HISTORY shows that unplanned, unanticipated discoveries in model organisms have had a major impact on the creation of new knowledge leading to a deeper understanding of human disease. By definition, unanticipated discoveries fall outside the boundaries of prospective road maps. I will use my own research path to illustrate the point, but not because my story is exceptional. It derives power from the fact that it is commonplace.
When I was a young scientist in the 1970s, one of my mentors made a statement I never forgot. It went something like this: “It does not matter where you start, even if the starting point seems mundane. With good training, plus a little luck, the avenues that you travel along will lead into unanticipated and unexplored territory. You will not know what the terrain looks like until you get there.” I took this to mean that I was authorized to practice science without a road map, although it was also pointed out that this was not the recommended formula for getting a fundable score on a federal grant and that navigating without a road map was the method of choice only for a small cadre of scientists called geneticists.
In 1975 I began my postdoctoral experience with Gerry Fink at Cornell University. I was shown a refrigerator with a handwritten sign on the door that said “Legacies.” I was told that I would find a project in that refrigerator. Inside was a box full of vials containing strains of Saccharomyces cerevisiae with a sign on it that said “Charnas.” I later co-authored an article with Larry Charnas (Culbertson et al. 1977), a Cornell undergraduate, but we never met. He had left for medical school before I arrived at Cornell. Larry made strains that carried mutations induced with ICR-170, a mutagen related to ICR-191, which was known to cause frameshift mutations in bacteria (Yourno and Health 1969). He also found extragenic suppressors mapping at two loci. I spent 3 years as a Jane Coffin Childs Fellow working on frameshift suppressors in the belief that this starting point could serve as an entrance into poorly understood mechanisms of post-transcriptional control of gene expression. I realized that translation did not take place in a vacuum and that gene expression was likely also to be regulated by translational mechanisms.
Beginning in 1978 as an assistant professor at the University of Wisconsin, Madison, I characterized frameshift mutations and suppressors. Several dozen ICR-induced mutations, mostly at the HIS4 locus, were cloned and sequenced. All of them were +1 insertions in runs of G/C base pairs (Mathison and Culbertson 1985). Twenty-seven suppressor genes were identified (Gaber et al. 1983), including tRNA and protein-coding genes. Three glycine and two proline tRNA multi-gene families were described. We learned a lot about the intricacies of coding by studying how altered tRNAs suppressed +1 insertions (Gaber and Culbertson 1984).
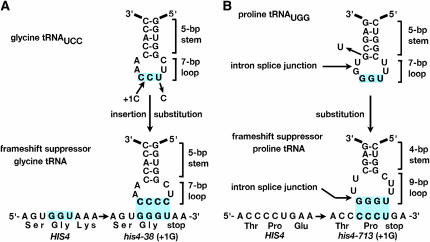
The glycine tRNA suppressors contained an extra nucleotide in the anticodon loop (Figure 1A). These tRNAs had 5 bp in the anticodon stem and eight unpaired bases in the anticodon loop instead of the normal seven bases (Gaber and Culbertson 1982; Mendenhall et al. 1987). The proline tRNAs revealed an interesting relationship between tRNA introns and frameshift suppression. One of the proline isoacceptors was produced from a redundant set of genes containing an intron in the anticodon loop that was spliced out to form the mature tRNA (Figure 1B) (Cummins et al. 1985). The suppressor alleles always produced a tRNA containing a base substitution in the bottom base pair of the anticodon stem rather than an insertion in the anticodon loop. This resulted in a shortened stem with 4 bp and an anticodon loop that, after splicing, contained nine unpaired bases. The anticodon loop was expanded without a base insertion, and this led to suppression (Winey et al. 1986). By analyzing suppressor tRNAs with eight- or nine-base anticodon loops in the context of structural models, the mechanism of suppression came to be understood in considerable detail (Culbertson et al. 1990). By inference, knowledge of the universal triplet code was enriched.
Figure 1.—
Altered tRNAs suppress frameshift mutations in yeast. (A) Glycine tRNA suppressors contain a base insertion in the anticodon loop, allowing them to suppress +1 insertions in glycine codons. (B) Proline tRNA suppressors derived from intron-containing genes contain base substitutions in the anticodon stem. This results in an expanded anticodon loop, allowing them to suppress +1 insertions in proline codons.
Some suppressor genes encoded proteins: the translation termination factor RF3, the global transcription factor Mbf1p, and ribosomal protein S3 (Wilson and Culbertson 1988; Hendrick et al. 2001). Some genes were not immediately found due to redundancy or because suppressor mutations were rare. Using specialized screens, frameshift suppressor mutations were found in TEF2, one of two redundant genes coding for EF1α, the elongation factor that controls translational proofeading (Sandbaken and Culbertson 1988), and EBS1, whose product inhibits translation initiation (Ford et al. 2006). In the accepted parlance of National Institutes of Health summary statements, useful information was forthcoming on mechanisms of translational fidelity, but all things considered, nothing earth shattering was discovered. However, the early work on frameshift suppressors led to future insights that shed light on human disease mechanisms.
Looking backward, the forks in the road that define my contributions were driven by curiosity. Major changes in direction were established by two exceptional graduate students in the mid-1980s (Figure 2). One, Mark Winey, was intrigued with tRNA splicing on the basis of what we knew about the intron-containing proline tRNAs. He devised an in vitro biochemical assay for tRNA-splicing endonuclease activity, which required scaling down a partial enzyme purification to a sample that could fit into a table-top centrifuge tube. He assayed 400 mutagenized strains from the “Hartwell collection,” the same strains that yielded the cell division cycle mutants, and found splicing-defective mutants defining two genes, SEN1 and SEN2 (Winey and Culbertson 1988). The latter turned out to encode a subunit of tRNA-splicing endonuclease (Trotta et al. 1997), whereas the product of the former was not part of the enzyme.
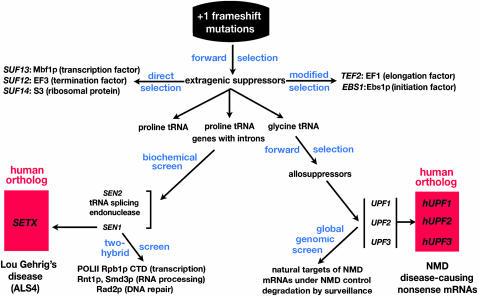
Figure 2.—
A road map from yeast to human disease. The forks in the road were made possible by curiosity-driven experiments.
I decided to pursue SEN1. It was more enigmatic because mutations in SEN1 conferred pleiotropic phenotypes (Ursic et al. 1997). It was later shown that Sen1p interacts with a wide variety of proteins, including the C-terminal domain of Rpb1p, the largest subunit of RNA polymerase II (Ursic et al. 2004). SEN1 mutations affect transcription, DNA repair, and RNA processing (Rasmussen and Culbertson 1998; Ursic et al. 2004; Steinmetz et al. 2006). Knowledge about Sen1p suddenly became important on a more global scale when it was reported that mutations in the human ortholog of yeast SEN1, called SETX (senataxin), were implicated in amyotrophic lateral sclerosis (ALS4), the syndrome made famous by the baseball player Lou Gehrig, who died of ataxia (Chen et al. 2004; Duquette et al. 2005).
Many of the human mutations reside in the N-terminal segment of the protein where we knew that domains for protein–protein interactions are located. In recent experiments, some of the human mutations were reconstructed in yeast SEN1 and shown to impair specific protein–protein interactions. Most notably, one mutation impairs the ability of Sen1p to bind to the c-terminal domain of Rpb1p (D. Ursic and M. R. Culbertson, unpublished results). The payoff is clear for years of work on yeast SEN1, performed without knowing when or how it would be useful. This fork in the road led directly to human disease. Basic knowledge about SEN1 function obtained through studies in yeast will guide future efforts to understand the specific biochemical defects in patients with SETX-related forms of ALS4.
When Mark Winey began work on SEN1 in the 1980s, someone who looked like a student, but older than average, came to my office, wanting to quit his job as a computer programmer and become a geneticist. He would pay his own way as a special student, take the required courses, and apply to graduate school. After the apprenticeship period for this student, I had to find a project for Peter Leeds, the computer programmer turned geneticist, who had a background in physics. I did not anticipate that he would make a ground-breaking discovery.
In my own postdoctoral studies, I observed that certain allelic combinations of frameshift mutations and suppressors like the one shown in Figure 1A were peculiar because suppression was temperature sensitive, a phenotype not typical of mutant tRNAs. I observed that, when a lawn of his4-38 SUF1 cells was left for a week at the restrictive temperature, colonies grew on the background of nongrowing cells. The variants harbored mutations at two unlinked loci, which I named UPF1 and UPF2 (for up-frameshift) (Culbertson et al. 1980). They were allosuppressors, requiring the presence of the suppressor tRNA to be phenotypically visible (Table 1). I saved the allosuppressors for almost 10 years in a box of “Legacies” in honor of Gerry Fink. I gave Peter the box and said that, if he could figure out what they were, he would have a thesis.
TABLE 1.
A curiosity-driven experiment leads to the discovery of the genes required for NMD in yeast
| Growth without histidine
|
||
|---|---|---|
| Genotype | 30° | 37° |
| Wild type | + | + |
| his4-38 suf1wt UPF1wt | − | − |
| his4-38 SUFIfs UPF1wt | + | − |
| his4-38 SUF1fs upf1Δ | + | + |
| his4-38 suf1wt upf1Δ | − | − |
The phenotypes of allosuppressors that were shown to be mutations in the yeast UPF genes are shown.
Peter observed that most of the his4 frameshift mutations brought a stop codon into register immediately after the site of the frameshift insertion. He found an article published from the laboratory of Francois Lacroute, showing that nonsense mutations in the URA3 gene caused reduced abundance of URA3 mRNA (Losson and Lacroute 1979). Losson and Lacroute (1979) hypothesized that nonsense mutations trigger accelerated mRNA decay. However, no genes were identified to link premature translation termination with mRNA decay. Putting the observations together, Peter argued that the normally out-of-frame stop codon following a +1 insertion might behave like the premature stop codons in URA3, triggering rapid mRNA decay and reduced abundance. The allosuppressors might be mutations in genes required for the decay pathway. This molecular explanation was consistent with the allosuppressor phenotype (Table 1). Growth at 37° resulted from the combined action of the frameshift suppressor tRNA and the increased abundance of the mRNA caused by mRNA stabilization in the absence of a functional decay pathway.
Peter cloned the UPF1 gene. At the same time, Mark Winey cloned SEN1. When they scanned the databases, no other genes that resembled UPF1 or SEN1 were found. In an act of serendipity, Peter and Mark compared UPF1 and SEN1 sequences with each other and found a long segment of close similarity due to the presence of conserved motifs in both proteins. SEN1 and UPF1 became the founding members of what are now called the superfamily I DNA/RNA helicases (Demarini et al. 1992; Koonin 1992; Leeds et al. 1992).
To test the model that UPF genes are required for mRNA decay, we collaborated with Allan Jacobson and colleagues at the University of Massachusetts Medical School because of their expertise in measuring RNA decay rates. The results from the collaboration and from Peter's thesis were published in 1991 and 1992 (Leeds et al. 1991, 1992). We demonstrated that the UPF genes were required for the phenomenon described earlier by Losson and Lacroute (1979) where premature termination of translation was thought to trigger rapid mRNA decay. The pathway that required the UPF genes came to be known as nonsense-mediated mRNA decay (NMD).
A colleague at the University of Wisconsin, Phil Anderson, and his student, Rock Pulak, also stumbled upon the NMD pathway in Caenorhabditis elegans with no premeditated intention of studying mRNA decay. They got there by a circuitous route similar to ours emanating from an interest in muscle development in worms (Hodgkin et al. 1989). I recall a joint lab meeting at which we realized that we were working on the same pathway in different organisms. They published a model in which NMD was an RNA surveillance pathway that prevented the accumulation of truncated proteins that might be deleterious (Pulak and Anderson 1993). This made sense for two reasons: (1) Errors in gene expression could produce deleterious truncated proteins and (2) diploid organisms carry potentially deleterious alleles in heterozygous condition that constitute what is called the hidden genetic load. NMD ensured the recessive nature of heterozygous nonsense alleles, thereby preventing negative effects when present in heterozygous condition (Grimson et al. 2004).
In C. elegans, seven genes required for NMD were identified (SMG-1–SMG-7) (Cali et al. 1999; Page et al. 1999; Anders et al. 2002), but in yeast only three UPF genes were found. The novel C. elegans genes included SMG-1, a PI3K kinase that phosphorylates SMG-2 (UPF1) and genes coding for components of protein phosphatase PP2A, which dephosphorylates SMG-2 (UPF1). The cycle of phosphorylation and dephosphorylation is required for NMD in C. elegans. In mammals, Lynn Maquat at the University of Rochester has worked since the early 1980s studying the phenomenon of rapid decay of nonsense mRNAs (Maquat et al. 1981). Her work was inspired by her mentor, Jeff Ross, a professor in the McArdle Labs at the University of Wisconsin, whom I recognize as the pioneer of biochemical studies on mRNA decay mechanisms. As sequences from mammalian genome projects became available, both human and mouse orthologs of the yeast UPF genes were identified (Applequist et al. 1996; Perlick et al. 1996; Lykke-Andersen et al. 2000; Serin et al. 2001). The genes coding for human kinase and phosphatase components involved in NMD were also identified by looking for orthologs of the C. elegans genes (Denning et al. 2001; Yamashita et al. 2001; Chiu et al. 2003).
The contributions from model organisms went beyond the identification of the genes. For example, to show that hUPF1 was required for NMD in an era before RNA interference methods were available, hUPF1 alleles carrying dominant-negative mutations were introduced into human cell lines to ask whether they conferred phenotypes expected for loss of function of the NMD pathway (Sun et al. 1998). The study was guided by dominant-negative mutations found earlier in yeast UPF1 by Peter Leeds (Leeds et al. 1992). The study showed that hUPF1 and yeast UPF1 not only were related by sequence comparison, but also were likely to be functionally equivalent.
In the mid-1990s, the cellular distribution and the association of Upf proteins with polyribosomes were established in yeast. It was suggested then that they might form subcomplexes that undergo sequential assembly on nonsense transcripts engaged in translation (Atkin et al. 1997). One of the yeast proteins, Upf3p, was found to shuttle between the nucleus and cytoplasm (Shirley et al. 1998, 2002), suggesting that it might be the first to assemble and might be involved in transcript selection and recruitment (Culbertson and Neeno-Eckwall 2005). When the human orthologs of the yeast UPF genes were identified, similar observations were made. Human Upf3p (which is encoded by two distinct genes), also a shuttling protein, was found to be a subunit of a protein complex that assembles at exon–exon junctions (called the exon junction complex, or the EJC) following splicing in the nucleus (Kim et al. 2001). The EJC exits the nucleus with mRNAs. Since most, if not all, exon junctions are marked with an EJC, multiple Upf3p molecules are distributed across the mRNAs at locations defined by the locations of the spliced introns. This fact caught the eye of a population geneticist at Indiana University, Michael Lynch, who published evidence suggesting a link between the evolution of introns and NMD (Lynch and Richardson 2002).
In the late 1990s, I got a call from Phil Hieter, who was then at Johns Hopkins University Medical School. He found that suppressors of a temperature-sensitive allele of CTF13, an essential gene coding for a kinetochore protein, were distributed among the three UPF genes required for NMD. We collaborated and found that the accumulation of wild-type CTF13 mRNA was dependent on the NMD pathway (Dahlseid et al. 1998). However, there were no in-frame premature stop codons in the protein-coding sequence. This was a hint that NMD was important for something in addition to RNA surveillance. There had been other earlier indications of this, but as a geneticist, I was convinced much more by the biological appearance of suppressors than by Northern blots.
Using DNA arrays, it was shown that NMD is part of the normal repertoire of gene expression in addition to its role in RNA surveillance (Lelivelt and Culbertson 1999; He et al. 2003). In a recent study, the decay rates of all RNAs in yeast were measured and compared in wild-type and Nmd− strains, using arrays (Guan et al. 2006). It was found that NMD performs a global RNA surveillance function by degrading nonfunctional transcripts, but it also controls the decay rate of >200 functional mRNAs and the expression level of several hundred more that respond indirectly to NMD. Several targeting mechanisms that involve special features of the selected mRNAs were described, such as the presence of a translated upstream open reading frame in the 5′ UTR that can trigger NMD (Gaba et al. 2005). The natural targets of NMD in yeast fall into two broadly defined functional groups: those affecting chromosome structure and behavior and those affecting cell surface dynamics, suggesting that NMD might play a role in the response of yeast to changes in the environment.
Contributions to NMD have also been forthcoming from other model organisms. In Arabidopsis thalania, it was recently shown that upf1 mutants cause phenotypes indicative of a role in plant development. Furthermore, NMD may be linked with RNA interference (Arciga-Reyes et al. 2006). In Drosophila melanogaster, UPF1, UPF2, and SMG-1 have been shown to play a role in development (Metzstein and Krasnow 2006). Studies on the function of UPF and SMG genes in processes in addition to RNA surveillance have been reported for mice and humans. Genetic rearrangements that occur during T-cell-mediated immunity generate nonsense transcripts that are degraded by NMD (Gudikote and Wilkinson 2002). Also, NMD degrades alternatively spliced transcripts that contain premature stop codons. Endogenous transposons and retroviruses are targets of NMD (Mendell et al. 2004; Pan et al. 2006).
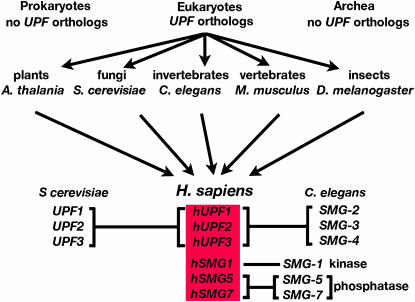
The UPF genes are ubiquitous in eukaryotes. In all likelihood, NMD is an ancient pathway that began to evolve when the ancestors of Archea and eukaryotes separated (Culbertson 2003). As our understanding of NMD in humans continues to emerge, what has been crystal clear is the reliance for new ideas on information from multiple model organisms representing diverse branches of the tree of life (Figure 3). Over the past few decades, contributions to our understanding of NMD have been made from model organisms representing plants, fungi, invertebrates, vertebrates, and insects.
Figure 3.—
A road map leading from parallel studies of NMD in representative model organisms to NMD in humans.
The way in which the NMD story has unfolded underscores the value of parallel studies in multiple model organisms to bring about a richer understanding of the biology of all organisms. The real power of genetics comes from a combination of phenotype-driven forward genetics in multiple model organisms coupled with candidate molecule-driven analyses of informative mutations in humans. One-third of all of the disease-causing alleles that have been sequenced from human patients with cancer or other genetic disorders are either frameshift or nonsense alleles (Culbertson 1999). Many of them trigger NMD. A diagnostic test for human nonsense mutations has been described (Noensie and Dietz 2001), clinical issues related to NMD discussed (Kuzmiak and Maquat 2006), and companies formed to exploit the possible therapeutic value of NMD. Who would have predicted that the study of frameshift mutations three decades ago would lead to a body of fundamental knowledge important in mechanisms of human disease?
In my research, there was no prospective road map leading directly from frameshift mutations to human disease. The road map looks clear enough in retrospect, but when broken into its component parts, my research path consisted of groups of hypothesis-driven experiments interspersed with curiosity-driven selections and screens where the goal was to get new mutants and/or find new genes. The branch points depended critically on the outcome of the curiosity-driven experiments, which frequently yielded unanticipated results. The hypothesis-driven experiments were merely the reductionist analyses of the biological gadgets uncovered by curiosity-driven, forward genetics experiments. When forward genetics is used as the primary driving force in research, it is possible to create new knowledge by navigating without a road map.
References
- Anders, K. R., A. Grimson and P. Anderson, 2002. SMG-5, required for nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 22: 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applequist, S. E., M. Selg, C. Raman and H. Jack, 1996. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 25: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciga-Reyes, L., L. Wootton, M. Kieffer and B. Davies, 2006. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47: 480–490. [DOI] [PubMed] [Google Scholar]
- Atkin, A. L., L. R. Schenkman, M. Eastham, J. N. Dahlseid, M. J. Lelivelt et al., 1997. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem. 272: 22163–22172. [DOI] [PubMed] [Google Scholar]
- Cali, B. M., S. L. Kuchma, J. Latham and P. Anderson, 1999. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 151: 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. Z., C. L. Bennett, H. M. Huynh, I. P. Blair, I. Puls et al., 2004. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am. J. Hum. Genet. 74: 1128–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, S. Y., G. Serin, O. O'Hara and L. E. Maquat, 2003. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson, M., 1999. RNA surveillance: unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 15: 74–80. [DOI] [PubMed] [Google Scholar]
- Culbertson, M. R., 2003. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13: 207–214. [DOI] [PubMed] [Google Scholar]
- Culbertson, M. R., and E. Neeno-Eckwall, 2005. Transcript selection and the recruitment of mRNA decay factors for NMD in Saccharomyces cerevisiae. RNA 11: 1333–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson, M. R., L. Charnas, M. T. Johnson and G. R. Fink, 1977. Frameshifts and frameshift suppressors in Saccharomyces cerevisiae. Genetics 86: 745–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson, M. R., K. M. Underbrink and G. R. Fink, 1980. Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics 95: 833–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson, M. R., P. Leeds, M. G. Sandbaken and P. G. Wilson, 1990. Frameshift Suppression. American Society for Microbiology, Washington, DC.
- Cummins, C. M., M. R. Culbertson and G. Knapp, 1985. Frameshift suppressor mutations outside the anticodon in yeast proline tRNAs containing an intervening sequence. Mol. Cell. Biol. 5: 1760–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlseid, J. N., J. Puziss, R. L. Shirley, A. L. Atkin, P. Hieter et al., 1998. Accumulation of mRNA coding for the Ctf13p kinetochore subunit of Saccharomyces cerevisiae depends on the same factors that promote rapid decay of nonsense mRNAs. Genetics 150: 1019–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini, D. J., M. Winey, D. Ursic, F. Webb and M. R. Culbertson, 1992. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 2154–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning, G., L. Jamieson, L. E. Maquat, E. A. Thompson and A. P. Fields, 2001. Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J. Biol. Chem. 276: 22709–22714. [DOI] [PubMed] [Google Scholar]
- Duquette, A., K. Roddier, J. McNabb-Baltar, I. Gosselin, A. St-Denis et al., 2005. Mutations in senataxin responsible for Quebec cluster of ataxia with neuropathy. Ann. Neurol. 57: 408–414. [DOI] [PubMed] [Google Scholar]
- Ford, A., Q. Guan, E. Neeno-Eckwall and M. R. Culbertson, 2006. Ebs1p: a global negative regulator of gene expression controlled by the Upf proteins in yeast. Eukaryot. Cell 5: 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba, A., A. Jacobson and M. S. Sachs, 2005. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell 20: 449–460. [DOI] [PubMed] [Google Scholar]
- Gaber, R. F., and M. R. Culbertson, 1982. The yeast frameshift suppressor gene SUF16–1 encodes an altered glycine tRNA containing the four base anti-codon 3′–CCCG–5′. Gene 19: 163–172. [DOI] [PubMed] [Google Scholar]
- Gaber, R. F., and M. R. Culbertson, 1984. Codon recognition during frameshift suppression in Saccharomyces cerevisiae. Mol. Cell. Biol. 4: 2052–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber, R. F., L. Mathison, I. Edelman and M. R. Culbertson, 1983. Frameshift suppression in Saccharomyces cerevisiae. VI. Complete genetic map of twenty-five suppressor genes. Genetics 103: 389–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson, A., S. O'Connor, C. L. Newman and P. Anderson, 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol. Cell. Biol. 24: 7483–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Q., W. Zheng, S. Tang, X. Liu, R. A. Zinkel et al., 2006. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2: 1924–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudikote, J. P., and M. F. Wilkinson, 2002. T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts. EMBO J. 21: 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F., X. Li, P. Spatrick, R. Casillo, S. Dong et al., 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12: 1439–1452. [DOI] [PubMed] [Google Scholar]
- Hendrick, J. L., P. G. Wilson, I. Edelman, M. G. Sandbaken, D. Ursic et al., 2001. Yeast frameshift suppressor mutations in the genes coding for transcription factor MBF1 and ribosomal protein S3: evidence for autoregulation of S3 synthesis. Genetics 157: 1141–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., A. Papp, R. Pulak, V. Ambros and P. Anderson, 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, V. N., N. Kataoka and G. Dreyfuss, 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293: 1832–1836. [DOI] [PubMed] [Google Scholar]
- Koonin, E. V., 1992. A new group of putative RNA helicases. Trends Biochem. Sci. 17: 495–497. [DOI] [PubMed] [Google Scholar]
- Kuzmiak, H. A., and L. E. Maquat, 2006. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol. Med. 12: 306–316. [DOI] [PubMed] [Google Scholar]
- Leeds, P., S. W. Peltz, A. Jacobson and M. R. Culbertson, 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5: 2303–2314. [DOI] [PubMed] [Google Scholar]
- Leeds, P., J. M. Wood, B. S. Lee and M. R. Culbertson, 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 2165–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelivelt, M. J., and M. R. Culbertson, 1999. Yeast Upf proteins required for RNA surveillance affect the global expression of the yeast transcriptome. Mol. Cell. Biol. 19: 6710–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson, R., and F. Lacroute, 1979. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc. Natl. Acad. Sci. USA 76: 5134–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J., M. D. Shu and J. A. Steitz, 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and A. O. Richardson, 2002. The evolution of spliceosomal introns. Curr. Opin. Genet. Dev. 12: 701–710. [DOI] [PubMed] [Google Scholar]
- Maquat, L. E., A. J. Kinniburgh, E. A. Rachmilewitz and J. Ross, 1981. Unstable beta-globin mRNA in mRNA-deficient b thalassemia. Cell 27: 543–553. [DOI] [PubMed] [Google Scholar]
- Mathison, L. M., and M. R. Culbertson, 1985. Suppressible and non-suppressible +1 G:C base-pair insertions induced by ICR–170 at the his4 locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 5: 2247–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell, J. T., N. A. Sharifi, J. L. Meyers, F. Martinez-Murillo and H. C. Dietz, 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Mendenhall, M. D., P. Leeds, H. Fen, L. Mathison, M. Zwick et al., 1987. Frameshift suppressor mutations affecting the major glycine transfer RNAs of Sacchacromyces cerevisiae. J. Mol. Biol. 194: 41–58. [DOI] [PubMed] [Google Scholar]
- Metzstein, M. M., and M. A. Krasnow, 2006. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2: 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noensie, E. N., and H. C. Dietz, 2001. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat. Biotechnol. 19: 434–439. [DOI] [PubMed] [Google Scholar]
- Page, M. F., B. Carr, K. R. Anders, A. Grimson and P. Anderson, 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Ceanorhabditis elegans and related to Upf1p in yeast. Mol. Cell. Biol. 19: 5943–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q., A. L. Saltzman, Y. K. Kim, C. Misquitta, O. Shai et al., 2006. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 20: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlick, H. A., S. M. Medghalchi, F. A. Spencer, R. J. Kenndzior and H. C. Dietz, 1996. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc. Natl. Acad. Sci. USA 93: 10928–10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak, R., and P. Anderson, 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7: 1885–1897. [DOI] [PubMed] [Google Scholar]
- Rasmussen, T. P., and M. R. Culbertson, 1998. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of snoRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 6885–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandbaken, M. G., and M. R. Culbertson, 1988. Mutations in elongation factor EF1–α affect the frequency of amino acid misincorporation and frameshifting in Saccharomyces cerevisiae. Genetics 120: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin, G., A. Gersappe, J. D. Black, R. Aronoff and L. E. Maquat, 2001. Identification and characterization of human orthologues to Saccharyomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol. 21: 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley, R. L., L. R. Schenkman, M. J. Lelivelt, J. N. Dahlseid and M. R. Culbertson, 1998. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell. Sci. 14: 3129–3143. [DOI] [PubMed] [Google Scholar]
- Shirley, R. L., A. S. Ford, R. Richards, M. Albertini and M. R. Culbertson, 2002. Nuclear import of Upf3p is mediated by importin-a/b and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast. Genetics 161: 1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz, E. J., C. L. Warren, J. N. Kuehner, B. Panbehi, A. Z. Ansari et al., 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell 24: 735–746. [DOI] [PubMed] [Google Scholar]
- Sun, X., H. A. Perlick, H. C. Dietz and L. E. Maquat, 1998. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci USA 95: 10009–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta, C. R., F. Miao, E. A. Arn, S. W. Stevens, C. K. Ho et al., 1997. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89: 849–858. [DOI] [PubMed] [Google Scholar]
- Ursic, D., K. L. Himmel, K. A. Gurley, F. Webb and M. R. Culbertson, 1997. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 25: 4778–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic, D., K. Chinchilla, J. S. Finkel and M. R. Culbertson, 2004. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 32: 2441–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, P., and M. R. Culbertson, 1988. SUF12 suppressor protein of yeast: a fusion protein related to the EF1 family of elongation factors. J. Mol. Biol. 199: 559–573. [DOI] [PubMed] [Google Scholar]
- Winey, M., and M. R. Culbertson, 1988. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics 118: 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., M. D. Mendenhall, C. M. Cummins, M. R. Culbertson and G. Knapp, 1986. Splicing of a yeast proline tRNA containing a novel suppressor mutation in the anticodon stem. J. Mol. Biol. 192: 49–63. [DOI] [PubMed] [Google Scholar]
- Yamashita, A., T. Ohnishi, I. Kashima, Y. Taya and S. Ohno, 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15: 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yourno, J., and S. Health, 1969. Nature of the hisD3018 frameshift mutation in Salmonella typhimurium. J. Bacteriol. 100: 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]