. J. Elledge and B. R. Brinkley, 1999. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 27: 3001–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., P. Ye, D. S. Yuan, X. Wang, J. S. Bader et al., 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081. [DOI] [PubMed] [Google Scholar]
- Panse, V. G., B. Kuster, T. Gerstberger and E. Hurt, 2003. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5: 21–27. [DOI] [PubMed] [Google Scholar]
- Panse, V. G., D. Kressler, A. Pauli, E. Petfalski, M. Gnadig et al., 2006. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic 7: 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau, J., and R. J. Wellinger, 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19: 4143–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova, G., S. Van Komen, S. Vergano, H. Klein and P. Sung, 1999. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem 274: 29453–29462. [DOI] [PubMed] [Google Scholar]
- Pouliot, J. J., C. A. Robertson and H. A. Nash, 2001. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells 6: 677–687. [DOI] [PubMed] [Google Scholar]
- Rallabhandi, P., K. Hashimoto, Y. Y. Mo, W. T. Beck, P. K. Moitra et al., 2002. Sumoylation of topoisomerase I is involved in its partitioning between nucleoli and nucleoplasm and its clearing from nucleoli in response to camptothecin. J. Biol. Chem. 277: 40020–40026. [DOI] [PubMed] [Google Scholar]
- Reindle, A., I. Belichenko, G. R. Bylebyl, X. L. Chen, N. Gandhi et al., 2006. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 119: 4749–4757. [DOI] [PubMed] [Google Scholar]
- Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior et al., 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15: 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher, M., B. Pfander, C. Hoege and S. Jentsch, 2006. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 8: 1284–1290. [DOI] [PubMed] [Google Scholar]
- Schwienhorst, I., E. S. Johnson and R. J. Dohmen, 2000. SUMO conjugation and deconjugation. Mol. Gen. Genet. 263: 771–786. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1986. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Soustelle, C., L. Vernis, K. Freon, A. Reynaud-Angelin, R. Chanet et al., 2004. A new Saccharomyces cerevisiae strain with a mutant Smt3-deconjugating Ulp1 protein is affected in DNA replication and requires Srs2 and homologous recombination for its viability. Mol. Cell. Biol. 24: 5130–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade, K., F. Vogel, I. Schwienhorst, B. Meusser, C. Volkwein et al., 2002. A lack of SUMO conjugation affects cNLS-dependent nuclear protein import in yeast. J. Biol. Chem. 277: 49554–49561. [DOI] [PubMed] [Google Scholar]
- Stelter, P., and H. D. Ulrich, 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191. [DOI] [PubMed] [Google Scholar]
- Strunnikov, A. V., L. Aravind and E. V. Koonin, 2001. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics 158: 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., T. Kahyo, E. A. Toh, H. Yasuda and Y. Kikuchi, 2001. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276: 48973–48977. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., V. Yong-Gonzalez, Y. Kikuchi and A. Strunnikov, 2005. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics 172: 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, J. A., S. H. Hall, K. G. Hamil, G. Grossman, P. Petrusz et al., 2002. Protein inhibitors of activated STAT resemble scaffold attachment factors and function as interacting nuclear receptor coregulators. J. Biol. Chem. 277: 16993–17001. [DOI] [PubMed] [Google Scholar]
- Therizols, P., C. Fairhead, G. G. Cabal, A. Genovesio, J. C. Olivo-Marin et al., 2006. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 172: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsalik, E. L., and M. R. Gartenberg, 1998. Curing Saccharomyces cerevisiae of the 2 micron plasmid by targeted DNA damage. Yeast 14: 847–852. [DOI] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson et al., 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Vance, J. R., and T. E. Wilson, 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA 99: 13669–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. C., 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3: 430–440. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel, J. A., E. S. Johnson, S. I. Reed and J. R. Yates, 3rd, 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279: 45662–45668. [DOI] [PubMed] [Google Scholar]
- Xhemalce, B., J. S. Seeler, G. Thon, A. Dejean and B. Arcangioli, 2004. Role of the fission yeast SUMO E3 ligase Pli1p in centromere and telomere maintenance. EMBO J. 23: 3844–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhemalce, B., E. M. Riising, P. Baumann, A. Dejean, B. Arcangioli et al., 2007. Role of SUMO in the dynamics of telomere maintenance in fission yeast. Proc. Natl. Acad. Sci. USA 104: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M. P., and G. Schatz, 1984. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA 81: 4819–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., and G. Blobel, 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., C. Y. Wu and G. Blobel, 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167: 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]