Abstract
Tob55 is the major component of the TOB complex, which is found in the outer membrane of mitochondria. A sheltered knockout of the tob55 gene was developed in Neurospora crassa. When grown under conditions that reduce the levels of the Tob55 protein, the strain exhibited a reduced growth rate and mitochondria isolated from these cells were deficient in their ability to import β-barrel proteins. Surprisingly, Western blots of wild-type mitochondrial proteins revealed two bands for Tob55 that differed by ∼4 kDa in their apparent molecular masses. Sequence analysis of cDNAs revealed that the tob55 mRNA is alternatively spliced and encodes three isoforms of the protein, which are predicted to contain 521, 516, or 483 amino acid residues. Mass spectrometry of proteins isolated from purified outer membrane vesicles confirmed the existence of each isoform in mitochondria. Strains that expressed each isoform of the protein individually were constructed. When cells expressing only the longest form of the protein were grown at elevated temperature, their growth rate was reduced and mitochondria isolated from these cells were deficient in their ability to assembly β-barrel proteins.
MITOCHONDRIA, chloroplasts, and gram-negative bacteria contain β-barrel proteins in their outer membranes (Gabriel et al. 2001; Tamm et al. 2001; Rapaport 2003; Schleiff et al. 2003; Wimley 2003). The mechanisms by which newly synthesized β-barrel precursor proteins are integrated into lipid bilayers and assembled into oligomeric structures are not yet fully understood for any of these systems (Rapaport 2003; Johnson and Jensen 2004; Voulhoux and Tommassen 2004; Paschen et al. 2005). In mitochondria, the precursors are initially recognized by the receptor components of the translocase of the outer mitochondrial membrane (TOM complex) and transferred through the TOM complex pore to the intermembrane space (IMS) side of the outer membrane (Rapaport and Neupert 1999; Krimmer et al. 2001; Model et al. 2001; Rapaport 2002). β-Barrel precursors are then relayed to the topogenesis of mitochondrial outer membrane β-barrel proteins (TOB) or to the sorting and assembly machinery (SAM) complex, which is also located in the outer membrane (Kozjak et al. 2003; Paschen et al. 2003; Wiedemann et al. 2003). During movement from the TOM to the TOB complex, β-barrel precursors interact with the small Tim protein complexes, which are soluble components of the IMS (Hoppins and Nargang 2004; Wiedemann et al. 2004; Habib et al. 2005). The TOB complex inserts the β-barrels into the outer mitochondrial membrane.
The major component of the TOB complex is Tob55 (also named Sam50/Omp85). Tob55 is a β-barrel protein itself and is essential for viability of yeast cells (Kozjak et al. 2003; Paschen et al. 2003; Gentle et al. 2004). Homologs of yeast Tob55 are found in virtually all eukaryotes and in almost all gram-negative bacteria (Paschen et al. 2003; Gentle et al. 2004; Dolezal et al. 2006). The group includes the bacterial Omp85/YaeT (Voulhoux et al. 2003; Wu et al. 2005), the plastid Toc75 (Eckart et al. 2002), and the mammalian Tob55/Sam50 proteins (Humphries et al. 2005). The N-terminal domain of Tob55 has been shown to recognize precursors of β-barrel proteins. This recognition may contribute to the coupling of the translocation of β-barrel precursors across the TOM complex to their interaction with the TOB complex (Habib et al. 2007). The TOB core complex contains two additional proteins, Tob38 (Tom38/Sam35) and Mas37 (Tom37/Sam37) (Wiedemann et al. 2003; Ishikawa et al. 2004; Milenkovic et al. 2004; Waizenegger et al. 2004). Both are peripherally associated with Tob55 on the cytosolic surface of the outer membrane. The functions of these two subunits are poorly defined.
Despite some progress in our understanding of the structure–function relationship of the TOB complex, many questions as to the functions of its subunits and their domains remain to be answered. In this report, we describe the characterization of Tob55 from the filamentous fungus Neurospora crassa and an analysis of its expression. We have shown that Tob55 is essential in N. crassa and that it interacts with β-barrel precursor proteins. Surprisingly, Tob55 is expressed as three isoforms that result from alternative splicing. Cells that express only the longest isoform display reduced growth rates at elevated temperature and at high salt concentrations. Mitochondria isolated from these cells have a reduced capacity to insert β-barrel precursors into the outer membrane. To our knowledge, this is the first component of the mitochondrial import machinery shown to exist in isoforms that arise from alternative splicing.
MATERIALS AND METHODS
Growth of N. crassa:
Growth and handling of N. crassa was as previously described (Davis and De Serres 1970). Unless stated otherwise, growth of cells was at 30°. For growth tests on plates, conidia were harvested and adjusted to a concentration of 107/ml. Serial 10-fold dilutions were made and 5 μl of each was spotted onto plates containing standard sorbose media with the appropriate supplements. The plates were incubated at the desired temperature for 1–3 days. The inhibitors p-fluorophenylalanine (fpa) and benomyl, at concentrations of 400 μm and 1 μg/ml, respectively, were used to shift nuclear ratios in sheltered heterokaryons. However, when cultures were grown to produce mitochondria for in vitro import experiments, the concentration of fpa was reduced to 250 μm to obtain mitochondria that were more robust for import reactions.
Strains used in this study are listed in Table 1. Several natural isolates were obtained from the Fungal Genetics Stock Center (FGSC). These are indicated by the genus and species name followed by the site of isolation in the wild.
TABLE 1.
Strains used in this study
| Strain | Genotype | Origin or source |
|---|---|---|
| NCN251 (also called 74A) | A | FGSC 2489 |
| 76-26 | his-3 mtrR a (mtrR imparts fpa resistance) | R. L. Metzenberg |
| 71-18 | pan-2 BmlR a (BmlR imparts benomyl resistance) | R. L. Metzenberg |
| HP1 | Heterokaryon of 76-26 plus 71-18 | Nargang lab |
| Tob55KO-1 | Sheltered heterokaryon. As HP1, but with replacement of the tob55 gene in 76-26 nucleus with a hygromycin resistance (hygR) cassette. | Transformation of HP1 with split marker fragments for tob55 knockout. |
| Tob55KO-3 | As Tob55KO-1 | As Tob55KO-1 |
| T55His6-1 | his-3 mtrR a Δtob55∷hygR contains an ectopic copy of genomic tob55 with N-terminal hexahistidinyl tag. Also bleomycin resistant. | Transformation of Tob55KO-3 with bleomycin resistance plasmid containing an N-terminal hexahistidinyl-tagged genomic tob55 gene. Selection for transformation of the his-3 mtrR nucleus. |
| T55His6-3 | As T55His6-1 | As T55His6-1 |
| ST55-2 | his-3 mtrR a Δtob55∷hygR contains an ectopic copy of tob55 cDNA specific for the short form. | Transformation of Tob55KO-3 with bleomycin resistance plasmid containing a cDNA version of tob55 specific for the short form. Selection for transformation of the his-3 mtrR nucleus. |
| IT55-8 | his-3 mtrR a Δtob55∷hygR contains an ectopic copy of tob55 cDNA specific for the intermediate form. | Transformation of Tob55KO-3 with bleomycin resistance plasmid containing a cDNA version of tob55 specific for the intermediate form. Selection for transformation of the his-3 mtrR nucleus. |
| LT55-2 | his-3 mtrR a Δtob55∷hygR contains an ectopic copy of tob55 cDNA specific for the long form. | Transformation of Tob55KO-3 with bleomycin resistance plasmid containing a cDNA version of tob55 specific for the long form. Selection for transformation of the his-3 mtrR nucleus. |
| cyt-2-1 | cyt-2-1 pan-2 a (may contain arom-9 or qa-2). | Nargang lab |
| [mi-3] | [mi-3] pan-2 A | H. Bertrand |
| N. crassa Mauriceville | A | FGSC 2225 |
| N. crassa Saratoga | A | FGSC 3226 |
| N. intermedia Fiji | A | FGSC 435 |
| N. intermedia Saratoga | a | FGSC 6605 |
| N. intermedia Varkud | a | FGSC 1822 |
| N. sitophila Everglades | a | FGSC 4202 |
| N. tetrasperma Hanalei | a | FGSC 2511 |
Creation of tob55 knockout strain:
The N. crassa Tob55 protein was identified as a homolog of the Saccharomyces cerevisiae protein (Paschen et al. 2003; Schmitt et al. 2006). A split marker approach, described previously (Colot et al. 2006), was used to replace the tob55 gene with a hygromycin resistance gene. Cosmid X22D7 of the pMOcosX library (Mccluskey and Kinsey 2000) was identified from the N. crassa genome project (Galagan et al. 2003) as containing tob55. The tob55 upstream and downstream regions were amplified by PCR using X22D7 as the template. The two portions of the split marker were transformed into heterokaryotic strain HP1 (Nargang et al. 1995) and several hygromycin-resistant transformants were purified and examined by Southern analysis for evidence of replacement of the tob55 gene in one nucleus.
Transformation of N. crassa:
DNA was transformed into N. crassa by electroporation of conidia as previously described (Margolin et al. 1997, 2000) with modifications (Tanton et al. 2003). To ensure that only pure homokaryotic strains were used following transformations, single transformant colonies were picked using sterile glass Pasteur pipettes and transferred to slants with Vogel's medium containing the appropriate nutritional requirements and the selective antibiotic(s). The slants were incubated at 30° until the surface of the agar was covered by the mycelium and then were removed to room temperature to conidiate. Conidia were streaked for single colonies onto plates identical to those used for the electroporation and incubated at 30° until colonies formed. These were picked to slants without the antibiotic for growth and conidiation.
When heterokaryotic strains were transformed and selected for integration into a single nucleus, the transformed strains were tested for their nutritional requirements following the purification process described above to ensure that the transformants were homokaryotic (Nargang et al. 1998).
In vitro import of radiolabeled proteins into isolated mitochondria:
Published procedures were used for the isolation of mitochondria (Mayer et al. 1993) and the import of mitochondrial preproteins (Harkness et al. 1994). Preproteins were produced in vitro by transcription and translation in rabbit reticulocyte lysate [Promega (Madison, WI) TnT reticulocyte lysate system] in the presence of [35S]methionine (ICN Biomedicals, Costa Mesa, CA). Import reactions were analyzed by sodium dodecylsulfate–polyacrylamide gel electrophoresis and viewed by autoradiography or a phosphorimager system. Quantification of the image from the latter was done using the Imagequant program [version 5.2, Molecular Dynamics (Eugene, OR)]. In vitro assembly of mitochondrial precursor proteins was studied using blue native gel electrophoresis (BNGE) and autoradiography (Rapaport et al. 2001).
Antibody production and affinity purification:
Antiserum was raised against a fusion protein composed of hexahistidinyl-tagged full-length mouse dihydrofolate reductase and residues 1–108 of the short isoform of the N. crassa Tob55 protein (see results). Following expression in Escherichia coli, the fusion protein was purified on a NiNTA column (QIAGEN, Mississauga, ON) in 8 m urea according to the manufacturer's instructions except that the protein was eluted in 0.1% SDS, 10 mm Tris–HCl, pH 7.4. The eluate plus adjuvant was injected into rabbits without further processing.
For affinity purification of antibody, the Tob55 fusion protein was purified on a NiNTA column (QIAGEN) in 8 m urea according to the manufacturer's instructions except that protein was eluted in 6 m guanidine, 100 mm NaH2PO4, 10 mm Tris–HCl, pH 4.5. A Centriplus centrifugal filter device (Millipore, Bedford, MA) with a molecular weight cutoff of 30 kDa was used to exchange the elution buffer to coupling buffer containing 0.1 m NaHCO3, 0.5 m NaCl, 6 m guanidine. The cycle of concentrating the sample to 1 ml by centrifugation at 2500 × g for 2 hr followed by addition of 15 ml of coupling buffer was repeated three times. The ligand coupling slurry, Affi-Gel 10 or Affi-Gel 15 (Bio-Rad, Hercules, CA), was prepared by washing with 5 volumes of distilled water. About 30 mg of fusion protein in coupling buffer was bound to 1 ml of Affi-Gel 10 or 15 by incubation with rocking at 4° overnight. The next day, the resin was washed with 5 vol of phosphate-buffered saline (10 mm phosphate, pH 7.4, 137 mm NaCl, 2.7 mm KCl) and 2 vol of 100 mm glycine, pH 2.5.
For purification of the antibody, 2 ml of serum was mixed with 100 μl 20× phosphate-buffered saline and incubated with 1 ml of the antigen-bound matrix for 4 hr at 4° with rocking. The resin was washed three times with 5 vol of phosphate-buffered saline. Antibody was eluted in 10 fractions of 200 μl with100 mm glycine, pH 2.5. Fractions were neutralized with 20 μl 1 m Tris–HCl, pH 10. The antibody-containing fractions were identified by mixing 2 μl of the eluant with 200 μl Bradford protein assay (Bio-Rad) in a microtiter plate.
BNGE and antibody supershifts:
Mitochondria (50 μg) were solubilized in 50 μl buffer N containing 1% digitonin in 20 mm Tris–HCl, pH 7.4, 0.1 mm EDTA, 50 mm NaCl, 1% glycerol (vol/vol), and 1 mm phenylmethylsulfonyl flouride. After gentle rocking at 4° for 15 min and a clarifying spin at 15,000 × g, the supernatant was added to 5 μl of sample buffer (5% Coomassie Brilliant Blue G-250 in 100 mm Bis–Tris, 500 mm 6-aminocaproic acid, pH 7.0) and gently mixed at 4°. Samples were analyzed on a 6–13% gradient blue native gel as previously described (Schägger and Von Jagow 1991; Schägger et al. 1994) except that electrophoresis was performed overnight (16–20 hr) at 4° between 40 and 60 volts and then for 1–1.5 hr at 500 V with buffer lacking Coomassie.
For antibody supershift assays, import reactions were performed in triplicate at 25° for 20 min using 20 μg of mitochondrial protein/reaction. Samples were processed as above except that, after the clarifying spin, 18 μl of buffer, 18 μl (6 μg protein) of affinity-purified Tob55 antibody, or 18 μl (6 μg protein) of an unrelated antibody was added. Following gentle rocking at 4° for 2 hr, sample buffer was added and the samples were subjected to BNGE. Affinity-purified antibody from rabbit serum, raised to a peptide of Drosophila melanogaster ATM protein (ataxia telangiectasia mutated), was used as an unrelated antibody for a negative control (Silva et al. 2004).
Mass spectrometry:
Isoforms of Tob55 were analyzed by mass spectrometry as described previously (Schmitt et al. 2006). Briefly, outer membrane vesicles were isolated, subjected to SDS–PAGE, and stained with Coomassie blue. Bands in the molecular weight range expected for the isoforms of Tob55 were excised and digested overnight with trypsin. Peptide masses in the range of 500–3500 Da were obtained by reflector matrix-assisted laser desorption ionization–time of flight (MALDI–TOF), whereas peptides in the mass range of 3500–6000 Da were analyzed by linear MALDI–TOF. Predicted peptide masses were obtained using the PeptideMass program set for isotope averaging (Wilkins et al. 1997; Gasteiger et al. 2005).
Other techniques:
Protein alignments were generated with the Kalign program (Lassmann and Sonnhammer 2005). The standard techniques of agarose gel electrophoresis, Southern and Northern blotting of agarose gels, preparation of radioactive probes, transformation of E. coli, and PCR were all performed as described (Ausubel et al. 1992). Additional procedures followed the supplier's recommendations or previously described techniques: isolation of plasmid DNA (QIAGEN), Western blotting (Good and Crosby 1989), detection of bands on Western blots using LumiGLO chemiluminescent substrate (Kirkegaard and Perry Laboratories, Gaithersburg, MD), genomic DNA extraction (Wendland et al. 1996), protein determination with the Coomassie dye binding assay (Bio-Rad), DNA sequencing using a DyeNamic sequencing kit (Amersham Biosciences) with a model 373 stretch sequencer separation system (Applied Biosystems, Foster City, CA). In some figures, irrelevant lanes were removed electronically.
RESULTS
Tob55 is essential in N. crassa:
To study the TOB complex in N. crassa, we constructed a knockout of the tob55 gene using a split marker approach (Colot et al. 2006). Since Tob55 was shown to be an essential protein in yeast (Paschen et al. 2003), the knockout was created in heterokaryon HP1 by sheltered disruption (Nargang et al. 1995). The component nuclei of this strain encode resistance to either fpa or benomyl and are auxotrophic for either histidine or pantothenate, respectively. These markers facilitate analysis and manipulation of nuclear ratios in the sheltered heterokaryon. Transformants were screened by Southern analysis (not shown) for the replacement event in one nucleus of the heterokaryon (Grotelueschen and Metzenberg 1995) and isolates Tob55KO-1 and Tob55KO-3 were chosen for further analysis. The growth rate of these strains was analyzed on various media. Growth of both knockouts was reduced on fpa plus histidine but was indistinguishable from controls on other media (Figure 1A), suggesting that the tob55 knockout occurred in the nucleus carrying fpa resistance (Figure 1B). The Tob55KO-3 strain was chosen for further work.
Figure 1.—
Sheltered disruption of tob55. (A) Split marker transformants of heterokaryon HP1 were isolated and purified as described in the text. Serial dilutions of conidia produced by the transformants were tested on the indicated media to determine which nucleus carried the tob55 disruption. (B) Genotype of the sheltered heterokaryons Tob55KO-1 and Tob55KO-3. The box symbolizes a heterokaryotic cell with circles representing its component nuclei. Genetic markers important for maintenance, selection, or manipulation of the strain are shown (tob55, topogenesis of outer membrane β-barrel proteins; his, histidine; pan, pantothenate; mtr, methyltryptophan resistance; Bml, benomyl resistance). A mutation in the mtr gene results in resistance to fpa. Certain mutations in the Bml gene result in resistance to benomyl. The nucleus with histidine auxotrophy and fpa resistance (mtr) carries the tob55 deletion.
To determine if tob55 is an essential gene in N. crassa, conidia from the Tob55KO-3 heterokaryotic strain were plated onto medium containing the nutritional requirements of both nuclei, and single colonies were picked to fully supplemented slants. Conidia that formed in these slants were examined for nutritional requirements. Testing of 200 isolates revealed that 125 were heterokaryons and 75 were pantothenate-requiring auxotrophs. The lack of histidine auxotrophs indicates that the tob55 gene is essential for viability in N. crassa. To prove that the lethality was specific for the loss of tob55, we transformed the Tob55KO-3 sheltered heterokaryon with a plasmid carrying bleomycin resistance and a genomic copy of tob55 encoding an allele for an N-terminal hexahistidinyl-tagged version of the protein. Transformants were selected on medium containing bleomycin, fpa, and histidine. Colonies appearing on the transformation plates were shown to be histidine auxotrophs, demonstrating that homokaryons of the histidine-requiring nucleus of the sheltered heterokaryon had been rescued.
Characterization of mitochondria with reduced levels of Tob55:
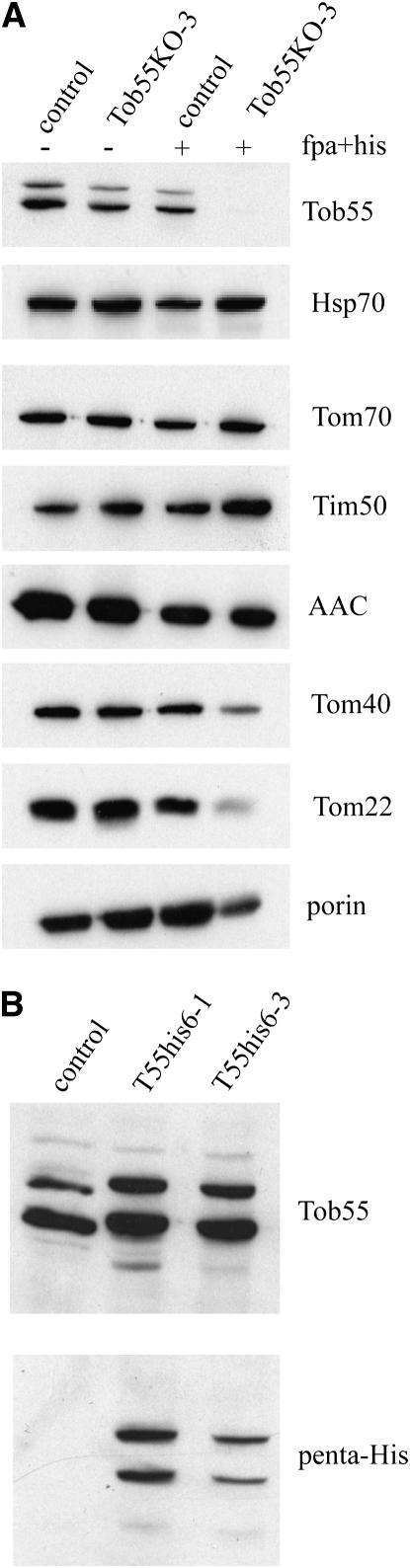
Examination of mitochondria in the heterokaryotic knockout strain Tob55KO-3 following growth in minimal medium revealed no alteration in the levels of any mitochondrial protein tested by Western blot analysis. However, mitochondria isolated from this strain following growth in the presence of histidine and fpa, which forces the knockout-bearing nucleus to predominate in the heterokaryon, contained very low levels of Tob55 (Figure 2A). We refer to such mitochondria as Tob55↓ mitochondria. The steady-state levels of two other outer membrane β-barrel proteins, Tom40 and porin, were also reduced in Tob55↓ mitochondria. Tom22 was also decreased. Although Tom22 is not a β-barrel protein, it is a component of the core TOM complex and its level is likely reduced because the deficiency of Tom40 does not allow proper assembly of Tom22 molecules. In support of this notion, Tom22 was previously observed to be reduced in a Tom40 mutant of N. crassa (Taylor et al. 2003). The levels of other mitochondrial proteins examined were unchanged by the reduction in Tob55 (Figure 2A).
Figure 2.—
N. crassa Tob55 is involved in biogenesis of β-barrel proteins. (A) Mitochondria were isolated from the control strain (HP1) and the tob55 knockout sheltered heterokaryon strain (Tob55KO-3) following growth in the absence (−) or presence (+) of 400 μm fpa and histidine. Mitochondrial proteins (20 μg) were separated by SDS–PAGE, blotted to nitrocellulose, and analyzed by immunodecoration with antisera against the indicated proteins. Tob55 is present as two bands of 62 and 58 kDa, respectively. (B) As in A except mitochondria were isolated from the control (HP1) and two strains (T55his6-1, T55his6-3) obtained by rescue of the Δtob55, histidine-requiring nucleus from strain Tob55KO-3 by a genomic tob55 gene encoding an N-terminal hexahistidinyl tag. Blots were immunodecorated with Tob55 antiserum or penta-His antiserum. his, histidine.
We consistently observed two Tob55 bands of apparent molecular weights 62 and 58 kDa following immunodecoration of Western blots. Both of these bands appear to represent Tob55 since both disappear in Tob55↓ mitochondria (Figure 2A) and both are detected by a penta-His antibody in mitochondria of strains rescued by a hexahistidinyl-tagged tob55 allele (Figure 2B). The nature of the different Tob55 bands is discussed further below.
Mitochondrial protein import in Tob55↓ mitochondria:
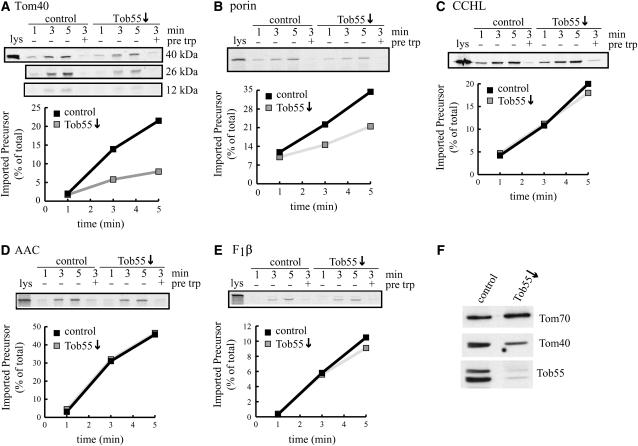
Tob55↓ mitochondria were examined for their ability to import mitochondrial precursor proteins. Import of the β-barrel proteins Tom40 and porin was reduced in Tob55↓ mitochondria (Figure 3, A and B). To exclude the possibility that the reduced levels of Tom40 and Tom22 observed in Tob55↓ mitochondria (Figure 2A) are the cause of the reduced import, we examined the import of other precursors known to be translocated via the TOM complex. Import of the non-β-barrel proteins cytochrome c heme lyase (CCHL), the ATP-ADP carrier (AAC), and the β-subunit of ATP synthase (F1β) to the IMS, the inner membrane, and the matrix, respectively, was not reduced in Tob55↓ mitochondria (Figure 3, C–E). Thus, the level of Tom40 in the mitochondria used for import (Figure 3F) is sufficient for in vitro import of these proteins. These data indicate that N. crassa Tob55 is involved in import of outer membrane β-barrel proteins.
Figure 3.—
Tob55-deficient mitochondria are defective in import of outer membrane β-barrel proteins. (A–E) Mitochondria were isolated from strain HP1 (control) and the tob55 knockout strain Tob55KO-3 (Tob55↓) following growth in the presence of 250 μm fpa plus histidine. For import assays, mitochondria were incubated with lysates containing radiolabeled mitochondrial precursors at 15° for the indicated times. Following a post-import proteinase K treatment, mitochondria were reisolated and subjected to SDS–PAGE. The gels were transferred to nitrocellulose and exposed to X-ray film and then a PhosphorImager screen. One sample from each strain was treated with trypsin prior to import (“pre trp”) to demonstrate receptor-dependent import. “lys” represents 33% of the total radioactivity added to each reaction. (F) Western blot showing the level of Tob55, Tom40, and Tom70 in mitochondria isolated from cultures grown in the presence of 250 μm fpa plus histidine.
The assembly of Tom40 and porin during import of the precursors into Tob55↓ mitochondria was assessed using BNGE. Analysis of Tom40 assembly by BNGE reveals two assembly intermediates of 250 and 100 kDa as well as the 400-kDa fully assembled TOM core complex (Rapaport and Neupert 1999; Model et al. 2001; Taylor et al. 2003). The level of all forms, but particularly the 250-kDa intermediate, was reduced during import of Tom40 precursor into Tob55↓ mitochondria (Figure 4A). Similarly, all complexes containing porin (240, 115, and 66 kDa) were reduced following import into Tob55↓ mitochondria (Figure 4B).
Figure 4.—
Assembly of Tom40 and porin in Tob55↓ mitochondria. Radiolabeled Tom40 (A) or porin (B) precursor were incubated for 20 min at the indicated temperature with mitochondria isolated from the control strain HP1 (Ct) or Tob55KO-3 (55↓), both grown in the presence of fpa (250 μm) and histidine. Mitochondria were washed with 80 mm KCl, reisolated, and lysed in blue gel sample buffer containing 1% digitonin. The samples were subjected to blue native gel electrophoresis, blotted to PVDF membrane, and analyzed by autoradiography. (C and D) Antibody supershift experiments. Import was performed as for A and B with Tom40 (C) and porin (D) precursor proteins using mitochondria isolated from a wild-type strain (NCN 251). Following import at 25°, mitochondria were lysed with 1% digitonin. Reactions were then incubated with buffer alone (“No Ab”), affinity-purified antibody to Tob55, or affinity-purified antibody to D. melanogaster ATM protein as a negative control for 2 hr at 4° and then subjected to BNGE. The gel was blotted to PVDF membrane and examined by autoradiography. Apparent sizes of the intermediates are shown on the left. The position of the supershifted products is indicated with an arrow on the right.
Antibody supershift assays were used to demonstrate that Tob55 is a component of assembly intermediates for Tom40 and porin. Affinity-purified Tob55 antibody was added to the import/assembly reactions of these proteins prior to analysis by BNGE. For Tom40, a shift of the 250-kDa assembly intermediate resulted from addition of the antibody (Figure 4C). This confirms that Tob55 is a component of the 250-kDa Tom40 assembly intermediate in N. crassa. The 250-kDa band is shifted to a position where it comigrates with a minor high-molecular-weight band that we have previously shown to be a nonproductive intermediate (Taylor et al. 2003). However, evidence for the shift is seen both by the reduced level of precursor in the 250-kDa band and by the increased intensity at the higher-molecular-weight position in the lane where the Tob55 antibody was added. For porin, addition of affinity-purified Tob55 antibody resulted in a shift in the 240-kDa complex (Figure 4D), establishing that this complex of the porin assembly pathway in N. crassa represents an assembly intermediate containing Tob55. This is the first direct evidence for the association of the porin precursor with the TOB complex.
The N. crassa tob55 mRNA is alternatively spliced:
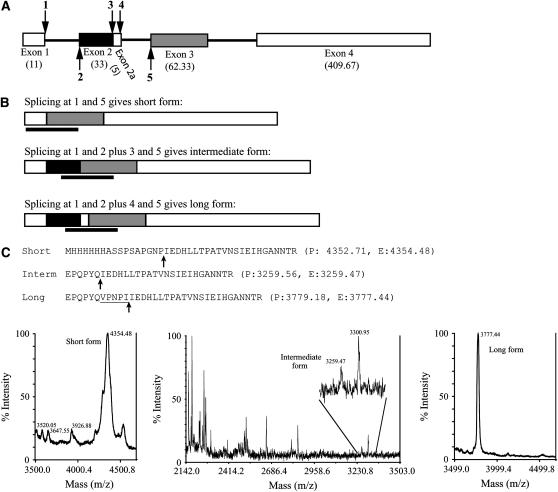
To obtain cDNA clones of tob55, we performed RT–PCR on RNA samples isolated from the wild-type strain NCN251. While examining one of the cloned cDNAs for PCR-induced sequencing errors, we discovered that exon 2 predicted by the N. crassa sequencing project (Galagan et al. 2003) was absent. Therefore, additional cDNA clones were sequenced. Analysis of these clones revealed that there were three different tob55 cDNAs that would encode Tob55 proteins of different lengths. The variability centers on alternative splice sites in and around the second exon of the structural gene with three possible 5′ splice sites and two possible 3′ splice sites (Figure 5A). We refer to the proteins generated from the three possible mRNAs identified as the long (521 residues), intermediate (516 residues), and short (483 residues) isoforms of Tob55 (Figure 5B). Sequence of 20 randomly selected cloned cDNAs revealed 10 clones representing the short form, 7 the intermediate form, and 3 the long form. Since only two bands were observed by Western analysis, we assume that the intermediate and long isoforms cannot be distinguished under the electrophoretic conditions used. To obtain evidence that the different forms exist at the protein level, outer membrane vesicles were purifed from strain T55His6-1. Proteins were separated by electrophoresis and individual bands of the size predicted for the Tob55 isoforms were analyzed by mass spectrometry. Peptides that define the long and intermediate forms were detected in the higher-molecular-weight band and a peptide defining the short form was detected in the lower-molecular-weight band (Figure 5C). Western blots using our Tob55 antibody suggest that the short form is more abundant than the intermediate and/or long forms. However, our antibody was produced to the N-terminal 108 residues of the short form so there may be more epitopes available to react with the antibody within this form of the protein. The levels of the two bands appear to be equal when the hexahistidinyl-tagged version of the protein is detected with the penta-His antibody (Figure 2B).
Figure 5.—
Intron/exon structure of the tob55 gene. (A) The positions of all possible exons (rectangular boxes) and introns (solid lines) of tob55 are indicated. The number of codons is given in parentheses below the number of each exon. Possible 5′ splice sites are shown above the line and possible 3′ splice sites are shown below. Potential splice sites are numbered in sequence. Exon 2a is not separated from exon 2 by an intron, but alternative splicing may remove exon 2a from the remainder of exon 2 in some cases. (B) The three forms of the Tob55 protein that arise from alternative splicing. Shading is coded to correspond to the different exons shown in A. The solid bars under each isoform indicate peptides predicted to be generated by tryptic digestion that would be unique for each isoform of the protein. (C) Mass spectrometry of Tob55 isoforms. The peptides predicted to be generated by tryptic digestion as indicated in B are shown. Six His residues occur after the initial Met residue on the short form because the analysis was done on a His-tagged version of the protein. Arrows under the peptide sequences indicate the splice point between exon 1 and exon 3 for the short form, between exon 2 and exon 3 for the intermediate (“interm”) form, and between exon 2a and exon 3 for the long form. The five residues that make up exon 2a are underlined in the long form. The sequence of each peptide is followed by its predicted (“P”) mass and the mass that was determined experimentally (“E”) by mass spectrometry. The mass of the predominant peak corresponding to the unique peptide for the short form shows that oxidation of the peptide, probably at the N-terminal Met residue, has occurred. The predicted mass includes this consideration. The predicted and experimentally determined masses for the short and long peptides differ by <2 Da. This is within the limits of accuracy for these large peptides whose masses were determined by linear MALDI–TOF. The tracings from the appropriate regions of the mass spectra for each peptide are shown below.
Do the different forms of Tob55 have specialized functions?
Since it is possible that the different forms of the protein might have specialized functions, we wanted to determine if the amounts of the different isoforms could be altered under various conditions. Wild-type cells were grown at three different temperatures but no differences in the Tob55 bands could be discerned. Similarly, no difference was observed in 15-hr-old mycelium, ungerminated conidia, or conidia germinated for 3 hr. It was conceivable that the isoforms were present only in certain strains of N. crassa and not in other strains or species. We obtained several natural isolates of Neurospora, representing four species: N. crassa, N. intermedia, N. sitophila, and N. tetrasperma. We also examined Tob55 in two mutant N. crassa strains: cyt-2-1, which carries a mutation in CCHL (Drygas et al. 1989) and [mi-3], which carries a mutation in the mitochondrially encoded Cox1 gene (Lemire and Nargang 1986). In all of these strains, both Tob55 bands were detected, suggesting that the alternative splicing occurs throughout the genus. Furthermore, the levels of the different forms do not appear to be altered in response to mutations affecting electron transport (supplemental data at http://www.genetics.org/supplemental/).
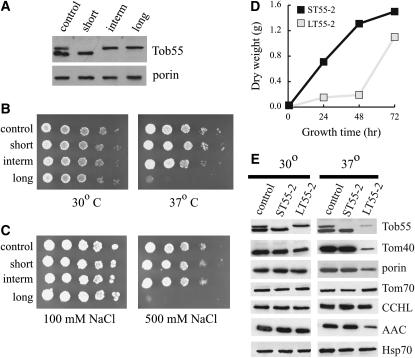
To further address the question of possible specialized function, strains were developed that expressed only one of the different isoforms of the protein. These strains were developed by transforming the knockout strain with plasmids carrying a bleomycin resistance marker and one of the different cDNA versions of the gene fused to the genomic tob55 promoter. Transformants were selected on medium containing histidine, bleomycin, and fpa. Isolates were examined for nutritional requirements, and the histidine-requiring homokaryons ST55-2 (short isoform), IT55-8 (intermediate isoform), and LT55-2 (long isoform) were chosen for further analysis (Figure 6A).
Figure 6.—
Strains expressing only the long isoform of Tob55 have growth defects. Constructs expressing cDNA versions of the three different Tob55 splicing variants from the endogenous tob55 promoter were used to rescue the Δtob55 nucleus from strain Tob55KO-3. This resulted in strains ST55-2, IT55-8, and LT55-2 expressing the short, intermediate (“interm”), and long isoforms of the protein, respectively. (A) Mitochondria (30 μg) isolated from these three strains and the control strain (76-26) following growth at 30° were examined on Western blots to demonstrate that only one form of the protein was expressed in each strain. Porin was used as the loading control. (B) Serial dilutions of conidia from the strains described in A were spotted onto solid sorbose-containing medium and grown at 30° or 37°. (C) As in B, but conidia were spotted on plates containing either 100 or 500 mm NaCl and were grown at 30°. (D) The short and long strains were grown at 37° in liquid cultures containing 250 ml of Vogel's medium. Each flask was inoculated with 106 conidia/ml. At the times indicated, the cultures were harvested and the pad of mycelium was dried and then weighed to give the dry weight of the culture. (E) The control, short, and long strains were grown in liquid medium at 30° and 37° for 24 hr. Mitochondria were isolated and examined for the presence of various mitochondrial proteins (indicated on the right) by Western blot analysis. Each lane contained 30 μg of mitochondrial protein.
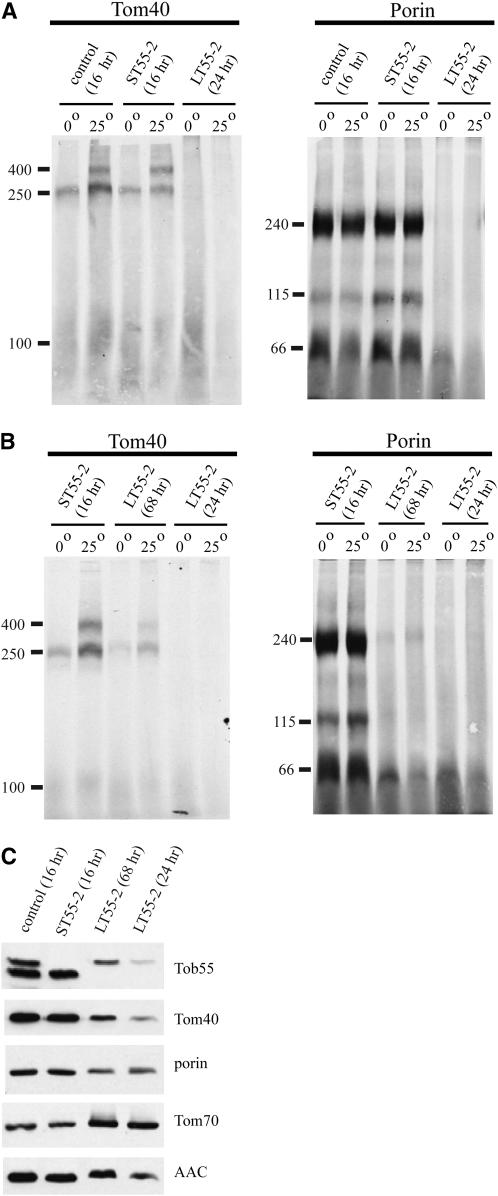
Each of the strains expressing an individual form of Tob55 was examined for growth rate under various conditions. No difference from the control strain was seen for any of the Tob55 strains on media containing chloramphenicol, an inhibitor of mitochondrial translation, or antimycin A, an inhibitor of electron transport chain complex III (not shown). However, growth on solid medium at 37° or in the presence of high concentrations of NaCl revealed a severe growth defect in strains expressing only the long isoform of Tob55 (Figure 6, B and C). To ensure that the growth defect in the long isoform was not due to a specific random integration event in an individual transformant, we examined eight additional long isoform strains and five additional short isoform strains. Results similar to those in Figure 6, B and C, were obtained for all the additional strains (not shown). In liquid medium, the strain expressing the long isoform has a peculiar phenotype at 37°. A small amount of growth occurs during the first 48 hr, but between 48 and 72 hr, substantial growth occurs (Figure 6D). Mitochondria isolated from strains expressing either the short or the long form of the protein following growth at 30° show no differences in the steady-state levels of any mitochondrial protein examined, including Tob55 (Figure 6E). However, in cells expressing only the long form grown for 24 hr at 37°, mitochondria were deficient in Tob55. There is also a slight deficiency in porin and a striking deficiency of Tom40. CCHL and Hsp70 appear to be present at normal levels, but levels of AAC are also reduced. Tom70 appears to be slightly increased. The deficiency of Tob55 observed in cells expressing only the long isoform seems to contradict the apparently unaltered ratio of the bands detected by Western blot analysis following growth of wild-type cells at high temperature (supplemental data at http://www.genetics.org/supplemental/). This suggests that the higher-molecular-weight isoform in the wild-type cells grown at high temperature is largely the intermediate isoform or that the long isoform can be assembled into the membrane at high temperature if mitochondria also contain the other isoforms. Once assembled, the long isoform might be stable, but inactive.
We also examined the assembly of the β-barrel proteins Tom40 and porin in mitochondria isolated from 30°- and 37°-grown cultures of wild-type cells and cells expressing only the short or long isoforms of Tob55. In mitochondria isolated from the 30° cultures, there were no differences observed between the strains (not shown). However, neither intermediates nor mature forms of Tom40 or porin were observed in assembly assays using mitochondria from cultures of the long isoform strain when grown at 37° for 24 hr, suggesting that little, if any, assembly of β-barrels occurs in these cells (Figure 7A). Since the growth of long isoform cultures at 37° appears to have a lag period of at least 48 hr (Figure 6D), we also examined assembly of β-barrels in mitochondria isolated from 68-hr cultures (Figure 7B). Assembly of Tom40 was observed in these mitochondria, but at reduced levels compared to the short isoform strain. A small amount of the 240-kDa porin intermediate was also observed in mitochondria from the 68-hr culture. Cultures expressing the long isoform were also examined for steady-state levels of various mitochondrial proteins after 68 hr of growth at 37° (Figure 7C). Interestingly, levels of Tob55 and Tom40 were higher in the 68-hr than in the 24-hr cultures, but porin did not increase. The level of AAC was also increased in the older cultures while the level of Tom70 appeared to remain slightly higher than in the wild-type control or the short Tob55 strain.
Figure 7.—
Assembly of β-barrel proteins in mitochondria isolated from 37° cultures. (A) Mitochondria were isolated from strain 76-26 (control) and ST55-2 following 16 hr growth at 37° (inoculum of 106 conidia/ml) and from LT55-2 after 24 hr at 37° using twice the amount of inoculum. Mitochondria were incubated with radiolabeled precursors of Tom40 or porin for 20 min at the indicated temperature and analyzed by BNGE. (B) As in A, except that LT55-2 was also grown for 68 hr with an inoculum of 106 conidia/ml. The size of assembly products is indicated on the left. (C) Western blots analysis of mitochondrial proteins from the strains indicated following growth at 37° for the indicated times.
Alignment of the long isoform with other Tob55 proteins:
To determine if residues in the long isoform were conserved, we compared the sequence at the N-terminal region of the N. crassa protein with Tob55 proteins from Homo sapiens, Arabidopsis thaliana, and the fungi Chaetomium globosum, Giberella zeae, Aspergillus nidulans, and S. cerevisiae (Figure 8). The alignment shows little conservation among the seven species in the region that defines the alternative forms, while similarity increases after the region. C. globosum, which is closely related to N. crassa (Suh and Blackwell 1999; Huhndorf et al. 2004), shares some identical residues within the region that defines the different isoforms, but no residues are shared with the five amino acids that are contained only in the long isoform of the N. crassa protein. Interestingly, predictions of protein-coding sequence from genome databases for other filamentous fungi (e.g., G. zeae and A. nidulans) give the long form as the product of the tob55 gene. However, our inspection of the tob55 genomic sequences shows that potential splice sites for removal of the second exon do exist (not shown), so that the short and long forms may be present in these organisms as well. We could not identify a consensus splice site that would give the intermediate form in these species.
Figure 8.—
Alignment of N-terminal region of Tob55. The position of the residues present in the N. crassa long isoform, but absent in the small isoform, are indicated by the thin line above the alignment. Residues absent in the intermediate isoform are shown by the thick line. The start of the POTRA domain (see discussion), as defined previously (Sanchez-Pulido et al. 2003), is indicated by a plus sign (+) above the alignment. Residues that are identical in six or more of the seven species are shown against a solid background. Residues identical in four or five species are shown against a shaded background. NcL, long isoform of N. crassa Tob55; Cg, C. globosum; Gz, G. zeae; An, A. nidulans; Sc, S. cerevisiae; At, A. thaliana; Hs, H. sapiens.
DISCUSSION
We have shown that reduced levels of N. crassa Tob55 result in slow growth as well as reduced import efficiency of β-barrel precursors to the outer mitochondrial membrane. Import of non-β-barrel mitochondrial proteins is not affected. We have also demonstrated that N. crassa Tob55 is present in a high-molecular-weight assembly intermediate with the precursors of Tom40 and porin. This marks the first direct demonstration of an association between the porin precursor and the TOB complex.
Surprisingly, our studies also demonstrated that there are three isoforms of the N. crassa Tob55 protein produced by alternative splicing. To our knowledge, this is the first description of alternative splicing producing isoforms of a protein involved in mitochondrial protein import. The observation that different isoforms occur in all strains and species of Neurospora examined suggests that they may serve some specialized purpose. It has been demonstrated that expression of isoforms for import components arising from multigene families in higher organisms can vary in certain conditions or tissues. For example, analysis of transcript levels for components of the import apparatus in Arabidopsis showed that, for the most part, one isoform was expressed most prominently in all tissues under normal conditions. However, treatment with inhibitors of the electron transport chain resulted in increased expression of the minor isoforms of several proteins, implying that the import apparatus may become functionally specialized in response to stress (Lister et al. 2004). Transcript and EST analysis of two Tom20 isoforms arising from separate genes in animals revealed that one isoform was specifically expressed in the testis of mice and fruit flies while the other form was ubiquitously expressed. Similarly, RNA interference knockdown experiments in Caenorhabditis elegans showed that one form of Tom20 is essential while the other appears to play a more specialized role (Likic et al. 2005). Our attempts to alter the expression of the Tob55 isoforms in N. crassa by varying temperature or treatment with chloramphenicol or antimycin A did not result in any obvious change in the ratio of the forms when examined by Western analysis, although it is possible that minor changes were not detected. Similarly, no change in the ratio was seen in conidiaspores or strains carrying mutations affecting electron transport.
Since strains expressing any isoform grow equally well at the permissive temperature, each form must be capable of importing all β-barrel proteins to some extent. At 37°, cells expressing only the long isoform of Tob55 are inefficient at assembling β-barrel proteins into the membrane. Since Tob55 is itself a β-barrel protein, it is difficult to establish if it is the assembly of the long isoform that is reduced or if the activity of the protein once assembled is compromised. Since long Tob55 cultures eventually begin to grow at 37° and have some capacity to assemble β-barrels, it appears that assembly of active long Tob55 gradually occurs. Perhaps other factors, such as chaperones, aid this process and become more abundant as the culture ages.
What function of the TOB complex is most likely disturbed by the extra residues in the long form at 37° or in the presence of high salt? The alternatively spliced region is near the N terminus of the protein, which is predicted to occur in the IMS (Paschen et al. 2003; Sanchez-Pulido et al. 2003). Since the other known proteins of the TOB complex (Mas37 and Tob38) are peripheral outer membrane proteins facing the cytosol, it seems unlikely that interactions between the long isoform of Tob55 and these proteins are affected by salt or high temperature. The TOB complex probably contains more than one molecule of Tob55 (Kozjak et al. 2003; Paschen et al. 2003; Waizenegger et al. 2004), and temperature-induced changes in structure in the IMS domain might affect interactions among Tob55 molecules in a single complex. However, it has been shown that yeast Tob55 lacking the 102 amino-terminal residues still assembles into the TOB complex (Habib et al. 2007).
A more likely cause for the defects in the long isoform is inefficient interaction with incoming β-barrel precursors in the IMS. Both prokaryotic and eukaryotic Tob55 homologs have been shown to contain one or more polypeptide-transport-associated (POTRA) domains in their N-terminal regions (Sanchez-Pulido et al. 2003). It has been suggested that POTRA domains may be involved in binding β-barrel precursors just prior to their insertion into the membrane (Bos and Tommassen 2004), and it has recently been demonstrated that the N-terminal region of yeast Tob55 does interact with β-barrel precursors (Habib et al. 2007). The extra residues in the long form of N. crassa Tob55 occur just prior to the beginning of the POTRA domain (Figure 8), and the defects caused by the long form of the protein in N. crassa at 37° or in the presence of high salt may be due to an effect on this domain. Further investigations will be aimed at confirming this possibility and on attempting to define specific roles for the different isoforms.
Acknowledgments
We are grateful to Ellen Homola and Shelagh Campbell for the donation of purified antibody to Drosophila ATM protein. We also thank the Protein Analysis Unit at the Adolf-Butenandt-Institut, Ludwig Maximilians University Munich, for mass spectrometry analysis. This work was supported by grants from the Canadian Institutes of Health Research to F.E.N. and from the Deutsche Forschungsgemeinschaft to D.R. and W.N. S.C.H. was supported by scholarships from the Natural Research Council of Canada and the Alberta Ingenuity Fund.
References
- Ausubel, R. A., R. Brent, R. E. Kingston, D. D. Moore and J. G. Seidman, 1992. Current Protocols in Molecular Biology. Greene and Wiley Interscience, New York.
- Bos, M. P., and J. Tommassen, 2004. Biogenesis of the Gram-negative bacterial outer membrane. Curr. Opin. Microbiol. 7: 610–616. [DOI] [PubMed] [Google Scholar]
- Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. Crew et al., 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R. H., and F. J. De Serres, 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17: 79–143. [Google Scholar]
- Dolezal, P., V. Likic, J. Tachezy and T. Lithgow, 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313: 314–318. [DOI] [PubMed] [Google Scholar]
- Drygas, M. E., A. M. Lambowitz and F. E. Nargang, 1989. Cloning and analysis of the Neurospora crassa gene for cytochrome c heme lyase. J. Biol. Chem. 264: 17897–17907. [PubMed] [Google Scholar]
- Eckart, K., L. Eichacker, K. Sohrt, E. Schleiff, L. Heins et al., 2002. A Toc75-like protein import channel is abundant in chloroplasts. EMBO Rep. 3: 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, K., S. K. Buchanan and T. Lithgow, 2001. The alpha and the beta: protein translocation across mitochondrial and plastid outer membranes. Trends Biochem. Sci. 26: 36–40. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. [DOI] [PubMed] [Google Scholar]
- Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins et al., 2005. Protein identification and analysis tool on the EXPASY server, pp. 571–607 in The Proteomics Protocols Handbook, edited by J. M. Walker. Humana Press, Clifton, NJ/Totowa, NY.
- Gentle, I., K. Gabriel, P. Beech, R. Waller and T. Lithgow, 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, A. G., and W. L. Crosby, 1989. Anaerobic induction of alanine aminotransferase in barley root tissue. Plant Physiol. 90: 1305–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotelueschen, J., and R. Metzenberg, 1995. Some property of the nucleus determines the competence of Neurospora crassa for transformation. Genetics 139: 1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib, S. J., T. Waizenegger, M. Lech, W. Neupert and D. Rapaport, 2005. Assembly of the TOB complex of mitochondria. J. Biol. Chem. 280: 6434–6440. [DOI] [PubMed] [Google Scholar]
- Habib, S. J., T. Waizenegger, A. Niewienda, S. Paschen, W. Neupert et al., 2007. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial β-barrel proteins. J. Cell Biol. 176: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkness, T. A. A., F. E. Nargang, I. Van der Klei, W. Neupert and R. Lill, 1994. A crucial role of the mitochondrial protein import receptor MOM19 for the biogenesis of mitochondria. J. Cell Biol. 124: 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins, S. C., and F. E. Nargang, 2004. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279: 12396–12405. [DOI] [PubMed] [Google Scholar]
- Huhndorf, S. M., A. N. Miller and F. A. Fernandez, 2004. Molecular systematics of the Sordariales: the order and the family Lasiosphaeriaceae redefined. Mycologia 96: 368–387. [PubMed] [Google Scholar]
- Humphries, A. D., I. C. Streimann, D. Stojanovski, A. J. Johnston, M. Yano et al., 2005. Dissection of the mitochondrial import and assembly pathway for human Tom40. J. Biol. Chem. 280: 11535–11543. [DOI] [PubMed] [Google Scholar]
- Ishikawa, D., H. Yamamoto, Y. Tamura, K. Moritoh and T. Endo, 2004. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J. Cell Biol. 166: 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A. E., and R. E. Jensen, 2004. Barreling through the membrane. Nat. Struct. Mol. Biol. 11: 113–114. [DOI] [PubMed] [Google Scholar]
- Kozjak, V., N. Wiedemann, D. Milenkovic, C. Lohaus, H. E. Meyer et al., 2003. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 278: 48520–48523. [DOI] [PubMed] [Google Scholar]
- Krimmer, T., D. Rapaport, M. T. Ryan, C. Meisinger, C. K. Kassenbrock et al., 2001. Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J. Cell Biol. 152: 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann, T., and E. L. Sonnhammer, 2005. Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire, E. G., and F. E. Nargang, 1986. A missense mutation in the oxi-3 gene of the [mi-3] extranuclear mutant of Neurospora crassa. J. Biol. Chem. 261: 5610–5615. [PubMed] [Google Scholar]
- Likic, V. A., A. Perry, J. Hulett, M. Derby, A. Traven et al., 2005. Patterns that define the four domains conserved in known and novel isoforms of the protein import receptor Tom20. J. Mol. Biol. 347: 81–93. [DOI] [PubMed] [Google Scholar]
- Lister, R., O. Chew, M.-N. Lee, J. L. Heazlewood, R. Clifton et al., 2004. A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol. 134: 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin, B. S., M. Freitag and E. U. Selker, 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44: 34–36. [Google Scholar]
- Margolin, B. S., M. Freitag and E. U. Selker, 2000. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation: correction. Fungal Genet. Newsl. 47: 112. [Google Scholar]
- Mayer, A., R. Lill and W. Neupert, 1993. Translocation and insertion of precursor proteins into isolated outer membranes of mitochondria. J. Cell Biol. 121: 1233–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey, K., and J. A. Kinsey, 2000. Fungal Genetics Stock Center catalogue of strains. Edition 8. Fungal Genet. Newsl. 47(Suppl.): 6–9. [Google Scholar]
- Milenkovic, D., V. Kozjak, N. Wiedemann, C. Lohaus, H. E. Meyer et al., 2004. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J. Biol. Chem. 279: 22781–22785. [DOI] [PubMed] [Google Scholar]
- Model, K., C. Meisinger, T. Prinz, N. Wiedemann, K. N. Truscott et al., 2001. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 8: 361–370. [DOI] [PubMed] [Google Scholar]
- Nargang, F. E., K.-P. Künkele, A. Mayer, R. G. Ritzel, W. Neupert et al., 1995. “Sheltered disruption” of Neurospora crassa MOM22, an essential component of the mitochondrial protein import complex. EMBO J. 14: 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargang, F. E., D. Rapaport, R. G. Ritzel, W. Neupert and R. Lill, 1998. Role of the negative charges in the cytosolic domain of TOM22 in the import of precursor proteins into mitochondria. Mol. Cell. Biol. 18: 3173–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen, S. A., T. Waizenegger, T. Stan, M. Preuss, M. Cyrklaff et al., 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426: 862–866. [DOI] [PubMed] [Google Scholar]
- Paschen, S., W. Neupert and D. Rapaport, 2005. Biogenesis of β-barrel membrane proteins of mitochondria. Trends Biochem. Sci. 30: 575–582. [DOI] [PubMed] [Google Scholar]
- Rapaport, D., 2002. Biogenesis of the mitochondrial TOM complex. Trends Biochem. Sci. 27: 191–197. [DOI] [PubMed] [Google Scholar]
- Rapaport, D., 2003. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 4: 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D., and W. Neupert, 1999. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J. Cell Biol. 146: 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D., R. Taylor, M. Käser, T. Langer, W. Neupert et al., 2001. Structural requirements of Tom40 for assembly into preexisting TOM complexes of mitochondria. Mol. Biol. Cell 12: 1189–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido, L., D. Devos, S. Genevrois, M. Vicente and A. Valencia, 2003. POTRA: a conserved domain in the FtsQ family and a class of β-barrel outer membrane proteins. Trends Biochem. Sci. 28: 523–526. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and G. von Jagow, 1991. Blue native electrophoresis for isolation of membrane complexes in enzymatically active form. Anal. Biochem. 199: 223–231. [DOI] [PubMed] [Google Scholar]
- Schägger, H., W. A. Cramer and G. von Jagow, 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217: 220–230. [DOI] [PubMed] [Google Scholar]
- Schleiff, E., L. A. Eichacker, K. Eckart, T. Becker, O. Mirus et al., 2003. Prediction of the plant β-barrel proteome: a case study of the chloroplast outer envelope. Protein Sci. 12: 748–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, S., H. Prokisch, T. Schlunck, D. G. Camp, II, U. Ahting et al., 2006. Proteome analysis of mitochondrial outer membrane from Neurospora crassa. Proteomics 6: 72–80. [DOI] [PubMed] [Google Scholar]
- Silva, E., S. Tiong, M. Pedersen, E. Homola, A. Royou et al., 2004. ATM is required for telomere maintenance and chromosome stability during Drosophila development. Curr. Biol. 14: 1341–1347. [DOI] [PubMed] [Google Scholar]
- Suh, S.-O., and M. Blackwell, 1999. Molecular phylogeny of the cleistothecial fungi placed in Cephalothecaceae and Pseudeurotiaceae. Mycologia 91: 836–848. [Google Scholar]
- Tamm, L. K., A. Arora and J. H. Kleinschmidt, 2001. Structure and assembly of β-barrel membrane proteins. J. Biol. Chem. 276: 32399–32402. [DOI] [PubMed] [Google Scholar]
- Tanton, L. L., C. E. Nargang, K. E. Kessler, Q. Li and F. E. Nargang, 2003. Alternative oxidase expression in Neurospora crassa. Fungal Genet. Biol. 39: 176–190. [DOI] [PubMed] [Google Scholar]
- Taylor, R., B. McHale and F. E. Nargang, 2003. Characterization of Neurospora crassa Tom40-deficient mutants and effect of specific mutations on Tom40 assembly. J. Biol. Chem. 278: 765–775. [DOI] [PubMed] [Google Scholar]
- Voulhoux, R., and J. Tommassen, 2004. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 155: 129–135. [DOI] [PubMed] [Google Scholar]
- Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols and J. Tommassen, 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299: 262–265. [DOI] [PubMed] [Google Scholar]
- Waizenegger, T., S. J. Habib, M. Lech, D. Mokranjac, S. A. Paschen et al., 2004. Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep. 5: 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, J., K. Lengeler and E. Kothe, 1996. An instant preparation method for nucleic acids of filamentous fungi. Fungal Genet. Newslett. 43: 54–55. [Google Scholar]
- Wiedemann, N., V. Kozjak, A. Chacinska, B. Schönfisch, S. Rospert et al., 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424: 565–571. [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., K. N. Truscott, S. Pfannschmidt, B. Guiard, C. Meisinger et al., 2004. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane. Intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 279: 18188–18194. [DOI] [PubMed] [Google Scholar]
- Wilkins, M. R., I. Lindskog, E. Gasteiger, A. Bairoch, J.-C. Sanchez et al., 1997. Detailed peptide characterisation using PEPTIDEMASS—a World-Wide-Web-accessible tool. Electrophoresis 18: 403–408. [DOI] [PubMed] [Google Scholar]
- Wimley, W. C., 2003. The versatile β-barrel membrane protein. Curr. Opin. Struct. Biol. 13: 404–411. [DOI] [PubMed] [Google Scholar]
- Wu, T., J. Malinverni, N. Ruiz, K. Seokhee, T. J. Silhavy et al., 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121: 235–245. [DOI] [PubMed] [Google Scholar]