Abstract
We consider inference for demographic models and parameters based upon postprocessing the output of an MCMC method that generates samples of genealogical trees (from the posterior distribution for a specific prior distribution of the genealogy). This approach has the advantage of taking account of the uncertainty in the inference for the tree when making inferences about the demographic model and can be computationally efficient in terms of reanalyzing data under a wide variety of models. We consider a (simulation-consistent) estimate of the likelihood for variable population size models, which uses importance sampling, and propose two new approximate likelihoods, one for migration models and one for continuous spatial models.
THERE are two common approaches to analyzing population genetic data. The first approach involves (i) inferring a genealogical or phylogenetic tree for the data and (ii) making inferences about demographic or other parameters conditional on this tree. Examples of this include inference of the demography (Underhill et al. 2001), nested clade analysis (Templeton et al. 1987), and phylogeographic and spatial analysis (Emerson and Hewitt 2005; French et al. 2005). Often this approach is applied informally, with the qualitative features of the inferred tree being used to suggest plausible demographic histories for the sample (e.g., Shen et al. 2000).
The second approach involves joint inference of the genealogical tree and the parameters. In many cases the genealogical tree is a nuisance parameter, and calculation of the likelihood for the parameters involves integrating out the unknown tree, for example, in inference about various demographic models under a coalescent prior, including variable population sizes (Griffiths and Tavaré 1994a; Kuhner et al. 1998; Drummond et al. 2005) and population structure (Bahlo and Griffiths 1998; Beerli and Felsenstein 1999), inference for selection (Coop and Griffiths 2004), dispersal of a population (Brooks et al. 2007), and inference for recombination rates (Griffiths and Marjoram 1996; Kuhner et al. 2000; Fearnhead and Donnelly 2002). (In the latter case the genealogical information is contained in a graph and not in a tree.)
The advantage of the second approach is that, assuming the model for the genealogical tree is reasonable, the uncertainty in this genealogy is correctly incorporated into the inference about the parameters of interest. This is particularly important for data where there is considerable uncertainty in the genealogy (which is common for many data sets). The first approach of conditioning on a single estimate of the genealogy can sometimes lead to biases in estimates and, more generally, to underestimates of the uncertainty in the parameters. These problems often mean that analysis conditional on the tree is often used primarily to test hypotheses (Templeton et al. 1987; French et al. 2005), rather than for estimating parameters of appropriate models.
However, implementing the second approach is considerably more challenging and generally requires the use of modern computationally intensive statistical methods (Stephens and Donnelly 2000). In particular, this often requires the development of customized programs to analyze the data under the specific model or models of interest, and the application of this approach can be limited by the availability of suitable software.
In this article we consider a new approach, which lies between these two approaches. The basic idea is (i) to perform inference for the genealogical/phylogenetic tree using a suitable Bayesian approach, obtaining a sample of trees from the posterior and (ii) to perform inference on the parameters of interest using this sample of trees. The idea is that by using a sample of trees in an appropriate way we can still take account of the uncertainty within the inference for the tree, but that this approach will be less computationally intensive and more widely applicable than the second approach above.
We consider inference under three different demographic models: (a) variable population size, (b) migration between discrete subpopulations, and (c) continuous spatial structure. For model a we present a simple importance-sampling approach that can reweight a sample of trees so that the resulting weighted sample approximates the posterior distribution of the genealogy under any variable population size model. For models b and c we propose approximate-likelihood functions based on specifying a probability model for the population or on spatial information of the sample given the genealogy.
Our aim is to evaluate the potential for this approach of postprocessing a sample of genealogical trees. As such we focus on the specific case of inference for a nonrecombining DNA region with infinite-sites data and known topology. The advantage of focusing on this special case is that there exists an algorithm for simulating directly from the posterior distribution of the coalescence times of the tree, under a specific prior (see methods). Thus we can focus on the computational and statistical efficiency of the postprocessing methods, without any need to take into account the possible effects of any inaccuracies in the method for generating the sample of trees. However, the ideas of postprocessing can be applied to the output of any MCMC or other approach for generating samples of trees from a known posterior distribution and thus are not restricted to the assumptions of infinite-sites data or known topology.
METHODS
Infinite-sites data and phylogenetic prior:
We focus on analyzing data from m chromosomes sampled from a population. We assume we have infinite-sites data from a nonrecombining region of the genome and that the topology of the genealogy is known. The infinite-sites data mean that we will know the number of mutations that have occurred on each branch of the genealogy. Our mutation model is that (for our chosen scaling of time) these mutations occur at a constant rate θ/2 along each branch of the genealogy.
We assume some labeling of the nodes in the genealogy and denote by t = (t1,…, tm−1) the coalescent times for these nodes. We take the usual convention of the current time being time 0 and time being measured backward into the past. We also introduce the notation t′ = (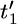 ,…,
,…, 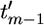 ) to denote the ordered coalescent times (so
) to denote the ordered coalescent times (so 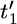 <
< 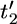 < … <
< … < 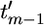 ). In the genealogy there are 2(m − 1) branches. The branch lengths are denoted by b = (b1,…, b2(m−1)), and sequence data can be summarized by the number of mutations on each branch: n = (n1,…, n2(m−1)). The branch lengths, b, are a linear function of the coalescent times, t; and to emphasize their interdependence we write b(t) and bi(t). The likelihood of the data, n, can be written as
). In the genealogy there are 2(m − 1) branches. The branch lengths are denoted by b = (b1,…, b2(m−1)), and sequence data can be summarized by the number of mutations on each branch: n = (n1,…, n2(m−1)). The branch lengths, b, are a linear function of the coalescent times, t; and to emphasize their interdependence we write b(t) and bi(t). The likelihood of the data, n, can be written as
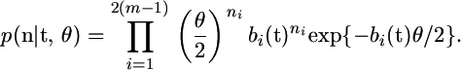 |
(1) |
Now we use the pure birth process prior of Rannala and Yang (1996) for the coalescent times, which assumes that the length of each branch has an exponential distribution with rate φ,
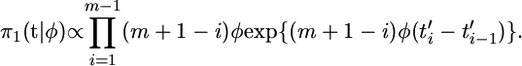 |
(2) |
Under this prior the posterior distribution for t (given φ and θ) is
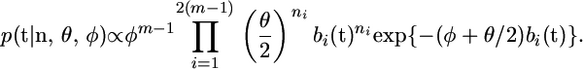 |
(3) |
Note that setting φ = 0 produces a posterior that is proportional to the likelihood function.
By introducing new variables s = (s1,…, sm−1), which satisfy si = (φ + θ/2)ti, we obtain
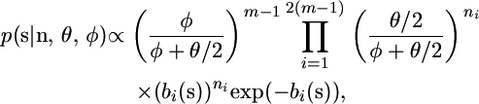 |
(4) |
where by the linear relationship between branch lengths and coalescent times bi(s) = (φ + θ/2)bi(t). Fearnhead and Meligkotsidou (2004) show how to draw independent and identically distributed (i.i.d.) samples from this density and hence (through rescaling) from the posterior (3). Furthermore this gives that the likelihood for φ is proportional to
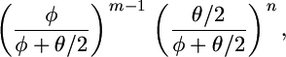 |
(5) |
where n is the total number of mutations.
Variable population size:
Consider a panmictic population of current effective population size N chromosomes, with time measured in units of N generations, and let the effective population size at time t in the past be N/λ(t). The distribution for the coalescence times for a random sample of m chromosomes from such a population (Griffiths and Tavaré 1994a) is
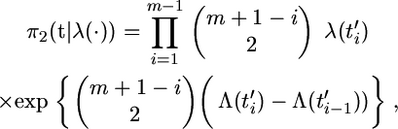 |
(6) |
where 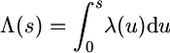 , and remember that the
, and remember that the 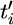 's are defined as ordered coalescent times.
's are defined as ordered coalescent times.
Interest lies in generating samples from the posterior distribution of the coalescent times p(t | λ(·), θ, n) and in calculating the marginal likelihood p(n | λ(·), θ). The former allows us to perform inference for a given demographic model, and the latter is required for choosing between different demographic models.
Both of these can be achieved through an algorithm that generates samples of the coalescent times from (3) and then reweights these samples. For example,
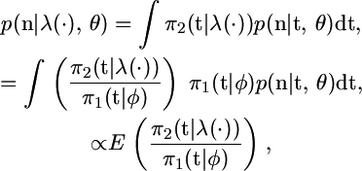 |
where the expectation is with respect to p(t | n, θ, φ), and the constant of proportionality is 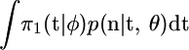 . The last step of working above uses
. The last step of working above uses 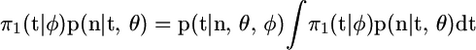 . A natural estimate of this expectation is based on the sample mean of π2(t | λ(·))/π1(t | φ) for an i.i.d. sample from p(t | n, θ, φ). In addition, the weighted sample will approximate p(t | λ(·), θ, n). This is a standard importance-sampling approach, and for more general details of this method see Srinivasan (2002).
. A natural estimate of this expectation is based on the sample mean of π2(t | λ(·))/π1(t | φ) for an i.i.d. sample from p(t | n, θ, φ). In addition, the weighted sample will approximate p(t | λ(·), θ, n). This is a standard importance-sampling approach, and for more general details of this method see Srinivasan (2002).
Specifically the algorithm is as follows:
Generate an i.i.d. sample of size K from (3) using the method of Fearnhead and Meligkotsidou (2004). Denote the sample as t(1), …, t(K).
For k = 1, …, K assign t(k) a weight wk = π2(t(k) | λ(.))/π1(t(k) | φ). Let
 .
.The weighted sample, t(1), …, t(K) with corresponding weights w1/C, …, wK/C, approximates the posterior p(t | λ(.), θ, n). Furthermore, an estimate of the marginal likelihood p(n | λ(.), θ) (up to a common constant of proportionality) is given by C/K.
The advantage of this approach is that the costly, in terms of CPU time, step of generating the sample of coalescent times in A is required only once. Calculating the importance-sampling weights in B has negligible CPU cost and thus can be repeated easily for a wide range of possible models for how the population size has varied through time. For informative data, the hope is that (3), which is closely related to the likelihood, will be a good proposal density for a wide range of λ(t)'s. However, the efficiency of this method is likely to depend crucially on the sample size m, which affects the dimension of t.
Migration models:
We now consider inference for a structured population model. We consider a model with D demes, each with constant population sizes N1, …, ND, respectively, and D × D backward migration matrix M = {Mij}. Under this model, backward in time a chromosome currently in deme i will migrate to deme j with rate Mij/2. The diagonal elements are defined so that rows of the matrix sum to zero, 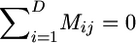 . We assume the population is at stationarity, so that the expected number of migrants leaving a deme is equal to the expected number entering, which corresponds to
. We assume the population is at stationarity, so that the expected number of migrants leaving a deme is equal to the expected number entering, which corresponds to 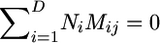 , and thus the model is parameterized by the migration matrix M, and the total population size
, and thus the model is parameterized by the migration matrix M, and the total population size 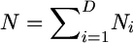 . Note that knowledge of the migration matrix and the total population size will define the population sizes of the individual demes.
. Note that knowledge of the migration matrix and the total population size will define the population sizes of the individual demes.
The data now include the deme in which each of the chromosomes was sampled. We propose an approximate-likelihood approach to estimating the migration rates. We first introduce an approximate likelihood function for the observed demes of the sample conditional on t. We denote this by 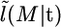 . The approximation that we use treats the deme that a chromosome belongs to in an equivalent way to an allele. This is an approximation as migration models assume strong density regulation, so that the population size of each deme is constant over time and a fixed proportion of chromosomes move from one deme to another in a single generation. By comparison our approximation is (by direct analogy to neutral Wright–Fisher models) equivalent to allowing the population size of these to fluctuate through time. Each chromosome in a given deme is choosing independently whether to migrate from its deme to another (with the probability of migrating and the deme to which it migrates being determined by the migration rates). For real-life populations, the truth is likely to lie in between these two extremes: with some degree of variation in population size of demes over time, but with density regulation restricting this variability.
. The approximation that we use treats the deme that a chromosome belongs to in an equivalent way to an allele. This is an approximation as migration models assume strong density regulation, so that the population size of each deme is constant over time and a fixed proportion of chromosomes move from one deme to another in a single generation. By comparison our approximation is (by direct analogy to neutral Wright–Fisher models) equivalent to allowing the population size of these to fluctuate through time. Each chromosome in a given deme is choosing independently whether to migrate from its deme to another (with the probability of migrating and the deme to which it migrates being determined by the migration rates). For real-life populations, the truth is likely to lie in between these two extremes: with some degree of variation in population size of demes over time, but with density regulation restricting this variability.
To define our approximate likelihood we first define γi = Ni/N for i = 1, …, D and introduce a forward migration matrix F whose entries satisfy Fij = NjMji/Ni, for i, j = 1, …, D. So the probability of a specific descendant of a chromosome in deme y being in deme x at a time t in the future is
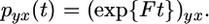 |
We introduce a vector x = (x1, …, x2m−1), where (x1, …, xm) denotes the deme of the m chromosomes in the sample, and (xm+1, …, x2m−1) are the demes of the internal nodes of the genealogy. We assume x2m−1 is the deme of the most recent common ancestor. Finally, for i = 1, …, 2m − 2, we let bi be the branch length connecting node i to its parent and yi be the deme of the parent of node i. Then we define a joint density
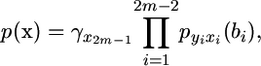 |
where the 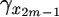 term comes from the stationary distribution of the migration process. Finally, the likelihood conditional on t is
term comes from the stationary distribution of the migration process. Finally, the likelihood conditional on t is
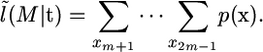 |
(7) |
Note that this likelihood is uninformative about the total population size N. Calculating (7) is possible using the peeling algorithm of Felsenstein (1981).
Our approximate likelihood is then obtained by averaging 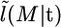 over samples of t from (3). So given a sample t(1), …, t(K) from (3), we get
over samples of t from (3). So given a sample t(1), …, t(K) from (3), we get
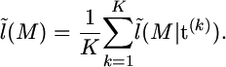 |
Note that a direct importance-sampling approach (similar to that used for the variable population size scenario) is not computationally feasible here. To calculate importance-sampling weights we need to know not only t but also the specific details of all migration events in the history of our sample. We have considered an importance-sampling approach that imputes the migration events, but the resulting method was highly inefficient because of the large space of possible migration events for any given data set.
Continuous spatial models:
Finally we consider inference for samples obtained across a continuous spatial habitat. We assume that the data now include a spatial location for each sampled chromosome. We focus on inference under an isolation-by-distance model.
For simplicity we first describe the model assuming a one-dimensional location. We assume that the displacement of the location of a chromosome from the location of its ancestor at time t in the past has a univariate Gaussian distribution, with zero mean and variance σ2t. First, condition on the genealogy of the sample. Furthermore, let μ be the location of the most recent common ancestor (MRCA), T be the time to the MRCA, and tij be the time back to the first common ancestor of chromosomes i and j. Then, conditional on this, the spatial data X = (X1, …, Xm) have a multivariate normal distribution with
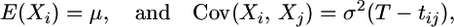 |
for all i, j = 1, …, m. The intuition here is that as dispersion is unbiased, the expected location of each sampled chromosome will be the location of the MRCA; whereas the covariance between the locations of two chromosomes is proportional to the amount of shared ancestry they have back to the most recent common ancestor. This model trivially extends to the case of two-dimensional locations where the dispersion in each direction is independent and identically distributed.
To perform inference we then introduce a prior distribution on the genealogy of the sample and a prior distribution on μ. We use (2) as the prior on the genealogy and we choose an improper uniform prior on μ. For this choice of prior on μ it is possible to analytically integrate out μ conditional on the genealogy (Rue and Held 2005). We write p(x | t, σ) to be the resulting conditional probability of the data, given just the genealogy and σ, and p(μ | x, t, σ) to be the corresponding conditional distribution for μ.
For many spatial genetic studies, samples are generated by first choosing the locations and then sampling chromosomes at those locations. Thus it makes sense to perform inference for σ under a conditional likelihood, where we condition on the spatial location. More generally, use of the conditional likelihood for σ means that inferences should depend less on the choice of prior on the genealogy (since in the limit as the mutation rate tends to 0, the conditional likelihood will become constant). If as before we denote the genetic data by n and the spatial data by x, then the conditional likelihood can be written as
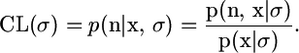 |
If we use the prior (2), but rather than specifying a value of φ use the uninformative hyperprior π(φ) ∝ 1/φ, then the denominator is constant as a function of σ (see the appendix), which greatly simplifies the calculation of this conditional likelihood.
We calculate CL(σ) by simulation as follows:
We simulate K i.i.d. samples of times, by repeatedly (i) simulating φ from its posterior and (ii) simulating t from (3) conditional on that φ. Denote the sample as t(1), …, t(K).
For k = 1, …, K assign t(k) a weight wk = p(x | t(k), σ). Let
 .
.- An estimate of CL(σ) is C/K, and the posterior distribution for μ is approximated by the mixture
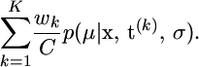
Simulation in part i of A is straightforward, as the posterior for φ is proportional to
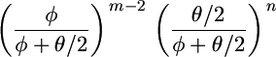 |
and can be related to a beta distribution through the transformation γ = φ/(φ + θ/2).
Simulation of continuous spatial data:
Simulating data under an appropriate continuous spatial model is difficult. There appear to be two approaches: first, those based on the isolation-by-distance model of Wright (1943), which ignores any regulation of population density and thus produces populations with infinite density (Felsenstein 1975), and, second, models that assume a constant population density (Wilkins and Wakeley 2002; Wilkins 2004) and require the population to live on some closed finite region.
As our inference model ignores any restriction on the location of chromosomes as required for these latter models, we simulated data under a version of the isolation-by-distance model of Wright (1943). In particular, we simulated the genealogical tree for our data under a coalescent model with exponential population growth and then conditional on this simulated the spread of the chromosomes from the model described above. The idea is to model a situation where the effect of population density regulation is less: that of a population growing in size to fill a new habitat. Note that we are simulating the data under a different model from that under which we are analyzing the data, as the distributions on the genealogy differ.
RESULTS
Variable population size:
The importance-sampling approach we propose for analyzing data under a range of variable population size scenarios is simulation consistent. That is, as the number of samples, K, of the coalescence times tends to infinity, then the estimate of the likelihood of a given scenario or the likelihood curve for a given set of parameters will converge to the true likelihood or likelihood curve. Similar results hold for the posterior distribution of the coalescence times. Thus the practicability and efficiency of the approach relies on the Monte Carlo error in these estimates and on how large K will need to be to obtain good estimates.
One way of empirically testing the accuracy of these estimates is to use the effective sample size (ESS) of Liu (1996) (see also Fearnhead and Donnelly 2001). The ESS is defined as
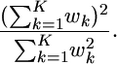 |
The ESS lies between 1 and K and has the interpretation that if an importance-sampling scheme has an ESS of E, then inference based on this scheme is roughly as accurate as inference based on E independent draws from the full posterior distribution. As a rough guide we would want E > 100 and preferably E > 1000 for the inferences to be reliable. (Increasing K by a factor should increase E by the same constant factor.)
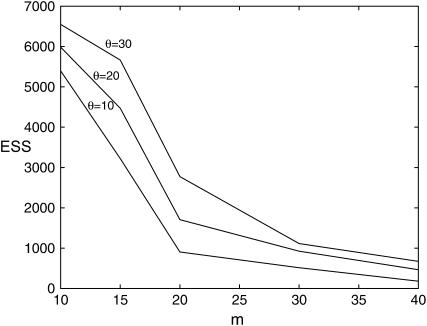
We investigated how the ESS of our method depends on the values of the mutation rate, θ, and the sample size, m. We simulated data from the exponentially growing population size model with rate of exponential growth β = 0.7 and various values of θ, namely θ = 10, 20, 30. Figure 1 shows the ESS values for analyzing data sets of size m = 10, 15, 20, 30, 40, using K = 10,000 weighted samples sampled from (3). (Here and below we set φ to the value that minimizes the likelihood in Equation 5, although results are insensitive to this choice.) It can be seen that the ESS decreases with m, but increases with θ. The results suggest that for θ = 10 analyzing sample sizes of up to 20–40 is reasonable, with slightly larger sample sizes possible for the larger θ-values. The speed of this approach means that analysis for larger values of m should be possible by increasing K.
Figure 1.—
ESS for analyzing data sets of size m = 10, 15, 20, 30, 40 simulated from the exponentially growing population size model with β = 0.7 and θ = 10, 20, 30.
To demonstrate the potential usefulness of our method we consider analyzing the data shown in Figure 2, under a variety of scenarios for the variable population size. We fix the parameters within our model (although our approach can equally be used to calculate likelihood surfaces for parameters of a given model). Our reason for focusing on different scenarios is that this is a situation where existing methods may not be able to be used (as existing software may allow analysis only for a certain class of models or would require being rerun for each model that is considered). Specifically, we consider the following models:
The constant population size model; for this model λ(t) = t.
The exponentially growing population size model; for this model λ(t) = eβt.
- The constant population size followed by exponential growth model; for this model we assume
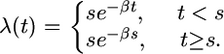
- The bottleneck model; for this model we assume
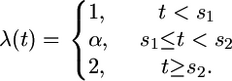
For the analysis below we fixed (a) θ = 15; (b) θ = 15 and β = 0.7; (c) θ = 15, s = 0.1, and β = −10 log(0.05); and (d) θ = 15, s1 = 0.165, s2 = 0.175, and α = 10. We focus on inferring the time to the most recent common ancestor (TMRCA) and in particular on looking at how robust these inferences are to the specific choice of model.
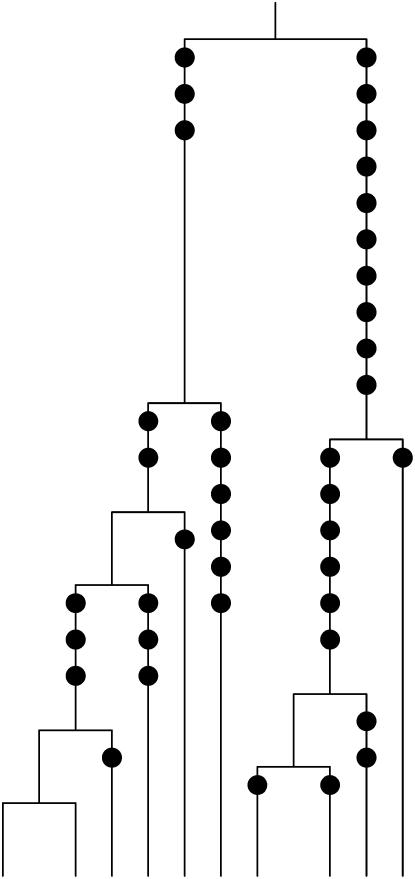
Figure 2.—
The coalescent tree for a sample of m = 10 chromosomes from the constant population size model. The mutations are depicted by solid circles on the branches of the tree.
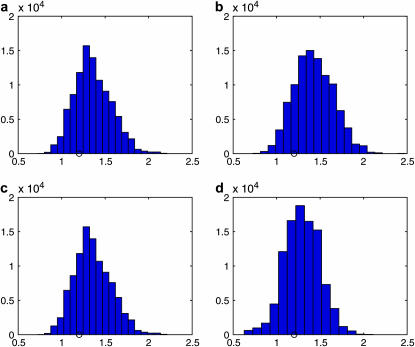
We simulated K = 10,000 sets of coalescence times from (3), which took <2 min on a Pentium 4 laptop PC with CPU of 3.20 GHz. Reweighting these sets of times took ∼1 sec for each model. The resulting histograms of the samples of the TMRCA for all models are shown in Figure 3, and the respective estimates of the marginal likelihood are (a) 0.4308, (b) 0.6248, (c) 0.0362, and (d) 2.4191 × 10−6. The ESSs of the weights were between 1000 and 5000 for models a–c and 98 for d. The histograms show that the estimate of the TMRCA appears robust across these different models.
Figure 3.—
Histograms of the samples of the TMRCA for the coalescent tree analyzed under (a) the constant population size model, (b) the exponentially growing population size model, (c) the constant population size followed by exponential growth model, and (d) the bottleneck model. The true value of the TMRCA is indicated in each plot by a circle.
Note that inference for the bottleneck model is more challenging than that for the other models as the importance-sampling weights depend crucially on the number of coalescences that lie within the period of the bottleneck and thus can have a large variance (and hence small ESS). The effect of a bottleneck depends primarily on its severity, defined as the product α(s2 − s1). Having a bottleneck with similar severity but larger α and smaller (s2 − s1) will lead to a more poorly behaved importance sampler.
Migration models:
Here we examine the performance of our approach at analyzing migration models. Note that we can estimate migration rates only relative to our choice of units for time, which is defined by our specification of the mutation rate θ. Therefore, we fix θ to its true value and look at estimates of the migration rates.
Our approach for migration models is based on an approximate likelihood, and first we need to check the validity of this approach. To do this we calculated the mean log-likelihood over a set of independent data. The shape of the mean log-likelihood governs the asymptotic behavior of the method, and in particular for an approximate likelihood method to produce consistent estimates it is required that the mean log-likelihood curve attains its maximum at the true value of the parameters (see Fearnhead 2003; Smith and Fearnhead 2005, for further discussion). Thus an important property of an approximate-likelihood method is that the mean log-likelihood curve attains its maximum at a value close to the true value.
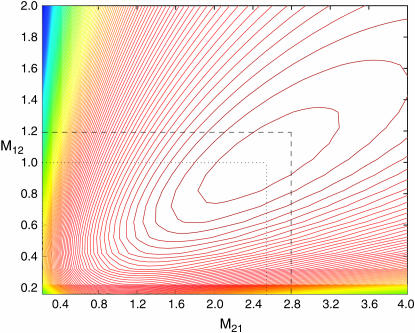
We simulated 100 coalescent trees with sample size of m = 10 from the migration model with D = 2 demes, N1 = 3000, N2 = 7000, M12 = 1.2, and M21 = 2.8. The mutation rate used was θ = 30. For each data set we based inferences on 2000 sets of coalescence times simulated from (3), again with φ set to the value that maximizes (5). We have estimated the mean log-likelihood at a grid of values of M12, M21. A contour plot of this log-likelihood surface is shown in Figure 4. The maximum of this curve is indeed close to the true parameter value (maximum at M12 = 1.02, M21 = 2.52). Similar results are obtained for a range of migration models (results not shown).
Figure 4.—
Contour plot of the mean log-likelihood surface of M12, M21 obtained from 100 simulated coalescent trees with sample size m = 10 under the migration model with D = 2 demes (each contour corresponds to 0.05 units of log-likelihood). The mutation rate used was θ = 30. Shown are the true parameter values, M12 = 1.2 and M21 = 2.8, and the values that maximize the surface, 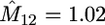 and
and 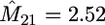 .
.
In Table 1 we present results on the performance of our approach, obtained from simulated data of size m = 10, 20 from the migration model with D = 2 demes for different values of the model parameters. We consider two sets of parameters: (a) N1 = N2 = 5000, M12 = M21 = 0.4 and (b) N1 = 3000, N2 = 7000, M12 = 1.2, M21 = 2.8. In each case we report the average of the most likely parameter values across 100 data sets, the standard errors of these estimates (in parentheses), and the associated coverage of the 95% likelihood-based confidence intervals (C.I.'s). The average CPU cost of analyzing a data set on our laptop PC is 30 sec for the m = 10 case and 50 sec for the m = 20 case.
TABLE 1.
Performance of our approximate-likelihood approach for simulated data under the migration model with D = 2 demes for scenarios (a) N1 = N2 = 5000, M12 = M21 = 0.4 and (b) N1 = 3000, N2 = 7000, M12 = 12, M21 = 28
| Case a
|
Case b
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| m | θ | 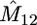 |
Coverage (%) | 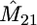 |
Coverage (%) | 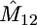 |
Coverage (%) | 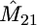 |
Coverage (%) |
| 10 | 15 | 0.46 | 100 | 0.48 | 100 | 1.02 | 92 | 2.50 | 89 |
| (0.26) | (0.26) | (0.64) | (1.30) | ||||||
| 10 | 30 | 0.42 | 100 | 0.46 | 100 | 1.08 | 95 | 2.62 | 97 |
| (0.22) | (0.26) | (0.62) | (1.22) | ||||||
| 20 | 15 | 0.36 | 99 | 0.38 | 99 | 1.04 | 87 | 2.42 | 82 |
| (0.24) | (0.24) | (0.72) | (1.46) | ||||||
| 20 | 30 | 0.38 | 97 | 0.38 | 97 | 1.06 | 90 | 2.66 | 88 |
| (0.30) | (0.30) | (0.70) | (1.36) | ||||||
In each case we report the estimates of the parameters based on 100 data sets, the standard errors (in parentheses), and the associated coverage of the 95% C.I.'s.
The method does have a bias, as can be seen in Figure 4 and Table 1; however, this bias is small compared to the standard error of the estimates and thus has a very small contribution to the mean square error of the estimator. The coverage properties of the confidence intervals vary notably between cases a and b; the reason for this is unclear. In this case it appears that the approximate-likelihood method performs much better and more robustly in terms of point estimates than in terms of assessing uncertainty in those estimates.
For comparison we reanalyzed the m = 10, θ = 15, M12 = M21 = 0.4 data sets using genetree (Griffiths and Tavaré 1994b; Bahlo and Griffiths 1998), which approximates the true likelihood curve. To use a single run of genetree required that we fix the relative population sizes in the two populations. So we ran genetree and reran our approach assuming that both θ and the relative population sizes were known and considered estimates of the single migration parameter. To implement genetree requires the choice of a driving value for the migration rate, and rather than choose a single value we ran genetree for five different values, ranging from 0.2 to 1.0, and averaged the likelihood curves across those obtained for each value. We ran genetree for 100,000 iterations for each driving value, which took around two orders of magnitude longer to run than our approach. The median of ESSs of the estimate of the likelihood at the true migration rate was 15 across the 100 simulations (in comparison with an ESS of >1000 for our method). The estimates from the two methods were highly correlated (correlation coefficient 0.75). The root mean square error of our estimates was ∼20% smaller than that of genetree. This suggests that for this case the Monte Carlo error within the genetree estimates of the likelihood curves is affecting the estimates of the migration rates more than the approximation error of our likelihood approach.
Continuous spatial models:
Finally we present results for the continuous spatial models. Again here we can estimate the parameters of the spatial model only relative to the mutation rate θ. Therefore, we fix the parameters of the demographic model to their true values and look at estimates of the spatial parameters.
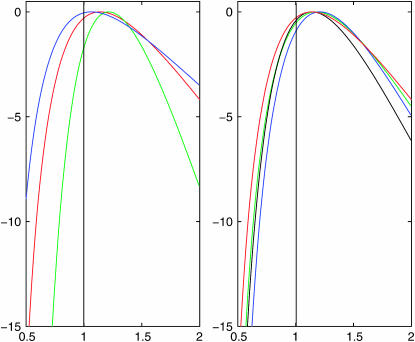
First, we check the validity of the approximate likelihood through calculating the mean log-likelihood for a range of parameters. For each set of parameters we simulated 100 data sets and then used our approximate approach with K = 5000 to estimate the likelihood curve of σ, the parameter governing the rate of spatial dispersion, and to obtain samples from the posterior distribution of the location of the MRCA. Combining information from all of the 100 simulated trees we estimated the average log-likelihood at a grid of values of σ. Figure 5 shows the resulting mean log-likelihood curves for a range of values of the sample size, m, the mutation rate, θ, and the population growth parameter, β. In each case σ = 1. The accuracy of the method appears to be primarily dependent on m, with the asymptotic bias of the method increasing as m increases (as the value of σ for which the maximum of the mean log-likelihood curve is attained gets further away from σ = 1 as m increases).
Figure 5.—
Plots of the log-likelihood surface of σ for a range of parameter values, each obtained from 100 simulated data sets. Left-hand plot: θ = 15, β = 1, and m = 10 (blue); m = 20 (red); and m = 40 (green). Right-hand plot: m = 20, θ = 30, and β = 1 (black); θ = 30, β = 2 (blue); θ = 15, β = 1 (red); and θ = 15, β = 2 (green).
In Table 2 we present a summary of the estimates of σ across the 100 data sets for each set of parameter values; and in Table 3 we give the root mean square error of the estimate of the position of the MRCA (these estimates had negligible bias); due to symmetry we show only the root mean square error for estimating one coordinate of the position.
TABLE 2.
Performance of our conditional-likelihood approach at estimating σ for the spatial model
| β = 1
|
β = 2
|
||||||
|---|---|---|---|---|---|---|---|
| m | θ | 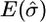 |
RMSE | Coverage (%) | 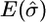 |
RMSE | Coverage (%) |
| 5 | 2 | 0.99 | 0.45 | 95 | 1.00 | 0.42 | 96 |
| 5 | 5 | 1.09 | 0.46 | 93 | 0.99 | 0.38 | 94 |
| 10 | 5 | 1.02 | 0.28 | 95 | 1.04 | 0.29 | 95 |
| 10 | 15 | 1.05 | 0.24 | 94 | 1.03 | 0.23 | 94 |
| 20 | 15 | 1.13 | 0.22 | 83 | 1.18 | 0.26 | 79 |
| 20 | 30 | 1.14 | 0.27 | 79 | 1.20 | 0.29 | 73 |
| 40 | 15 | 1.22 | 0.31 | 57 | 1.23 | 0.30 | 51 |
| 40 | 30 | 1.22 | 0.30 | 45 | 1.28 | 0.32 | 40 |
We report the mean of the estimates of σ (truth σ = 1), the root mean square error of the estimates, and the coverage probability of 95% approximate confidence intervals. (The grid of σ-values ranged from 0 to 4 for m = 5 and m = 10 and from 0 to 2 for m > 10.)
TABLE 3.
Performance of our conditional likelihood (CL) method and the sample mean (SM) at estimating the position of the MRCA
| β = 1
|
β = 2
|
||||
|---|---|---|---|---|---|
| m | θ | CL | SM | CL | SM |
| 5 | 2 | 0.62 | 0.73 | 0.48 | 0.52 |
| 5 | 5 | 0.54 | 0.62 | 0.51 | 0.56 |
| 10 | 5 | 0.67 | 0.70 | 0.51 | 0.53 |
| 10 | 15 | 0.66 | 0.66 | 0.55 | 0.56 |
| 20 | 15 | 0.61 | 0.66 | 0.52 | 0.58 |
| 20 | 30 | 0.56 | 0.62 | 0.52 | 0.59 |
| 40 | 15 | 0.66 | 0.69 | 0.46 | 0.51 |
| 40 | 30 | 0.68 | 0.71 | 0.55 | 0.62 |
Numbers show root mean square error for inferring a single coordinate of the position. By symmetry, the results for the other coordinate are the same.
We see that the estimates of σ are accurate for values of m up to 10, and any bias is small relative to the standard error of the estimator; beyond this we note a bias in our estimates, and the root mean square error actually increases when we move from m = 10 to m = 40. Coverage properties also appear good for values of m up to 10; but beyond this the confidence intervals are substantially anti-conservative. The values of β and θ appear to have little effect on the results. These results are consistent with those from Figure 5, with the bias of the estimator starting to dominate its performance for m = 20 and particularly for m = 40.
For comparison with our estimate of the position of the MRCA, we also calculated a simple unbiased estimate for each data set, which is obtained by taking the average of the locations of the sample. The root mean square error of one coordinate of the position is also shown in Table 3. Our approach is uniformly more accurate—with quite notable reduction in root mean square error for m = 20 and m = 40. Note that the estimates are more accurate for β = 2 than for β = 1 due to the tree being shorter and thus the spatial spread of the data being less.
One approach to reduce the bias of estimates for the m = 20 and m = 40 cases is to use a composite likelihood. We tried a simple approach: For the m = 20 case we split the data into two disjoint subsamples of size 10; and for the m = 40 case we split the data into four disjoint subsamples of size 10. For both cases we then calculated the log-likelihood for each subsample of size 10 and averaged this log-likelihood across the two, or the four, subsamples. We calculated our estimate of σ as the value that maximized this average log-likelihood curve. (Confidence intervals were calculated by treating the average log-likelihood curve as a standard log-likelihood curve.) The results are shown in Table 4. The bias and root mean square error of the estimates are substantially reduced for this approach, and also the coverage probabilities are much closer to 95%.We investigated using more, but nondisjoint, subsamples and found no improvement in the estimates. We also tried using smaller subsamples (e.g., four disjoint subsamples of size 5 for the m = 20 case), but obtained worse performance in this case.
TABLE 4.
Performance of our composite-likelihood approach at estimating σ for the spatial model
| β = 1
|
β = 2
|
||||||
|---|---|---|---|---|---|---|---|
| m | θ | 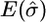 |
RMSE | Coverage (%) | 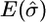 |
RMSE | Coverage (%) |
| 20 | 15 | 1.01 | 0.22 | 96 | 1.02 | 0.25 | 93 |
| 20 | 30 | 0.98 | 0.19 | 95 | 1.00 | 0.20 | 95 |
| 40 | 15 | 0.94 | 0.23 | 92 | 0.99 | 0.24 | 95 |
| 40 | 30 | 0.96 | 0.21 | 91 | 0.93 | 0.18 | 95 |
We report the mean of the estimates of σ (truth σ = 1), the root mean square error of the estimates, and the coverage probability of 95% approximate confidence intervals. (We split the data into disjoint subsamples of size 10. The grid of σ-values ranged from 0 to 2.)
To demonstrate the advantage of postprocessing a sample of genealogical trees, rather than conditional analysis based on a single tree, we considered the alternative approach of inferring σ given a single estimate of the genealogy. Such an approach (i) obtains an estimate of the coalescent times 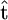 using the genetic data and (ii) bases inference on the conditional likelihood
using the genetic data and (ii) bases inference on the conditional likelihood 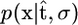 . We used the maximum-likelihood estimator of
. We used the maximum-likelihood estimator of 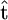 (which for these models can be calculated using the method of Meligkotsidou and Fearnhead 2005).
(which for these models can be calculated using the method of Meligkotsidou and Fearnhead 2005).
Here we present results for the m = 2 and m = 5 cases, although similar results are obtained for larger values of m. One difficulty with using the maximum-likelihood estimate of t is that the estimate of the coalescence time for two identical sequences is 0, which is inconsistent with chromosomes sampled from distinct locations. Thus in our analysis below we simulate data conditional on a sample having no identical sequences. We do not take account of this conditioning when analyzing the data.
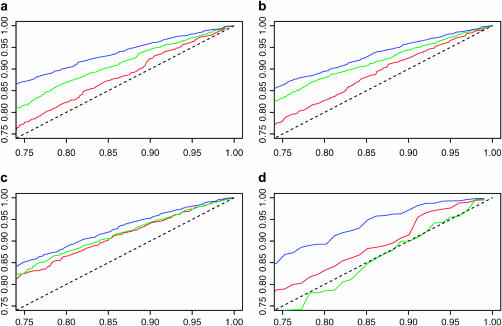
Figure 6 gives probability–probability (PP) plots of the likelihood-ratio statistics for testing σ = 1 against draws from a chi-square distribution with 1 d.f. We show this plot as this PP plot is related to the coverage properties of confidence intervals for the parameter, and if the likelihood-ratio statistic is approximately distributed as a chi-square distribution with 1 d.f., then it shows that the likelihood method is correctly quantifying the uncertainty in the parameter. This analysis is slightly complicated for the m = 2 case, as the sample size is too small for the asymptotic limit of the likelihood-ratio statistic to be a very good approximation—thus we also show the PP plot for the likelihood-ratio statistic conditional on knowing the true coalescence time. For each value of θ we give PP plots for the new approximate-likelihood method, the conditional analysis for the data sets with at least one segregating site. For smaller values of θ the approach that conditions on the maximum-likelihood estimate (MLE) for the coalescence time substantially underestimates the uncertainty of the estimate for σ. As θ increases the distribution of the likelihood-ratio (LR) statistic approaches the distribution of the LR statistic conditional on the true value of the coalescence time.
Figure 6.—
Probability–probability (PP) plots of a 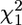 -distribution against the likelihood-ratio (LR) statistics for (red) our conditional likelihood method, (blue) analysis conditional on the maximum-likelihood estimate of the coalescence times, and (green) analysis conditional on the true coalescence times. a–c are based on 1000 data sets, with m = 2; β = 1; and (a) θ = 1, (b) θ = 2, and (c) θ = 4. d is based on 100 data sets with m = 5, θ = 2, and β = 1. We simulated all data sets conditional on there being no identical sequences in the data set.
-distribution against the likelihood-ratio (LR) statistics for (red) our conditional likelihood method, (blue) analysis conditional on the maximum-likelihood estimate of the coalescence times, and (green) analysis conditional on the true coalescence times. a–c are based on 1000 data sets, with m = 2; β = 1; and (a) θ = 1, (b) θ = 2, and (c) θ = 4. d is based on 100 data sets with m = 5, θ = 2, and β = 1. We simulated all data sets conditional on there being no identical sequences in the data set.
The effect of conditioning on the MLE of the times is less pronounced on the point estimates of σ. For the m = 5 case, the two sets of MLEs are highly correlated (correlation = 0.96) and give almost identical root mean square errors, although conditioning on the MLE appears to give slight underestimates of σ. A measure of the efficiency of our approach can be seen by looking at the correlation of the estimates from our method with those conditional on the true coalescence times; this again is high (correlation = 0.80). A related idea is used for inferring species trees in Edwards et al. (2007).
DISCUSSION
We have considered postprocessing of samples of genealogies, in particular to learn about the demographic parameters for a sample and the robustness of inference to changes in the demographic model. While in our applications we have considered infinite-sites data from a nonrecombining region of DNA, but the ideas can be applied much more generally. (For example, for the variable population size analysis, changing the method of simulating the data will affect only step B of the algorithm, with the denominator of the importance-sampling weights being the prior of the model under which the sample of genealogies was generated.) All that is required is that there is computational machinery (e.g., MCMC algorithms) that can produce samples of genealogies for the data. For example, analysis of more general mutation models is possible using the Bayesian phylogenetic packages such as MrBayes (Ronquist and Hulsenbeck 2000) and Bambe (Larget and Simon 1999), while analysis of (recombining) bacterial multilocus sequence typing data is possible using ClonalFrame (Didelot and Falush 2007).
We first considered inference for a variable population size and robustness of inference of coalescence times to changes in the model for the population size. An importance-sampling approach, which is “exact” in the limit as the computational cost increases, is possible here. In practice the efficiency of this method will depend on the sample size and the mutation rate, with efficiency decreasing as sample size increases or mutation rate decreases. Our results suggest that this approach is practicable for sample sizes of up to 50 chromosomes. The advantage of this postprocessing is that it enables a data set to be analyzed quickly under a range of different models. As such we view that this approach will be useful in terms of a preliminary analysis of a potentially large data set. We can first subsample an appropriate number of chromosomes (of the order of 10–50) and analyze these under a variety of models. This will help inform us as to what are the appropriate models for analyzing the complete data (using a more dedicated/computationally intensive approach) and also give insights as to how robust the results about the coalescence times of the tree will be.
We also considered inference in structured populations: both discrete subpopulations and continuous spatial models. There are similarities in the approximate-likelihood approach we consider for both of these cases. We first simulate a sample of genealogies and then average over the conditional likelihood of the spatial data given the genealogy. (For the migration model we also use an approximate conditional likelihood, which is equivalent to allowing the population sizes in the demes to fluctuate through time.) This approach implicitly assumes a conditional independence structure to the data: that the spatial and genetic data are conditionally independent given the genealogy. As such our model assumes a prior for the genealogy and then conditional models for the spatial/genetic data given the genealogy. The prior for the genealogy is that assumed within our computational method for producing the sample of genealogies, in our case the phylogenetic prior described in methods; although alternative methods for simulating the genealogies could be used that assume different priors. For the continuous spatial model, if we had chosen our prior to be that used to simulate the data (coalescent under exponential growth), then our approach gives a simulation-consistent approach for calculating the true likelihood of the data. The results we presented thus give an idea of the robustness of our approximate-likelihood method to the choice of the wrong prior. For practical applications, where the true choice of the prior (or equivalently model) for the genealogy is not known, the robustness of any method to the choice of this prior will be of paramount importance. In most scenarios that we examined the bias of the approximate-likelihood method is small compared to the standard error of the estimate. In general as m increases, biases increase. This is because as m increases the genealogical prior we use does not correctly capture the distribution of some of the coalescence times, and this then starts to introduce notable biases into the method.
For implementation of our approximate-likelihood method to new data and models it is important to know for what sample sizes the method will produce good statistical properties, such as small biases and appropriate coverage probabilities for confidence intervals. Currently, to evaluate this accurately will require some form of simulation study chosen to be appropriate for the models and data being considered. The results we have presented give insight into for what sample sizes the method will perform well. Our method can be applied to large data sets using a composite-likelihood approach. A large data set can be split into smaller subsamples (with the possibility of each chromosome appearing in many subsamples), with the approximate log-likelihood calculated for each subsample, and these approximate log-likelihood curves can be combined through averaging them together. An estimate of the parameter(s) is given by the value(s) that maximize this composite log-likelihood. The performance of such a method is governed by the shape of the mean of the log of the approximate likelihood, as shown in Figure 5 (see Fearnhead 2003). We tested out one implementation of this composite-likelihood approach for the continuous spatial model and found it to perform well using subsamples of size 10.
In particular, a pairwise-likelihood approach is likely to be a simple and flexible method for analyzing continuous spatial data sets (currently there are few methods for analyzing such models). For such a pairwise approach it is simple to allow for quite general models of the spatial spread of the population through time; all that is required is the specification of a family of densities, p(x1, x2; t), for the probability of two chromosomes that share a common ancestor at time t in the past being located at positions x1 and x2.
Acknowledgments
This work was supported by Engineering and Physical Sciences Research Council grant R91724 and by an Engineering and Physical Sciences Research Council Springboard Fellowship to P.F.
APPENDIX
The prior (2) can be obtained by simulating s from the prior with φ = 1 and then letting t = φs. Thus if we define S and sij's to satisfy T = φS and tij = φsij, so they are the respective times obtained from s, we get that
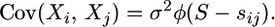 |
Thus the intuition behind the result is that, as under the prior, the data are solely informative about the product σ2φ, and using the scale invariance prior for φ will result in no information about σ.
Formally we use the fact that
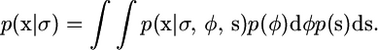 |
We consider the integral with respect to φ, assuming a given s, and demonstrate that this does not depend on σ, from which the fact that p(x | σ) does not depend on σ follows. For notational simplicity we assume μ = 0 in the following.
Now, for our given s, let Σ be the covariance matrix obtained when σ = φ = 1, so Σij = (S − sij) for i, j = 1, …, m. Further let Q = Σ−1 and A = xTQx/2. Then
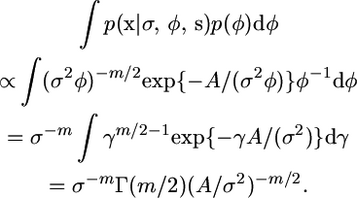 |
For the second equality we used the transformation γ = 1/φ. The final expression does not depend on σ as required.
References
- Bahlo, M., and R. C. Griffiths, 1998. Inference from gene trees in a subdivided population. Theor. Popul. Biol. 57: 79–95. [DOI] [PubMed] [Google Scholar]
- Beerli, P., and J. Felsenstein, 1999. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152: 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, S. P., I. Manopoulou and B. C. Emerson, 2007. Assessing the affect of genetic mutation: a Bayesian framework for determining population history from DNA sequence data, pp. 25–50 in Bayesian Statistics 8, edited by J. M. Bernardo, M. J. Bayarri, J. O. Berger, A. P. Dawid, D. Heckerman, A. F. M. Smith and M. West. Oxford University Press, Oxford.
- Coop, G., and R. C. Griffiths, 2004. Ancestral inference on gene trees under selection. Theor. Popul. Biol. 66: 219–232. [DOI] [PubMed] [Google Scholar]
- Didelot, X., and D. Falush, 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, A. J., A. Rambaut, B. Shapiro and O. G. Pybus, 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22: 1185–1192. [DOI] [PubMed] [Google Scholar]
- Edwards, S. V., L. Liu and D. K. Pearl, 2007. High-resolution species trees without concatenation. Proc. Natl. Acad. Sci. USA 104: 5936–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, B. C., and G. M. Hewitt, 2005. Phylogeography. Curr. Biol. 15: 367–371. [DOI] [PubMed] [Google Scholar]
- Fearnhead, P., 2003. Consistency of estimators of the population-scaled recombination rate. Theor. Popul. Biol. 64: 67–79. [DOI] [PubMed] [Google Scholar]
- Fearnhead, P., and P. Donnelly, 2001. Estimating recombination rates from population genetic data. Genetics 159: 1299–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead, P., and P. Donnelly, 2002. Approximate likelihood methods for estimating local recombination rates (with discussion). J. R. Stat. Soc. Ser. B 64: 657–680. [Google Scholar]
- Fearnhead, P., and L. Meligkotsidou, 2004. Exact filtering for partially-observed continuous-time Markov models. J. R. Stat. Soc. Ser. B 66: 771–789. [Google Scholar]
- Felsenstein, J., 1975. A pain in the torus: some difficulties with the model of isolation by distance. Am. Nat. 109: 359–368. [Google Scholar]
- Felsenstein, J., 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17: 368–376. [DOI] [PubMed] [Google Scholar]
- French, N. P., M. Barrigas, P. Brown, P. Ribiero, N. J. Williams et al., 2005. Spatial epidemiology and natural population structure of campylobacter jejuni colonising a farmland ecosystem. Environ. Microbiol. 7: 1116–1126. [DOI] [PubMed] [Google Scholar]
- Griffiths, R. C., and P. Marjoram, 1996. Ancestral inference from samples of DNA sequences with recombination. J. Comput. Biol. 3: 479–502. [DOI] [PubMed] [Google Scholar]
- Griffiths, R. C., and S. Tavaré, 1994. a Sampling theory for neutral alleles in a varying environment. Philos. Trans. R. Soc. Lond. Ser. B 344: 403–410. [DOI] [PubMed] [Google Scholar]
- Griffiths, R. C., and S. Tavaré, 1994. b Simulating probability distributions in the coalescent. Theor. Popul. Biol. 46: 131–159. [Google Scholar]
- Kuhner, M. K., J. Yamato and J. Felsenstein, 1998. Maximum likelihood estimation of population growth rates based on the coalescent. Genetics 149: 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner, M. K., J. Yamato and J. Felsenstein, 2000. Maximum likelihood estimation of recombination rates from population data. Genetics 156: 1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larget, B., and D. L. Simon, 1999. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol. Biol. Evol. 16: 750–759. [Google Scholar]
- Liu, J. S., 1996. Metropolised independent sampling with comparisons to rejection sampling and importance sampling. Stat. Comput. 6: 113–119. [Google Scholar]
- Meligkotsidou, L., and P. Fearnhead, 2005. Maximum likelihood estimation of coalescence times in genealogical trees. Genetics 171: 2073–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala, B., and Z. Yang, 1996. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43: 304–311. [DOI] [PubMed] [Google Scholar]
- Ronquist, F., and J. P. Hulsenbeck, 2000. MrBayes3: Bayesian phylogenetic reconstruction under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rue, H., and L. Held, 2005. Gaussian Markov Random Fields: Theory and Applications. CRC Press/Chapman & Hall, Cleveland; Boca Raton, FL/London; New York.
- Shen, P., F. Wang, P. A. Underhill, C. Franco, W. Yang et al., 2000. Population genetic implications from sequence variation in four Y chromosome genes. Proc. Natl. Acad. Sci. USA 97: 7354–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N. G. C., and P. Fearnhead, 2005. A comparison of three estimators of the population-scaled recombination rate: accuracy and robustness. Genetics 171: 2051–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, R., 2002. Importance Sampling. Springer, New York.
- Stephens, M., and P. Donnelly, 2000. Inference in molecular population genetics (with discussion). J. R. Stat. Soc. Ser. B 62: 605–655. [Google Scholar]
- Templeton, A. R., E. Boerwinkle and C. F. Sing, 1987. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in Drosophila. Genetics 117: 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill, P. A., G. Passarino, A. A. Lin, P. Shen, M. Lahr et al., 2001. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann. Hum. Genet. 65: 43–62. [DOI] [PubMed] [Google Scholar]
- Wilkins, J. F., 2004. A separation-of-timescales approach to the coalescent in a continuous population. Genetics 168: 2227–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins, J. F., and J. Wakeley, 2002. The coalescent in a continuous, finite, linear population. Genetics 161: 873–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1943. Isolation by distance. Genetics 28: 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]