Abstract
The importance of a gene's natural chromatin environment for its normal expression is poignantly illustrated when a change in chromosome position results in variable gene repression, such as is observed in position effect variegation (PEV) when the Drosophila melanogaster white (w) gene is juxtaposed with heterochromatin. The Enhancer of variegation 3-9 [E(var)3-9] gene was one of over a hundred loci identified in screens for mutations that dominantly modify PEV. Haploinsufficiency for E(var)3-9 enhances wm4 variegation, as would be expected from increased heterochromatin formation. To clarify the role of E(var)3-9 in chromosome structure, the gene has been cloned and its mutant alleles characterized. The involvement of E(var)3-9 in structure determination was supported by its reciprocal effects on euchromatic and heterochromatic PEV; E(var)3-9 mutations increased expression of a variegating heterochromatic gene in two tissue types. E(var)3-9 mutations also had a recessive phenotype, maternal effect lethality, which implicated E(var)3-9 function in an essential process during embryogenesis. Both phenotypes of E(var)3-9 mutations were consistent with its proposed function in promoting normal chromosome structure. The cloning of E(var)3-9 by classical genetic methods revealed that it encodes a protein with multiple zinc fingers, but otherwise novel sequence.
EUKARYOTIC genomes exhibit differential packaging, and distinctions in chromosome structure are associated with differing genetic properties, such as transcriptional activity, centromere function, chromosome pairing, and replication timing. Position effect variegation (PEV) in Drosophila melanogaster has long been a model system for studying determinants of chromosome structure (reviewed by Weiler and Wakimoto 1995; Schotta et al. 2003). PEV results from chromosome rearrangements with breakpoints in cytologically distinct chromosome regions, the densely staining heterochromatin, which is rich in repetitive sequences but gene-poor, and the lightly staining euchromatin characterized by a high density of genic DNA. Euchromatic genes in the vicinity of the breakpoint are subject to inactivation when brought into proximity of heterochromatin, although this repression exhibits cell-to-cell variability. The early cytological studies of polytene chromosomes from individuals bearing variegation-inducing rearrangements first indicated that mosaic inactivation was due to the variable spreading of a heterochromatic chromatin structure into the formerly euchromatic region and have since been supported by molecular assays (Hayashi et al. 1990; Wallrath and Elgin 1995; Sun et al. 2000). More recently, a role for histone modifications and small RNAs in heterochromatin formation and heterochromatin-mediated silencing has been established (Grewal and Rice 2004). However, many questions remain about the mechanism(s) of heterochromatin formation, silencing and spreading, the role of transinteractions, and the gene products that regulate the euchromatic and heterochromatic states. PEV remains a valuable tool to address these issues.
Several large-scale genetic screens were conducted to isolate dominant genetic modifiers of PEV as a means to identify factors affecting the formation of heterochromatic and euchromatic domains (Reuter and Wolff 1981; Sinclair et al. 1983; Reuter et al. 1987; Locke et al. 1988; Wustmann et al. 1989; Dorn et al. 1993; reviewed by Schotta et al. 2003). These studies utilized the archetype of PEV, the X chromosome inversion white-mottled four [In(1)wm4], which results in a fly with red and white variegated eyes due to the juxtaposition of the w+ gene next to heterochromatin (Muller 1930). Mutants were identified as showing an increase or decrease in the fraction of red ommatidia. The genetic modifiers that suppressed the variegation, Su(var) mutations, were postulated to encode the structural components of heterochromatin and the enzymes that act upon them. Characterization of several Su(var) genes has validated this prediction. The Su(var)3-9 gene encodes the chromo and SET domain protein responsible for histone H3 lysine 9 (H3K9) methylation, an epigenetic modification associated with transcriptional silencing (Schotta et al. 2002). The Su(var)205 gene encodes the chromodomain and heterochromatin-associated protein HP1, which binds to methylated H3K9 (James and Elgin 1986; Eissenberg et al. 1990; Bannister et al. 2001). The Su(var)3-3 gene was recently determined to encode a histone H3 lysine 4 demethylase (Rudolph et al. 2007).
The genetic modifiers that enhanced wm4 variegation, E(var) mutations, were postulated to encode proteins that antagonize heterochromatin formation or promote euchromatin formation. However, decreased expression of the wm4 allele is a phenotype that would be expected from mutation of the transcriptional machinery as well. Indeed, the early finding that several E(var) genes encode proteins that might best be described as transcriptional activators [e.g., Trl (Farkas et al. 1994), brm (Tamkun et al. 1992), Asx (Sinclair et al. 1998), and E2F (Seum et al. 1996)] indicated that mutants with more global effects on chromosome structure would be a subset of those identified in the initial genetic screens. In an attempt to distinguish those E(var) mutants that have a more widespread role in chromatin structure from those required for expression of a subset of genes including the w gene, I carried out a supplementary genetic screen of the E(var) class of mutants (Weiler and Wakimoto 2002). In this pilot study, I tested several E(var) mutants for the ability to suppress PEV of the heterochromatic light (lt) gene and was able to discriminate between them. Normal expression of the lt gene requires a heterochromatic environment, and chromosomal rearrangements that displace it to distal euchromatin cause it to variegate (Wakimoto and Hearn 1990). The rationale for this test was that mutations that shifted the euchromatin–heterochromatin balance in favor of heterochromatin formation should increase expression of a variegating heterochromatic gene, whose expression requires and is sensitive to the level of surrounding heterochromatin. On the contrary, mutations that enhanced wm4 variegation via required transcription factors should similarly enhance or have no effect on lt variegation. Two uncharacterized complementation groups representing genes E(var)3-4 and E(var)3-5 were shown to have a reciprocal effect on euchromatic and heterochromatic PEV and thus are likely involved in regulating the balance between heterochromatin and euchromatin formation.
Only a small fraction of the E(var) and Su(var) genes have been molecularly defined, with the uncharacterized majority holding promise to reveal important insights into the regulation of higher order chromatin structure. This article describes the cloning of one of these loci—the E(var)3-9 gene. The E(var)3-9 gene was identified in the same genetic screen for dominant E(var) mutations as were the E(var)3-4 and E(var)3-5 genes (Dorn et al. 1993). As described below, I found that E(var)3-9 mutations suppressed lt gene variegation as well. However, unlike these two genes, the E(var)3-9 mutations were not associated with recessive lethality but rather conferred recessive female sterility of the maternal effect class. The coincidence of the chromatin and sterility phenotypes prompted the further investigation of this gene, as it was suggestive of an essential role for E(var)3-9 in the establishment of a normal chromosome structure during embryogenesis.
MATERIALS AND METHODS
Drosophila stocks and culture conditions:
Stocks were maintained at 25° on cornmeal–malt medium described as standard medium by the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu). The E(var)3-91 and E(var)3-92 mutations were isolated in the E(var) screens of Dorn et al. (1993) and generously provided by R. Dorn and G. Reuter. The P{Δ2-3}-1C X chromosome insertion stock was a gift from the laboratory of W. Engels. The Df(3R)BSC24 deletion was created at the Bloomington Drosophila Stock Center using the hybrid element insertion (HEI) strategy (Gray et al. 1996; Preston et al. 1996; Parks et al. 2004). The Exelixis deficiencies Df(3R)Exel6152 and Df(3R)Exel6151 (Parks et al. 2004) and DrosDel deficiencies Df(3R)ED5301 and Df(3R)ED5331 (Ryder et al. 2004) were synthesized using flipase (FLP) recombinase and FRT-bearing transposon insertions (Golic and Golic 1996). Other mutations and strains described in this study are described in FlyBase (http://flybase.indiana.bio.edu).
Larvae for the Malpighian tubule assays were cultured at 25° under identical conditions of low crowding for each experiment. To eliminate potential sex differences, only female larvae were used for the assays. The larvae were derived from Df(2L)ltX10 Bc/SM1, lt16 females to effectively eliminate any maternal contribution to Malpighian tubule pigmentation. The paternal parent carried the T(2;3)TSTL, Cy Tb balancer for chromosomes 2 and 3, the lt-variegating rearrangement In(2LR)ltG10, and either a wild-type or E(var)3-9−-bearing third chromosome. The In(2LR)ltG10/Df(2L)ltX10 Bc and In(2LR)ltG10/Df(2L)ltX10 Bc; E(var)3-9−/+ larval progeny were identified as Bc− Tb+ individuals.
The pigment assays were performed on flies cultured at 25°. To examine the effects on lt variegation, simultaneous crosses were established between lt1 stw3 females and males bearing the TSTL, Cy Tb balancer for chromosomes 2 and 3, the lt-variegating rearrangement In(2L)ltx2, and either a wild-type or E(var)3-9−-bearing third chromosome. The In(2L)ltx2/lt1 or In(2L)ltx2/lt1; E(var)3-9−/+ offspring of each cross were identified as Cy+ Tb+ progeny. The effect on variegation of wm4 was determined by crossing in parallel wm4 females to males having the E(var)3-9− or wild-type chromosome balanced with TM3, Sb. The E(var)3-9+ chromosome was derived from precise excision of the P{wHy}CG11971DG08508 transposon.
Assays of Malpighian tubule pigmentation:
Wandering third instar female larvae of the appropriate genotype were collected as described above. The Malpighian tubules were dissected in 0.7% NaCl and stained in 0.1 μg/ml DAPI/0.7% NaCl on a multiwell slide. For each larva, 30 contiguous cells from each posterior arm of the tubules were scored for the presence of pigment granules, using UV illumination and 100× magnification. Data for each genotype were accumulated from a minimum of two experiments. Statistical analysis was performed using Statview 4.5 (Abacus Concepts, Berkeley, CA).
Drosophila red eye pigment assay:
Quantitation of eye pigments was performed according to the method of Ephrussi and Herold (1944) as described in Weiler and Wakimoto (2002). In brief, newly eclosed flies were sorted by sex and genotype, aged at 25° for five days and stored at −70°. Red pigment from aliquots of 10 heads per genotype was extracted in acidified ethanol, and the samples split into two tubes for duplicate processing and measurement of optical Absorbance at 480 nm. The duplicate readings for each sample were then averaged.
Stubble variegation assay:
A reciprocal translocation between the second and third chromosomes places the Stubble (Sb) gene, having the Sb1 mutation, under the repressive influence of the chromosome 2 heterochromatin in the T(2;3)SbV strain. Silencing of Sb1 effects a wild-type bristle, while its expression results in the Stubble phenotype. Crosses were performed between T(2;3)SbV/+; TM3, Ser e females and w; E(var)3-9 e/TM3, Ser e males cultured at 25°. The E(var)3-9 e/ T(2;3)SbV progeny were identified as Ser+ e+. The TM3, Ser e/ T(2;3)SbV siblings were identified as Ser− e+ and used as controls. No PEV modifying effect has been observed for the TM3, Ser e chromosome. To eliminate the potential influence of sex on variegation, only female progeny were scored. Fourteen bristles: the anterior and posterior sternopleurals, the upper and lower humerals, the anterior and posterior scutellars, and the posterior dorsocentrals, were scored for a Sb− or wild-type phenotype.
Assessment of female fertility:
Females were cultured at 25° with a supplement of live yeast–water paste to promote egg laying, prior to embryo collection using apple-grape–juice agar plates. Collected embryos were monitored over several days at 25° for hatching, and any resulting first instar larvae were transferred to culture medium for further development.
Meiotic recombination mapping:
E(var)3-9 was mapped on the third chromosome with reference to genetic markers of the ru1 h1 th1 st1 pp cu1 sr1 ca1 chromosome. Initially, E(var)3-91/ ru1 h1 th1 st1 pp cu1 sr1 ca1 females were crossed to ru1 h1 th1 st1 cu1 sr1 es Pr1 ca1/TM6B, Tb Hu ca e males, and Pr− male offspring bearing a recombinant chromosome were crossed to E(var)3-92/TM3, Sb females. The Pr+ Sb+ female offspring were tested for complementation of E(var)3-9− female sterility. Later experiments followed the same scheme but utilized a ru1 h1 Ki1 roe1 pp cu1 sr1 es ca1/TM3, Sb strain to detect recombinant chromosomes and map E(var)3-9 with respect to pp.
P element-mediated male recombination mapping:
The technique of P element-mediated male recombination was used to map E(var)3-9 mutations relative to the following P element insertions within the 85CD region: P{lacW}l(3)L6332L6332, P{lacW}neurj6B12, P{PZ}pum01688, P{PZ}Aats-trp03559, P{lacW}l(3)L4092L4092, P{EP}D1EP743, P{EP}EP3068EP3068, and P{SUPor-P}pumKG02259. For the first set of experiments ru1 h1 E(var)3-91 cu1 sr1 es/P insertion males bearing the P{Δ2-3}-1C transposase source on the X chromosome were crossed to ru1 h1 th1 st1 cu1 sr1 es Pr1 ca1/TM3, Sb females, and Pr− male offspring were screened for ru+ e− and ru− e+ recombinants. The frequency of recombinants ranged from 0 to 0.45% of males scored (1308 ≤ n ≤ 1963). Females bearing the recombinant chromosome heterozygous with E(var)3-92 were tested for fertility to ascertain if the E(var)3-91 allele was present on the recombinant chromosome. Segregation of the E(var)3-91 allele with the left chromosome arm marker (ru) indicated it was proximal to the P element, whereas segregation with distal (right arm) marker (e) indicated E(var)3-9 was distal to the P element. Subsequent mapping experiments utilized the CyO, H{PDelta2-3}Hop2.1 chromosome as the transposase source, which appeared to increase the recovery of recombinants (0.4–0.8%, for those crosses in which the males were counted). In addition, different combinations of proximal and distal phenotypic markers were used for the detection of recombinants. For two experiments, the male offspring of +/ CyO, H{PDelta2-3}Hop2.1; ru1 h1 E(var)3-91 cu1 sr1 es/Gl1 P insertion males crossed to w; TM3, Sb e / TM6B, Tb Hu e females were scored for Gl− e− and Gl+ e+ phenotypes. However, all Gl− e− males were found to be sterile, precluding the complementation test. For the remaining experiments, the st and ca mutations were recombined onto the P element-bearing chromosome, and st+ ca− and st− ca+ recombinant males were isolated from the offspring of +/CyO, H{PDelta2-3}Hop2.1; ru1 h1 E(var)3-91 cu1 sr1 es/st1 P ca1 or +/CyO, H{PDelta2-3}Hop2.1; E(var)3-92 e/st1 P ca1 insertion males crossed to st1 Sbsbd−1 es ro1 ca1 females.
Mobilization of P{wHy}CG11971DG08508:
Male flies bearing the P{wHy}CG11971DG08508 element and third chromosome P transposase source TMS, Sb1 P{Δ2-3}99B were crossed to w− females, and w− Sb+ males were recovered and stocked. Flies homozygous for the excision chromosome were analyzed by PCR for the retention of P element sequence within the E(var)3-9 gene. Single fly DNA was isolated for PCR by the method of Gloor et al. (1993). PCR reactions employed primers that hybridized to the E(var)3-9 genomic sequence proximal and distal to the insertion site [E(var)3-9 2261F primer, 5′-GCCGAACTGCTCCTGTGTCT-3′ and E(var)3-9 2660R primer, 5′-GTCGCTTTGTGGAACGGATT-3′, respectively] in combination with the universal P element end primer, Pendout2 (5′-CGACGGGACCACCTTATGTT-3′) (Mohr and Gelbart 2002) to allow precise excision events to be distinguished from imprecise excision events. The absence of the two P insertion-specific PCR products and the presence of a 400-bp genomic PCR product identified precise excision events. Both a precise excision line, which complemented E(var)3-9 mutants, and a w− line retaining both P element ends and failing to complement E(var)3-9 mutants, were retained.
P{wHy} mutagenesis and PCR analysis:
The genetic scheme of Huet et al. (2002) was followed to generate deletions flanking the P{wHy}CG11971DG08508 insertion. In brief, y1 w67c; Gla/CyO, P{hs-H\T-2} females carrying the heat-shock inducible hobo transposase were crossed to males bearing the P{wHy}CG11971DG08508 hybrid element insertion, and the progeny were heat-shocked for thirty minutes at 37° on days 2, 4, and 6 of development. Male offspring of genotype y1 w67c/Y; +/CyO, P{hs-H\T-2}; +/P{wHy}CG11971DG08508 were crossed to y1 w67c; D1/TM3, Sb Ser females. The progeny of this G1 cross were screened for w+ y− and w− y+ exceptional males. The w+ y− and w− y+ third chromosomes of these exceptional males were stocked prior to analysis by PCR. A maximum of one exceptional male per phenotypic class per vial was retained, ensuring that all isolates were independent.
PCR analysis of single fly DNA was performed as described above to assess if either P element end was retained within the exceptional chromosome. A rearrangement induced by replicative local hopping of the hobo element within the hybrid element followed by recombination between hobo elements would result in the deletion of a portion of the hybrid element including one P end, with hobo sequence delimiting the truncation. Given the orientation of the hybrid element, proximal deletions and distal duplications would be recovered within the w− y+ class, and distal deletions and proximal duplications within the w+ y− class.
To determine the nature of each recombination event, the genomic sequence flanking the deleted hybrid element end was amplified by inverse PCR (iPCR). The method of E. J. Rhem, Berkeley Drosophila Genome Project (BDGP), (http://www.fruitfly.org/about/methods/inverse.pcr.html) was followed using AluI to digest the genomic DNA and the hobo end primers specified by Mohr and Gelbart (2002). The PCR product was used directly for sequence determination following treatment with ExoSAP-IT (United States Biochemical, Cleveland) for most isolates. When secondary amplification products were present, the most abundant PCR product was gel purified (QIAGEN Gel Extraction kit, QIAGEN, Valencia, CA) prior to sequencing. In two cases in which the amplified sequence was too short to identify a unique genomic location, digestion with PstI and Sau3AI yielded larger iPCR products.
Genomic sequence analysis of E(var)3-9 alleles:
The insertion site of the P{wHy}CG11971DG08508 insertion was determined by PCR amplifying the 5′ and 3′ P flanking regions using primers E(var)3-9 2261F and Plac1 (5′-CACCCAAGGCTCTGCTCCCACAAT-3′, BDGP) for the proximal sequence and E(var)3-9 2660R and Pry4 (5′-CAATCATATCGCTGTCTC ACTCA-3′, BDGP) for the distal sequence, followed by sequence analysis of the ExoSAP-IT (United States Biochemical) treated products.
The coding region of the E(var)3-91, E(var)3-92, and E(var)3-93 alleles was amplified using a single fly genomic DNA template and two primer sets that produce overlapping PCR products: E(var)3-9 577F (5′-TTGGCAGGCCAGTGGAAA-3′) and E(var)3-9 1748R (5′-CCGCACCCATCTCCTCAA-3′) and E(var)3-9 1569F (5′-ACTGCCTGCTTGTGCTCCTT-3′) and E(var)3-9 2660R. The PCR products were purified using ExoSAP-IT (United States Biochemical) and sequenced from each end.
RESULTS
E(var)3-9 mutations have dominant reciprocal effects on euchromatic and heterochromatic PEV:
Two E(var)3-9 mutant alleles were recovered in the pUChsneory+ transposon mutagenesis screen of Dorn et al. (1993) for dominant mutations that enhanced wm4 variegation. However, there was a high rate of recovery of spontaneous mutations in the experiments and, like the majority of isolates, the E(var)3-91 and E(var)3-92 alleles were not associated with a P insertion (Dorn et al. 1993). An additional mutant allele E(var)3-93 is present on the balancer chromosome TM6B (as indicated in the stock list of G. Reuter in file reuter2.csv available at http://chervil.bio.indiana.edu:7092/stocks/labs/lab-info.html).
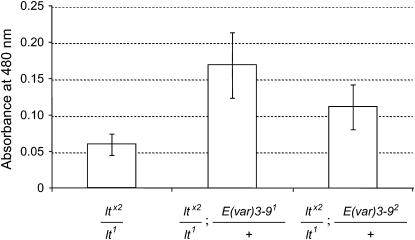
If enhancement of wm4 variegation by E(var)3-9 mutations reflects the conversion of portions of the genome into a more heterochromatic chromatin state, the variegation of heterochromatic genes might be expected to be suppressed (Hearn et al. 1991; Weiler and Wakimoto 2002). The effect of E(var)3-9 mutations on variegation of the heterochromatic lt gene was assessed by monitoring lt-dependant pigmentation of the adult eye and the larval Malpighian tubules. For measuring eye pigmentation, the flies were heterozygous for the lt-variegating inversion ltx2 and the hypomorphic lt1 allele, which is required for adult viability. As illustrated in Figure 1, extraction and quantification of eye pigment clearly showed that both the E(var)3-91 and E(var)3-92 mutations suppressed the variegation of ltx2. The E(var)3-93 allele similarly suppressed ltx2 variegation, although the results were not quantified by pigment assay. For assessing the effects of E(var)3-9 mutations on lt variegation in the Malpighian tubules, the larvae were heterozygous for the lt-variegating inversion ltG10 and the deficiency ltx10. As illustrated in Table 1, variegation of ltG10 was suppressed by both the E(var)3-91 and the E(var)3-92 alleles.
Figure 1.—
E(var)3-9 mutations suppress lt variegation in the adult eye. The eye pigmentation of 5-day-old ltx2/lt1; TM3/+ females was compared with that of sibling ltx2/lt1; E(var)3-91/+ and ltx2/lt1; E(var)3-92/+ females. Red eye pigment was extracted from groups of 10 heads and the mean absorbance value (± one standard deviation) for 9–10 groups is shown. The pigment values for both E(var)3-9 classes were significantly higher than that of the control (P < 0.001) using a Student's t-test.
TABLE 1.
E(var)3-9 mutations suppress lt-variegated pigmentation of the Malpighian tubule cells
| No. of larvae showing different percentages of pigmented Malpighian tubule cellsa
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | 5 | 10 | 15 | 20 | 25 | 30 | 35 | 40 | 45 | 50 | 55 | 60 | 65 | 70 | 75 | 80 | 85 | 90 | 95 | 100 | Meanb |
| ltG10/Df;+ | 31 | 5 | 3 | 1 | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 2.44 |
| ltG10/Df; E(var)3-91/+ | 20 | 8 | 5 | 3 | 1 | 1 | — | — | — | — | — | — | — | — | — | 1 | — | — | — | — | 5.77 |
| ltG10/Df; E(var)3-92/+ | 10 | 16 | 7 | 4 | 2 | 2 | 1 | — | — | — | — | — | — | — | — | — | — | 1 | — | — | 7.60 |
The number of larvae in each percentage pigmentation level, with column headings indicating the upper limit of the interval. Sixty cells were scored per individual.
The change in the mean percentage of pigmented cells due to mutation of E(var)3-9 was highly significant (P < 0.001 for each allele) as determined by a Mann-Whitney U-test.
The modifying effect of E(var)3-9 mutations did not appear to extend to cells of the adult bristle lineage. The generality of the enhancement of euchromatic gene variegation was investigated by testing the E(var)3-91 and E(var)3-92 mutations for a dominant effect on the variegation of the euchromatic Sb gene in the adult bristles. The T(2:3)SbV translocation carries the dominant Sb1 allele, which is variably inactivated due to its juxtaposition near to heterochromatin, and results in flies with a mixture of normal and Sb− bristles. Consequently, enhancement of PEV is viewed as a decrease in Sb− bristles, while suppression of PEV is observed as an increase in Sb− bristles. The average number of Sb− bristles (out of 14 per fly) was not significantly different between E(var)3-91 (8.5 ± 2.3; n = 75), E(var)3-92 (8.7 ± 2.1; n = 78), and the control sibling classes (8.0 ± 2.2; n = 76 and 8.6 ± 2.0; n = 74, respectively).
E(var)3-9 mutations cause recessive female sterility:
E(var)3-9 mutations act recessively to confer a female sterile phenotype. E(var)3-9 mutant females lay morphologically normal eggs that fail to hatch, placing them into the maternal effect lethal class of female sterile mutants. The hatching rate of embryos produced by E(var)3-91/E(var)3-92 heteroallelic females was determined to be 0.07% (Table 2). For 400 of these embryos, none of which hatched, Canton-S was the male parent, indicating that the sterility is not rescued by zygotic E(var)3-9+ expression. In other experiments, the embryos were examined under light mineral oil using bright field microscopy to confirm that the vast majority was fertilized (Wieschaus and Nusslein-Volhard 1998, data not shown). Similar results were obtained when the mutant alleles were heterozygous with the deficiency Df(3R)BSC24, which is deleted for E(var)3-9 (see below). The small percentage of embryos that hatched died at all subsequent developmental stages; some survived to adulthood with no obvious phenotypic abnormalities.
TABLE 2.
Viability of embryos produced by E(var)3-9 mutant females
| No. of embryos
|
||||
|---|---|---|---|---|
| Maternal genotype | Paternal genotype | Trial | Total | Hatched |
| E(var)3-91/E(var)3-92 | Mixeda | 1 | 210 | 0 |
| E(var)3-91/E(var)3-92 | E(var)3-91/E(var)3-92 | 1 | 180 | 0 |
| 2 | 592 | 1 | ||
| E(var)3-91/E(var)3-92 | +/+ | 1 | 400 | 0 |
| E(var)3-91/Df(3R)BSC24 | Mixedb | 1 | 628 | 7 |
| 2 | 380 | 0 | ||
| 3 | 375 | 0 | ||
| E(var)3-92/Df(3R)BSC24 | Mixedb | 1 | 597 | 10 |
| 2 | 294 | 1 | ||
The females were permitted to mate with their siblings, which consisted of E(var)3-91/E(var)3-92, E(var)3-91/TM3, Sb, and E(var)3-92/TM3, Sb males.
The females were permitted to mate with their siblings, which consisted of E(var)3-9/Df(3R)BSC24, E(var)3-9/TM3, Ser, and Df(3R)BSC24/TM3, Sb males.
The E(var)3-9 gene corresponds to predicted gene CG11971:
Meiotic mapping of E(var)3-9 was undertaken with respect to its recessive female sterile phenotype. Recombination with the multiply marked ru1 h1 th1 st1 pp cu1 sr1 ca1 chromosome revealed that the E(var)3-91 allele was located between p (3–48) and cu (3–50) at ∼48.8 map units on the right arm of chromosome 3. Four chromosomes having a recombination event in the p–cu interval, three retaining the E(var)3-91 allele and one not, were tested for enhancement of wm4 variegation. A perfect correlation was observed between the female sterility and the E(var) phenotype.
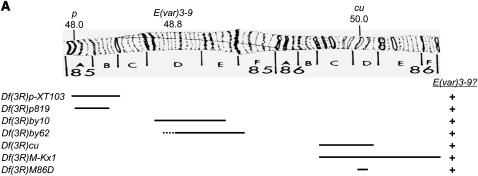
The results of meiotic mapping indicated that E(var)3-9 resides within the 85A–86D intervals of the cytogenetic map. Only seven deficiencies were publicly available at the time and spanned the p–cu region: Df(3R)p-XT103, Df(3R)p819, Df(3R)by10, Df(3R)by62, Df(3R)cu, Df(3R)M-Kx1, and Df(3R)M86D. As illustrated in Figure 2A, complementation analysis of E(var)3-91 with these deficiencies revealed that all complemented the recessive female sterility. Together, they deleted intervals 85A2–87B5, with the exception of intervals 85C1-2–85D8 and 85F6–86C1. The meiotic recombination data were most consistent with E(var)3-9 localizing within the 85C1-2–85D8 interval, as delineated by Df(3R)p-XT103 and Df(3R)by10.
Figure 2.—
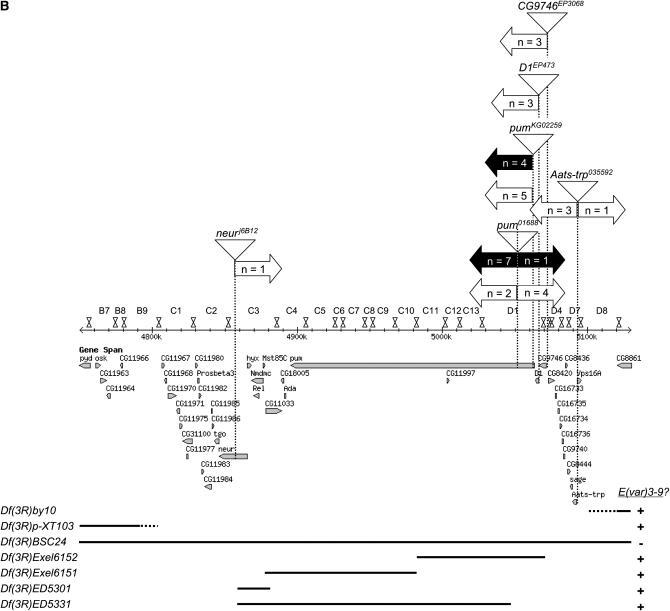
Complementation analyses with deficiencies spanning the 85 and 86 cytological intervals localize the E(var)3-9 gene to 85C1-3 or 85D2-8. (A) A drawing of cytological intervals 85 and 86 is shown above depictions of the alignment of seven deficiencies, with the approximate deleted region illustrated as a black line. The results of a complementation test between each deficiency and E(var)3-91, for female fertility, is shown at right. (B) The chromosome map of the 85B7–85D8 region is demarcated by numbered band and illustrated as it corresponds to the genomic sequence and genes (http://www.flybase.org, Release 5.1). Vertical dotted lines indicate the positions of six P-element insertions (illustrated as triangles) used for male recombination mapping. (Two insertions yielding no data are omitted.) The right and left block arrows projecting from each dotted line reflect the relative position of the E(var)3-9 gene with respect to that P insertion as indicated by the phenotype of the male recombinants, of the number specified by the value of n. The data from multiple experiments are combined. White block arrows depict the results of male recombination mapping with the E(var)3-91 allele, and black arrows depict the results of male recombination mapping with the E(var)3-92 allele. Shown below the map are depictions of seven deficiencies as they correspond to this region. The deleted sequences and results of complementation analysis with E(var)3-91 and/or E(var)3-92 are illustrated as in A.
The technique of P element-mediated male recombination mapping (Preston and Engels 1996; Chen et al. 1998) was employed to better localize E(var)3-9 within the 85CD region. The results of a series of mapping experiments are illustrated in Figure 2B. Initial data (i.e., a subset of the data set presented in Figure 2B) suggested that the E(var)3-91 mutation mapped within the pumilio (pum) gene. However, the results of complementation tests with pum mutant alleles, P{nos-pum} transgene rescue experiments, and genomic sequence analysis failed to support this hypothesis (data not shown). Additional mapping experiments with both the E(var)3-91 and E(var)3-92 alleles yielded conflicting results for the localization of E(var)3-9 with respect to the P{PZ}pum01688 insertion. This avenue of gene mapping was not pursued further.
The recent availability of several relatively small deletions engineered via in vivo manipulation of P elements allowed a further narrowing of the region of interest. The results of complementation analyses are illustrated in Figure 2B. The Df(3R)BSC24 deletion presumably spans from the pyd gene (85B7) to the Fps85D gene (85D15). Df(3R)BSC24 failed to complement the female sterility of E(var)3-9 mutations as anticipated. The deficiencies Df(3R)Exel6152, Df(3R)Exel6151, Df(3R)ED5301, and Df(3R)ED5331 are predicted to collectively delete the region from neuralized (85C3) through CG9746 (85D2). All four deficiencies complemented E(var)3-9 mutations. These data, in combination with the previous deficiency data, identify two candidate regions for E(var)3-9 corresponding approximately to cytological intervals 85C1-3 and 85D2-8.
The 85D2-8 cytological interval is defined proximally by the Df(3R)6152 distal breakpoint and distally by the proximal breakpoint of Df(3R)by10. Although the by10 deficiency has not been mapped molecularly, its cytological characterization would suggest that 11 genes, 3 characterized (sage, Aats-trp, and Vps16A) and 8 predicted, compose this interval.
The 85C1-3 cytological interval is defined proximally by the Df(3R)p-XT103 distal breakpoint and distally by the common breakpoint of Df(3R)ED5301 and Df(3R)ED5331, within the neuralized gene. The breakpoint of the Df(3R)p-XT103 deficiency was molecularly mapped by Kim-Ha et al. (1991). An examination of their published data suggests that the breakpoint maps between predicted genes CG11966 and CG11967. Fifteen genes, two characterized (tango and Proβ3) and thirteen predicted, compose this interval, which extends from CG11967 to tango. As the P element-mediated male recombination mapping data (Figure 2B) strongly suggested that E(var)3-9 localized proximal to 85D (26 of 32 recombinants), further analyses focused upon this interval.
There are no mutant alleles or transposable element insertions in the CG11967 to tango region that have been reported to confer a female sterile phenotype. However, three lethal insertions have been reported. Although the E(var)3-91 and E(var)3-92 alleles were homozygous viable, it remained a formal possibility that they were hypomorphic alleles of an essential gene. Complementation analysis was performed between lethal P insertion alleles in the tango and CG11985 genes and the E(var)3-91 and E(var)3-92 alleles. In all cases, complementation was observed. As the P insertion allele of CG11984 is balanced with TM6B, which bears the E(var)3-93 allele, complementation of this lethal allele was assumed.
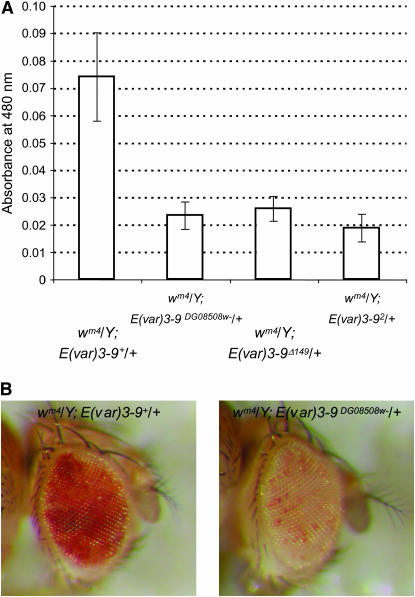
A viable P{wHy} insertion that localizes within the 85C1-3 interval was obtained with the aim of isolating a nested set of deletions using the P{wHy} hybrid transposable element-based method (Huet et al. 2002, see below). Females homozygous for the chromosome bearing the P{wHy}CG11971DG08508 insertion were discovered to be sterile. A complementation test with E(var)3-9 mutant alleles revealed that the chromosome carried a mutant allele of E(var)3-9. To determine if the female sterility was due to the P{wHy} insertion, the transposon was mobilized and excision events identified by loss of the white+ (w+) eye color marker. Precise excision of P{wHy}CG11971DG08508 was found to restore female fertility. In addition, the location of the element within predicted gene CG11971 was verified by determination of the genomic sequence flanking both the 5′ and 3′ P ends. As illustrated in Figure 3, a strain bearing a w− derivative of the P{wHy}CG11971DG08508 insertion, in comparison to the precise excision line, enhanced variegation of wm4. Similar levels of pigmentation were observed for flies bearing the E(var)3-92 allele, the w− P insertion allele P{wHy}CG11971DG08508w−, and the deletion allele E(var)3-9Δ149, which is missing much of the coding sequence (see below). These results confirmed that E(var)3-9 corresponds to gene CG11971.
Figure 3.—
E(var)3-9 mutations enhance wm4 variegation in the adult eye. (A) Extracted pigment from groups of 10 males, aged 5 days, was quantified by spectrophotometry. The mean absorbance value (± one standard deviation) for 10 groups per genotype is shown. The decrease in pigmentation for the E(var)3-9DG08508w−, E(var)3-9Δ149 and E(var)3-92 alleles with respect to the wild-type E(var)3-9 allele, obtained by precise excision of the P{wHy}DG08508 element, is highly significant (P < 0.001 for each allele) as determined by a Student's t-test. Enhancement was also observed in females, which had a much higher overall pigmentation level (data not shown). (B) The extent of wm4 variegation is illustrated for a representative male fly bearing the E(var)3-9+ (left) or E(var)3-9Δ149 allele (right).
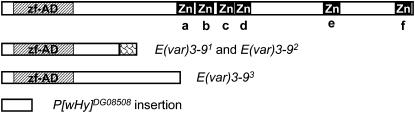
The E(var)3-9 gene product is a zinc finger protein. The C-terminal half of the protein was predicted to have minimally two, but up to six, zinc fingers of the C2H2 type. As illustrated in Figure 4, three zinc fingers were identified when the E(var)3-9 protein sequence was queried against the Pfam and Prosite databases, two in common and one unique to each search. The third zinc finger identified by Prosite was found as a potential hit by Pfam. Two novel zinc fingers, in addition to the four already noted, were found in a query against the SMART database. A zinc finger-associated domain was found at the N terminus (Pfam). These features implicated the E(var)3-9 protein in nucleic acid binding, perhaps as a dimer. Numerous potential phosphorylation sites were also predicted.
Figure 4.—
The 588 amino acid E(var)3-9 protein has multiple predicted zinc fingers and a zinc finger-associated domain. As illustrated in a diagram of the coding sequence, high confidence matches to a C2H2 type zinc finger (Zn) motif were found when the E(var)3-9 protein sequence was queried against the Pfam database (c, d, and f), the Prosite database (c, e, and f), and the SMART database (a–f). An N-terminal zinc finger-associated domain (zf-AD) was also detected in a Pfam search. The potential protein products of E(var)3-9 mutant alleles are illustrated below the full-length protein. The frameshift mutation present in the E(var)3-91 and E(var)3-92 alleles and the nonsense mutation present in the E(var)3-93 allele would terminate protein synthesis prior to translation of the zinc fingers. An appendage of 24 amino acids due to out-of-frame translation is depicted for the E(var)3-91 and E(var)3-92 gene products. The position of the P{wHy}DG08508 insertion with respect to the coding region is shown.
Isolation of deletions and duplications using the P{wHy} hybrid element:
The P{wHy} mutagenesis (Huet et al. 2002) was carried out using the P{wHy}CG11971DG08508 insertion within the E(var)3-9 gene with the intent of isolating a deletion allele. Deletions are generated using P{wHy} by inducing the local replicative transposition of the hobo (H) element carried within the hybrid transposon, using ectopically expressed hobo transposase. When the transposed hobo insertion is in the same orientation as the original element, recombination between hobo elements will result in duplication or deletion of the intervening region. These events can be detected through loss of expression of one of the marker genes that flank the hobo element within the hybrid transposon, w+ or yellow+ (y+). As the P{wHy}CG11971DG08508 insertion is oriented with the w+ gene proximal and the y+ gene distal to H, proximal deletions should appear w− y+ and distal deletions should appear w+ y−.
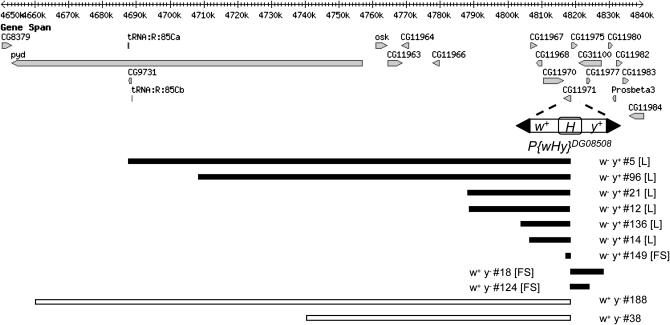
Males bearing the P{wHy}CG11971DG08508 element and a transgene containing the hobo transposase under the control of the heat shock promoter were heat shocked to induce hobo transposition and mated individually to y w females (as described in materials and methods). Among the progeny of 195 fertile males, 44 independent fertile w− y+ or w+ y− exceptional males were identified. PCR analysis of the P{wHy} hybrid element in lines derived from these males revealed that 16 isolates retained either the 5′ or 3′ P end, but not both, indicating that a deletion or duplication event had occurred. iPCR was carried out to amplify the genomic sequence flanking the truncated transposon end (i.e., the hobo element), for the purpose of DNA sequencing. The result of sequence mapping, which was successful for 11 lines, is shown in Figure 5. Two lines were lost prior to iPCR, and three failed to yield a single unique-sequence iPCR product. The deletion in line 149 (E(var)3-9Δ149) is internal to the E(var)3-9 coding sequence.
Figure 5.—
Molecularly mapped deletions and duplications isolated by P{wHy}DG08508 mutagenesis. The chromosome 3R genomic sequence and genes (http://www.flybase.org, Release 5.1) in the region affected by the deletions and duplications isolated in this study are illustrated. The P{wHy} hybrid transposon was inserted within predicted gene CG11971, identified herein as E(var)3-9, at position 3R:4,818,459 (first nucleotide of the 8-bp duplication) in the orientation shown. As described in the results, replicative hopping of the hobo (H) element followed by a recombination between the original and transposed H elements resulted in deletions (black bars) and duplications (white bars) of sequence flanking the P{wHy} insertion. The events were detected through loss of expression of the w+ or y+ marker genes of the transposon. Isolates having a homozygous lethal [L] or female sterile [FS] phenotype are indicated. The size of the deletion or duplication for each isolate was as follows: 5 (131,065 bp); 96 (110,076 bp); 21 (30,322 bp); 12 (29,841 bp); 136 (14,739 bp); 14 (12,090 bp); 149 (1065 bp); 18 (9940 bp); 124 (5598 bp); 188 (158,344 bp); 38 (78,061 bp).
Sequence analysis of the three spontaneous E(var)3-9 mutant alleles:
The E(var)3-9 mutant alleles isolated by Dorn et al. (1993) were characterized by sequence analysis of PCR-amplified genomic DNA isolated from homozygous individuals. For each allele, the E(var)3-9 coding sequence matched the published genomic sequence (Adams et al. 2000; FlyBase Release 5.1) with one exception. Both alleles possessed identical frame shift mutations resulting from the replacement of 11 bp of genomic sequence with an unrelated 16 bp of sequence. As a consequence of this mutation, the predicted mutant protein product should be identical with the wild-type protein (of 588 amino acids in length) for 168 amino acids and then terminate following an additional 24 amino acids resulting from out-of-frame translation. Sequence analysis of the E(var)3-93 allele of TM6B was performed using genomic DNA from an individual heterozygous for E(var)3-93 and the deletion allele E(var)3-9Δ149, since TM6B homozygotes are not viable. One primer of each primer pair used for PCR amplification corresponded to the deleted region, resulting in specific amplification of the E(var)3-93 template. A nonsense mutation was identified that should cause premature termination of the E(var)3-9 protein after 255 amino acids. The potential translation products of these mutants, as they relate to features of the E(var)3-9 protein, is illustrated in Figure 4.
DISCUSSION
The results described herein demonstrate that E(var)3-9 is a zinc finger protein and corresponds to predicted gene CG11971. A BLAST search with the E(var)3-9 protein sequence against the nonredundant protein database fails to reveal a homolog outside of the genus Drosophila. Rather, the significant hits are primarily to zinc finger proteins and show limited homology in regions within the C-terminal half of the protein. Consistent with this result, E(var)3-9 was also identified in the screen for fast-evolving genes of Schmid and Tautz (1997) as anon1E9. Another modifier of PEV Su(var)3-7 encodes a heterochromatin-associated protein with multiple zinc fingers (Reuter et al. 1990; Cleard et al. 1995). Intriguingly, it too is a fast-evolving gene (Jaquet et al. 2006). At this time there is little functional information about the E(var)3-9 gene product with which to interpret this characteristic. However, one of three potential explanations for why proteins evolve rapidly noted by Schmid et al. (1999) is involvement in reproduction. This report shows that E(var)3-9 is required for female fertility, and mutations appear to decrease male fertility as well (unpublished observation).
The mutation identified in alleles E(var)3-91 and E(var)3-92 derived from an insertion–deletion (indel) event, strongly suggesting that it arose spontaneously during the mitotic divisions of gametogenesis (i.e., premeiotically) and/or was present within the population of flies used for the mutagenesis. I was unable to obtain new copies of the two strains to verify that they harbored the same mutation. Although it is formally possible that the alleles were confused during years of stock maintenance, DNA samples that were isolated from each strain in the year 2000 were recently sequenced and both found to harbor this indel allele. The recombinant lines produced from meiotic recombination mapping using the E(var)3-91 allele prior to this date similarly possessed the indel allele.
The E(var)3-9Δ149 allele isolated in this study was generated using the P{wHy} deletion-generator technique. The deletion extends 1065 bp from the P{wHy} insertion site and includes part of the first and the entire second and third exons of the gene. This technique proved an efficient means to isolate deletions of an extensive size range. Approximately 8% of the fertile G1 crosses yielded a putative deletion or duplication as compared to frequencies of ∼11% and 15% for the two P{wHy} insertions employed by Huet et al. (2002) and between 12% and 14% for the three insertions employed by Mohr and Gelbart (2002). Although different P{wHy} insertions may be expected to differ in efficiency, the slightly lower recovery rate noted here may also result from the fact that a maximum of one w− y+ and/or w+ y− exceptional offspring per G1 male was retained in this study. Similar to Huet et al. (2002), the majority of the sequence-mapped events (9 of 11) were deletions. The deletions have not been confirmed by genetic criteria, which would only be possible for the 2 largest deletions due to an absence of mutant alleles for the majority of genes. From this small number of deletions, several conclusions can be drawn about uncharacterized genes in the vicinity of E(var)3-9. Predicted genes CG11975, CG31100, and CG11977 are apparently not essential for viability, as isolates 18 and 124 were homozygous viable. On the other hand, deletion of predicted genes CG11970, CG11968, and CG11967 (isolates 12, 14, and 136) was homozygous lethal indicating that at least one of these genes is essential.
The E(var)3-9 gene is a member of the E(var) class of dominant PEV modifiers, which were isolated as a means to identify factors that inhibit heterochromatin formation and/or promote a euchromatic chromatin state. In support of this proposed function, this report illustrates that mutations in E(var)3-9 have the reciprocal effect on (i.e., suppress) variegation of the heterochromatic lt gene. This property would be expected for mutations that alter the balance of heterochromatin and euchromatin within the nucleus (Weiler and Wakimoto 2002). The majority of PEV modifier mutations studied to date are loss-of-function alleles, suggesting that the wild-type product has a concentration-dependent function (Schotta et al. 2003). Is this also true of E(var)3-9 mutations? The three spontaneous mutations create truncated proteins that could conceivably interfere with the function of the wild-type protein, within a protein complex. It is not known if these abnormal proteins are stable. However, the P{wHy} hybrid element is inserted within the first exon of the E(var)3-9 gene following the 43rd codon (see Figure 4). Even if a gene product were produced, it would bear little resemblance to the E(var)3-9 protein. Therefore, enhancement of PEV by E(var)3-9 mutations is surely due to haploinsufficiency of the locus. However, the spontaneous mutations could also be antimorphic.
Both the maternal effect lethal phenotype and the E(var) phenotype of E(var)3-9 mutations could be due to an alteration of chromatin structure. The maternal effect lethal phenotype could reflect a role for E(var)3-9 in establishing an appropriate chromatin structure essential for early embryonic development. As zygotic E(var)3-9 expression is not required for viability, the maintenance of this particular chromatin structure is either not essential or, if vital, can be carried out by other proteins. Nevertheless, E(var)3-9 mutations do have phenotypic effects during the larval and adult stages, as evidenced by the modification of euchromatic and heterochromatic PEV. In fact, all of the assays for modification of PEV described in this study involve paternal transmission of the E(var)3-9 mutant allele. These data indicate that a decrease in E(var)3-9 protein function later in development is consequential and implicate E(var)3-9 in a nonessential and/or partially redundant chromatin maintenance function.
There does not appear to be an absolute requirement for maternally provided E(var)3-9 for viability. It is curious that a small fraction of embryos produced by E(var)3-9 mutant females hatch and variably develop as far as adulthood. Perhaps other embryonic gene products are occasionally able to compensate for the absence of E(var)3-9. While a slight difference in hatching rate was observed between E(var)3-91/E(var)3-92 heteroallelic females in comparison with each allele heterozygous with a deficiency, the significance and cause is uncertain. The E(var)3-91 and E(var)3-92 chromosomes may share second-site mutations that affect the hatching rate. Or, as noted above, the allele could also be an antimorph. Another variable in this study was the genotype of the males. A portion of the embryos assayed for viability in the E(var)3-9−/Df(3R)BSC24 experiments would have been heterozygous for a wild-type E(var)3-9 allele. This fraction would have been substantially smaller in the E(var)3-91/E(var)3-92 experiments, where over half of the embryos derived from a cross to E(var)3-91/E(var)3-92 males. Intriguingly, the single embryo to hatch from the cross between E(var)3-91/E(var)3-92 males and females died as a young first instar larva. Further investigation will be required to determine if the zygotic genotype has a small effect.
E(var)3-9 is not the first example of an association between female fertility and PEV. Dominant mutations in the Su(var)2-1, Su(var)3-3, and Su(var)3-5 genes are recessive female sterile (Reuter et al. 1982; Reuter et al. 1986). Interestingly, mutations of all three genes exhibit a recessive sensitivity to the histone deacetylase inhibitor butyrate. An increase in the level of histone H4 acetylation was observed in mutant larvae bearing the Su(var)2-11 mutation (Dorn et al. 1986). Although the Su(var)2-1 and Su(var)3-5 genes have yet to be cloned, Su(var)3-3 was recently determined to encode the Drosophila homolog of the human histone demethylase LSD1, which can be found in a protein complex that includes the histone deacetylase Rpd3 (Rudolph et al. 2007). Data showing a key role for Su(var)3-3 in chromatin packaging of the germline serve to connect the PEV and fertility phenotypes in this example (Rudolph et al. 2007). By further example, mutations in the piwi, Spindle-E (a.k.a. homeless), and aubergine genes result in female sterility and have been shown to suppress the silencing of a P{hsp70-w+} transgene insertion into pericentric heterochromatin (Pal-Bhadra et al. 2004). These gene products are part of the RNAi machinery and illustrate a link between small RNAs and heterochromatin formation in Drosophila (reviewed by Kavi et al. 2005). As a zinc finger protein, the E(var)3-9 gene product could have a role in either histone modification or small RNA biogenesis, or an as yet unrecognized facet of chromatin formation. Just as the phenotype of suppression of variegation has been a rewarding tool for identifying gene products and mechanisms involved in the formation of heterochromatin, the analysis of enhancers of variegation such as E(var)3-9 should be fruitful in revealing the important processes and components that antagonize it.
Acknowledgments
The author thanks R. Kellum for suggestions on the manuscript. This work was supported by a National Science Foundation grant (award nos. MCB0131604 and MCB0531808).
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas et al., 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. [DOI] [PubMed] [Google Scholar]
- Chen, B., T. Chu, E. Harms, J. P. Gergen and S. Strickland, 1998. Mapping of Drosophila mutations using site-specific male recombination. Genetics 149: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard, F., M. Matsarskaia and P. Spierer, 1995. The modifier of position-effect variegation Suvar(3)7 of Drosophila: there are two alternative transcripts and seven scattered zinc fingers, each preceded by a tryptophan box. Nucleic Acids Res. 23: 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn, R., S. Heymann, R. Lindigkeit and G. Reuter, 1986. Suppressor mutations of position-effect variegation in Drosophila melanogaster affecting chromatin properties. Chromosoma 93: 398–403. [Google Scholar]
- Dorn, R., J. Szidonya, G. Korge, M. Sehnert, H. Taubert et al., 1993. P transposon-induced dominant enhancer mutations of position-effect variegation in Drosophila melanogaster. Genetics 133: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg, J. C., T. C. James, D. M. Foster-Hartnett, T. Hartnett, V. Ngan et al., 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 87: 9923–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi, B., and J. L. Herold, 1944. Studies of eye pigments of Drosophila. I. Methods of extraction and quantitative estimation of the pigment components. Genetics 29: 148–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas, G., J. Gausz, M. Galloni, G. Reuter, H. Gyurkovics et al., 1994. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature 371: 806–808. [DOI] [PubMed] [Google Scholar]
- Gloor, G. B., C. R. Preston, D. M. Johnson-Schlitz, N. A. Nassif, R. W. Phillis et al., 1993. Type I repressors of P element mobility. Genetics 135: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, K. G., and M. M. Golic, 1996. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 144: 1693–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, Y. H., M. M. Tanaka and J. A. Sved, 1996. P-element-induced recombination in Drosophila melanogaster: hybrid element insertion. Genetics 144: 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal, S. I., and J. C. Rice, 2004. Regulation of heterochromatin by histone methylation and small RNAs. Curr. Opin. Cell Biol. 16: 230–238. [DOI] [PubMed] [Google Scholar]
- Hayashi, S., A. Ruddell, D. Sinclair and T. Grigliatti, 1990. Chromosomal structure is altered by mutations that suppress or enhance position effect variegation. Chromosoma 99: 391–400. [DOI] [PubMed] [Google Scholar]
- Hearn, M. G., A. Hedrick, T. A. Grigliatti and B. T. Wakimoto, 1991. The effect of modifiers of position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics 128: 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet, F., J. T. Lu, K. V. Myrick, L. R. Baugh, M. A. Crosby et al., 2002. A deletion-generator compound element allows deletion saturation analysis for genomewide phenotypic annotation. Proc. Natl. Acad. Sci USA 99: 9948–9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, T. C., and S. C. Elgin, 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6: 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquet, Y., M. Delattre, J. Montoya-Burgos, A. Spierer and P. Spierer, 2006. Conserved domains control heterochromatin localization and silencing properties of SU(VAR)3–7. Chromosoma 115: 139–150. [DOI] [PubMed] [Google Scholar]
- Kavi, H. H., H. R. Fernandez, W. Xie and J. A. Birchler, 2005. RNA silencing in Drosophila. FEBS Lett. 579: 5940–5949. [DOI] [PubMed] [Google Scholar]
- Kim-Ha, J., J. L. Smith and P. M. Macdonald, 1991. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66: 23–35. [DOI] [PubMed] [Google Scholar]
- Locke, J., M. A. Kotarski and K. D. Tartof, 1988. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics 120: 181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, S. E., and W. M. Gelbart, 2002. Using the P[wHy] hybrid transposable element to disrupt genes in region 54D–55B in Drosophila melanogaster. Genetics 162: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1930. Types of visible variations induced by X-rays in Drosophila. J. Genet. 22: 299–335. [Google Scholar]
- Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra et al., 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Preston, C. R., and W. R. Engels, 1996. P-element-induced male recombination and gene conversion in Drosophila. Genetics 144: 1611–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, C. R., J. A. Sved and W. R. Engels, 1996. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics 144: 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, G., and I. Wolff, 1981. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol. Gen. Genet. 182: 516–519. [DOI] [PubMed] [Google Scholar]
- Reuter, G., R. Dorn and H. J. Hoffmann, 1982. Butyrate sensitive suppressor of position-effect variegation mutations in Drosophila melanogaster. Mol. Gen. Genet. 188: 480–485. [DOI] [PubMed] [Google Scholar]
- Reuter, G., R. Dorn, G. Wustmann, B. Friede and G. Rauh, 1986. Third chromosome suppressors of position-effect variegation loci in Drosophila melanogaster. Mol. Gen. Genet. 202: 481–487. [Google Scholar]
- Reuter, G., J. Gausz, H. Gyurkovics, B. Friede, R. Bang et al., 1987. Modifiers of position-effect variegation in the region from 86C to 88B of the Drosophila melanogaster third chromosome. Mol. Gen. Genet. 210: 429–436. [DOI] [PubMed] [Google Scholar]
- Reuter, G., M. Giarre, J. Farah, J. Gausz, A. Spierer et al., 1990. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature 344: 219–223. [DOI] [PubMed] [Google Scholar]
- Rudolph, T., M. Yonezawa, S. Lein, K. Heidrich, S. Kubicek et al., 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3–3. Mol. Cell 26: 103–115. [DOI] [PubMed] [Google Scholar]
- Ryder, E., F. Blows, M. Ashburner, R. Bautista-Llacer, D. Coulson et al., 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K. J., and D. Tautz, 1997. A screen for fast evolving genes from Drosophila. Proc. Natl. Acad. Sci. USA 94: 9746–9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K. J., L. Nigro, C. F. Aquadro and D. Tautz, 1999. Large number of replacement polymorphisms in rapidly evolving genes of Drosophila. Implications for genome-wide surveys of DNA polymorphism. Genetics 153: 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann et al., 2002. Central role of Drosophila SU(VAR)3–9 in histone H3–K9 methylation and heterochromatic gene silencing. EMBO J 21: 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta, G., A. Ebert, R. Dorn and G. Reuter, 2003. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Biol. 14: 67–75. [DOI] [PubMed] [Google Scholar]
- Seum, C., A. Spierer, D. Pauli, J. Szidonya, G. Reuter et al., 1996. Position-effect variegation in Drosophila depends on dose of the gene encoding the E2F transcriptional activator and cell cycle regulator. Development 122: 1949–1956. [DOI] [PubMed] [Google Scholar]
- Sinclair, D. A., T. A. Milne, J. W. Hodgson, J. Shellard, C. A. Salinas et al., 1998. The Additional sex combs gene of Drosophila encodes a chromatin protein that binds to shared and unique Polycomb group sites on polytene chromosomes. Development 125: 1207–1216. [DOI] [PubMed] [Google Scholar]
- Sinclair, D. A. R., R. C. Mottus and T. A. GRIGLIATTI, 1983. Genes which suppress position effect variegation in Drosophila melanogaster are clustered. Mol. Gen. Genet. 191: 326–333. [Google Scholar]
- Sun, F. L., M. H. Cuaycong, C. A. Craig, L. L. Wallrath, J. Locke et al., 2000. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. USA 97: 5340–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun, J. W., R. Deuring, M. Scott, M. Kissinger, A. M. Pattatucci et al., 1992. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SW12. Cell 68: 561–572. [DOI] [PubMed] [Google Scholar]
- Wakimoto, B. T., and M. G. Hearn, 1990. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath, L. L., and S. C. Elgin, 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9: 1263–1277. [DOI] [PubMed] [Google Scholar]
- Weiler, K. S., and B. T. Wakimoto, 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577–605. [DOI] [PubMed] [Google Scholar]
- Weiler, K. S., and B. T. Wakimoto, 2002. Suppression of heterochromatic gene variegation can be used to distinguish and characterize E(var) genes potentially important for chromosome structure in Drosophila melanogaster. Mol. Genet. Genomics 266: 922–932. [DOI] [PubMed] [Google Scholar]
- Wieschaus, E., and C. Nusslein-Volhard, 1998. Looking at embryos, pp. 179–214 in Drosophila: A Practical Approach, edited by D. B. Roberts. Oxford University Press, New York.
- Wustmann, G., J. Szidonya, H. Taubert and G. Reuter, 1989. The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol. Gen. Genet. 217: 520–527. [DOI] [PubMed] [Google Scholar]