Abstract
Here we show that in the nematode Caenorhabditis elegans mutational inactivation of two autophagy genes unc-51/atg1 and bec-1/atg6/beclin1 results in small body size without affecting cell number. Furthermore, loss-of-function mutations in unc-51 and bec-1 suppress the giant phenotype of mutant animals with aberrant insulin-like growth factor-1 (insulin/IGF-1) or transforming growth factor-β (TGF-β) signaling. This function for unc-51 and bec-1 in cell size control and their interaction with these two growth modulatory pathways may represent a link between the hormonal and nutritional regulation of cell growth.
IN multicellular organisms, the regulation of cell size is intimately linked to nutrient and growth factor availability and requires a well-controlled balance between macromolecule synthesis and degradation (Klionsky and Emr 2001; Saucedo and Edgar 2002; Oldham and Hafen 2003; Danielpour and Song 2005; Leevers and Mcneill 2005). Cells divide only after they reach a critical size. Thus, cell growth is a prerequisite of cell proliferation and may be dysregulated in human malignancies. The insulin-like growth factor-1 (insulin/IGF-1) and transforming growth factor-β (TGF-β) signaling cascades are known as major regulatory systems for cell growth, proliferation, and differentiation (Saltiel and Kahn 2001; Derynck and Zhang 2003; Oldham and Hafen 2003). However, little is known about cellular pathways that mediate these processes.
In Caenorhabditis elegans, both insulin/IGF-1 and TGF-β signaling pathways affect body length by controlling cell size (Krishna et al. 1999; Suzuki et al. 1999; Morita et al. 1999, 2002; Mcculloch and Gems 2003). For example, loss-of-function mutations in the gene daf-2, which encodes the nematode IGF-1 receptor, extend body length, compared to the wild type (Mcculloch and Gems 2003). The type I TGF-β receptor SMA-6 also influences body size in nematodes (Krishna et al. 1999). SMA-6 is activated by the growth factor DBL-1 and represses the expression of the PR-related protein LON-1, which is a novel negative regulator of cell growth (Suzuki et al. 1999; Morita et al. 1999, 2002). Mutations that eliminate the activity of LON-1 increase body length by 1.5-fold, whereas animals with elevated DBL-1 activity are also longer than the wild type.
The insulin/IGF-1 and TGF-β hormonal systems also control reproductive growth in this organism (Riddle and Albert 1997). Mutant nematodes with decreased DAF-2/IGF-1 receptor activity enter into a state of developmental diapause called dauer, which is an arrested larval form specialized to survive unfavorable conditions. In addition, mutations that inactivate the type I and type II TGF-β receptors, DAF-1 and DAF-4, respectively, result in constitutive dauer development independently of environmental cues (Estevez et al. 1993; Gunther et al. 2000). It was shown that dauer development in insulin/IGF-1- and TGF-β-signaling mutant nematodes requires the function of autophagy genes, and that normal dauer morphogenesis is associated with increased autophagy (Meléndez et al. 2003). Autophagy is a highly regulated cellular pathway used by eukaryotic cells to degrade parts of their contents during development and to survive nutrient deprivation (Klionsky and Emr 2001). Autophagic degradation of cytosolic materials is a major route for turnover of cellular macromolecules and organelles, in particular proteins and mitochondria. In this study, we investigate whether unc-51 and bec-1, which are mutationally characterized C. elegans autophagy genes, are required for maintaining normal cell size.
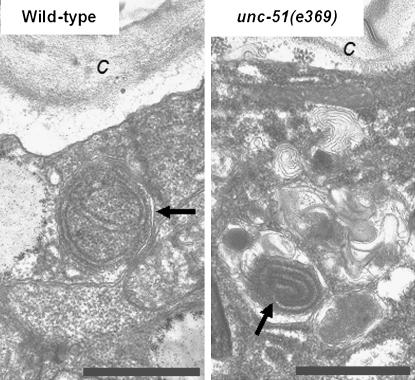
The wild-type C. elegans strains display a characteristic body length of 1.2 mm (Brenner 1974). We examined unc-51 loss-of-function mutant nematodes at a well-defined developmental stage (see Table 1) and found that they show a marked shortening in mean body size. For example, body length at the young adult stage was 0.89 ± 0.04 mm in unc-51(e369) mutants vs. 1.25 ± 0.05 mm in wild-type animals (unpaired t-test; N = 250, P < 0.0001) (Figure 1 and Table 1). unc-51 encodes a serine/threonine kinase similar to the yeast autophagy protein Atg1 (Ogura et al. 1994), which is a key regulator of autophagosome formation (Klionsky 2005). Moreover, the autophagic process appeared to be defective in unc-51 mutants. According to our electron microscopic observations, autophagic vacuoles were present almost exclusively in lateral hypodermal cells, and their membrane was excessively whorled (Figure 2). This may be indicative of defective autophagic vacuole formation in these animals. We also monitored the effects of mutations in another C. elegans autophagy gene, bec-1, on body length. bec-1 is an essential gene that is the nematode ortholog of the human tumor suppressor gene Beclin1 and yeast atg6 (Meléndez et al. 2003; Takács-Vellai et al. 2005). Because bec-1 loss-of-function mutations arrest development at different stages (Takács-Vellai et al. 2005), we rescued the lethality of bec-1 mutants by an unstable (extrachromosomal) transgene array containing wild-type copies of bec-1 (bec-1(−); Ex[pbec-1∷BEC-1∷GFP + rol-6(su1006)]). On average, randomly selected adults of bec-1(−); Ex[bec-1(+)] genotype were significantly shorter than wild-type nematodes (Figure 1 and Table 1). The reduced body length of certain bec-1(−); Ex[bec-1(+)] adults may result from incomplete rescue of bec-1 in somatic cells. The possibility that this is due to an unlinked mutation that is not rescued by the array is unlikely because both bec-1 alleles ok691 and ok700 behaved similarly. In good agreement with the body length data of bec-1(−); Ex[bec-1(+)] and unc-51 mutant adults, their body volume was also markedly reduced as compared with the wild type (Table 2). Together, these results suggest that bec-1(−); Ex[bec-1(+)] and unc-51 mutant animals display a characteristic small body size (Sma) phenotype.
TABLE 1.
Mutational inactivation of unc-51 or bec-1 reduces body length in C. elegans
| Genotype | Body length (mm) | N | Phenotype |
|---|---|---|---|
| Wild type | 1.28 ± 0.07 | 241 | Wild-type |
| unc-51(e369) | 0.87 ± 0.09 | 266 | Sma |
| unc-51(e1189) | 0.9 ± 0.08 | 304 | Sma |
| bec-1(ok691); Ex[bec-1(+)] | 0.99 ± 0.08 | 157 | Sma |
| bec-1(ok700); Ex[bec-1(+)] | 1.04 ± 0.14 | 154 | Sma |
| lon-1(e185) [TGF-β]a | 1.72 ± 0.08 | 228 | Lon |
| lon-2(e678) [TGF-β]a | 1.51 ± 0.09 | 243 | Lon |
| dbl-1(+++) [TGF-β]a | 1.38 ± 0.09 | 252 | Lon |
| daf-2(e1370) [insulin/IGF-1]a | 1.36 ± 0.08 | 185 | Lon |
| unc-51(e1189); lon-1(e185) | 1.12 ± 0.1 | 288 | Sma |
| unc-51(e1189); dbl-1(+++) | 1.05 ± 0.06 | 279 | Sma |
| unc-51(e1189); daf-2(e1370) | 0.98 ± 0.06 | 250 | Sma |
| unc-51(e369); lon-1(e185) | 1.07 ± 0.06 | 225 | Sma |
| unc-51(e369); lon-2(e678) | 1.16 ± 0.11 | 253 | Non-Lon |
| unc-51(e369); dbl-1(+++) | 1.05 ± 0.09 | 265 | Sma |
| bec-1(ok691); Ex[bec-1(+)]; lon-2(e678) | 1.1 ± 0.22 | 47 | Sma |
| bec-1(ok691); Ex[bec-1(+)]; dbl-1(+++) | 1.04 ± 0.17 | 58 | Sma |
| bec-1(ok700); Ex[bec-1(+)]; lon-2(e678) | 1.08 ± 0.2 | 47 | Sma |
| bec-1(ok700); Ex[bec-1(+)]; dbl-1(+++) | 1.12 ± 0.13 | 42 | Sma |
| bec-1(ok700); Ex[bec-1(+)]; lon-1(e185) | 1.08 ± 0.1 | 65 | Sma |
| bec-1(ok700); Ex[bec-1(+)]; daf-2(e1370) | 1.02 ± 0.09 | 89 | Sma |
Loss-of-functions mutations in unc-51 or bec-1 reduce body length and suppress, at least partially, the Lon phenotype of insulin/IGF-1 and TGF-β mutants. dbl-1(+++) indicates the DBL-1-overexpressing strain. Body-length measurements were performed as follows. Well-fed, young adult nematodes, which on the day when they become gravid, i.e., start to produce embryos, were anesthetized with 10 mm sodium-azide solution for 5 min. This treatment allows for coiled unc-51 mutants to be relaxed. Body length of anesthetized worms was measured with a dissecting microscope and micrometer. Note that unc-51 mutant animals are defective in movement, therefore, only those individuals were selected for body-length measurement that had remained in the bacterium layer and thus appeared well fed. This criterion may help to avoid the potential effect of restricted food intake on cell growth. Data are given as mean ± standard deviation. For single mutant animals, P < 0.0001 (unpaired t-test, body length of single mutants were compared with that of wild type). For double mutant genotypes, P < 0.0001 (unpaired t-test, body length of double mutants were compared with that of the corresponding “Lon” single mutants). Sma, small body size; Lon, long body length.
daf-2 encodes the IGF-1 receptor, whereas dbl-1, lon-1, and lon-2 encode components of the TGF-β signaling axis.
Figure 1.—
Body size of mutant nematodes with reduced autophagy. The TGF-β genetic pathway component lon-1 encodes a negative regulator of cell size, whereas unc-51 and bec-1 are two autophagy genes. Bars, 0.1 mm.
Figure 2.—
The autophagic process appears to be defective in unc-51 mutant animals. (Left) Autophagic vacuole with normal double isolation membrane (arrow) in the hypodermis from a wild-type animal. (Right) Abnormal autophagic vacuole (arrow) with strongly myelinated isolation membrane is embedded in a region of the cytoplasm abundant in membrane whorls in a hypodermal seam cell from unc51(e369) mutant. c, cuticule; bars, 1 μm. For fixation and embedding of transmission electron microscopic samples, the nematodes were treated individually. They were cut open under a dissecting microscope in a drop of fixative composed of 0.2% glutaraldehyde and 3.2% formaldehyde in 0.15 m cacodylate buffer. After an overnight fixation at 4°, the fixative was changed to washing buffer (0.1 m cacodylate buffer), and the samples were embedded in agar, post-fixed with 0.5% cacodylate-buffered OsO4, stained with 2% uranyl acetate, dehydrated in ethanol and propylene oxide, and embedded in Durcupan (Fluka Chemical, Buchs, Switzerland). Thereafter the samples were cut along the longitudinal body axis with Reichert-Jung Ultracut-E type ultramicrotome, stained with lead citrate, and examined in a JEM100CX II electron microscope.
TABLE 2.
unc-51 and bec-1 mutant adults have decreased body volume
| Genotype | Body length (mm) | Diameter (mm) | Volume (mm3) | % body volume | N |
|---|---|---|---|---|---|
| Wild type | 1.2 ± 0.02 | 0.083 ± 0.0015 | 0.00619 ± 0.0002 | 100 | 8 |
| unc-51(e1189) | 0.86 ± 0.07 | 0.076 ± 0.0013 | 0.0036 ± 0.00019 | 58.17 | 11 |
| bec-1(ok700); Ex[bec-1(+)] | 1.02 ± 0.15 | 0.083 ± 0.001 | 0.0041 ± 0.0003 | 66.88 | 11 |
Body diameter and volumes were determined as described (Kammenga et al. 2007). For mutant body volumes, P < 0.001 (unpaired t-test). Data are given as mean ± standard deviation.
unc-51 and bec-1 mutant animals also exhibited delay in development (data not shown), but had wild-type cell numbers as revealed by scoring different cell types expressing reporters labeled with green fluorescent protein. For example, we used the tra-1∷gfp and prk-1∷gfp markers to visualize intestinal cells; mec-7∷gfp, which is expressed in certain Q-cell descendants; and cdh-3∷gfp and ajm-1∷gfp to label lateral seam cells. We found twenty GFP-positive intestinal cells in both wild type as well as in unc-51(e369) and bec-1(ok691); Ex[bec-1(+)] mutant animals (Figure 3 and results not shown). Furthermore, mutations in unc-51 and bec-1 also did not affect the number of Q-cell progeny and hypodermal seam cells. We next monitored the mean longitudinal diameter of gut cells by Nomarski microscopic analysis of strains expressing prk-1∷gfp (Figure 3D). For example, mean gut cell diameter was only 41.9 ± 1.9 μm in unc-51(e369) mutants (N = 35 animals), as compared with 56.1 ± 3.2 μm in wild-type animals (N = 30). We also measured the volume of the intestine and the length and area of seam cells, as described previously (Wang et al. 2002; Hirose et al. 2003). Our data shown in Tables 3 and 4 indicate that the reduced body size of unc-51 mutants was due to a decrease in cell size (mutants deficient for BEC-1 were not examined for this trait). Thus, autophagy genes, or at least some of them, are required for normal cell growth.
Figure 3.—
Visualization of intestinal cells in adult nematodes. (A) TRA-1∷GFP is expressed in the nucleus of gut cells from wild-type adult. (B) TRA-1∷GFP expression in unc-51(e369) mutant adult. (C) Expression of PRK-1∷GFP in the intestine of unc-51(e369) animal. (D) Expression of PRK-1∷GFP in individual intestinal cells of wild-type (top) and unc-51(e369) mutant (bottom) animals. Brackets indicate the longitudinal border of the first four individual intestinal cells. Both images were made with the same magnification.
TABLE 3.
Cell size measurements in unc-51 mutant animals
| Genotype | Seam cell length (μm) | Seam cell area (μm2) | N |
|---|---|---|---|
| ajm-1∷gfp | 33.2 ± 5.1 | 1.25 ± 0.12 | 55 |
| unc-51(e369); ajm-1∷gfp | 27.4 ± 4.0 | 0.95 ± 0.2 | 39 |
The ajm-1∷gfp marker localizes to the adherens junctions surrounding the seam cells (Mohler et al. 1998). Seam cell measurements were carried out at the L3 stage. P < 0.001 (unpaired t-test). Data are given as mean ± standard deviation.
TABLE 4.
Organ volume measurements in unc-51 mutant animals
| Genotype | Volume of the intestine (nl) | % relative volume | N |
|---|---|---|---|
| prk-1∷gfp | 0.95 ± 0.27 | 100 | 20 |
| unc-51(e369), prk-1∷gfp | 0.76 ± 0.33 | 78 | 20 |
For intestinal volume measurements, animals were selected at young adulthood. P < 0.001 (unpaired t-test). Data are given as mean ± standard deviation.
Next, we evaluated the effects of inhibiting the function of UNC-51 and BEC-1 on the long body size (Lon) phenotype of mutant strains defective in insulin/IGF-1 or TGF-β signaling. We found that daf-2(e1370), lon-1(e185), and lon-2(e678) mutants as well as DBL-1-overexpressing nematodes, which as single mutant animals are each long (Krishna et al. 1999; Suzuki et al. 1999; Morita et al. 1999, 2002; Mcculloch and Gems 2003), displayed small or wild-type body size when they also carried a loss-of-function mutation in unc-51 or bec-1 (Table 1). Note that double mutants carrying the daf-2 mutation e1370 were grown at 15° until they reached the L2 larval stage and then transferred at 25° to develop further. This temperature shift prevented these worms from arrest development as abnormal dauer larvae (Meléndez et al. 2003). Suppression of the Lon phenotype in insulin/IGF-1 and TGF-β pathway mutant animals by mutations in unc-51 and bec-1 suggests that autophagy genes interact with and, possibly, act downstream of these hormonal systems, as well as suggesting that autophagy may be implicated in cell growth control. In other words, unc-51 and bec-1 are epistatic to daf-2, dbl-1, and lon-1 to influence body size. However, in certain double mutant combinations unc-51 and bec-1 mutant alleles did not completely suppress body lengths of lon-1, lon-2, and daf-2 mutants, as well as DBL-1-overexpressing animals, i.e., the size of these double mutants was intermediate between the corresponding single mutants (Table 1). Alternatively, this implies that parallel pathways might exist in which unc-51 and bec-1 control body size independently from insulin/IGF-1 and/or TGF-β signaling. If autophagy genes mediate the effects of both signal transduction axes in the control of cell growth, then unc-51 and bec-1 should function both in parallel and downstream of either of these growth modulatory pathways, explaining intermediate body sizes observed (see below).
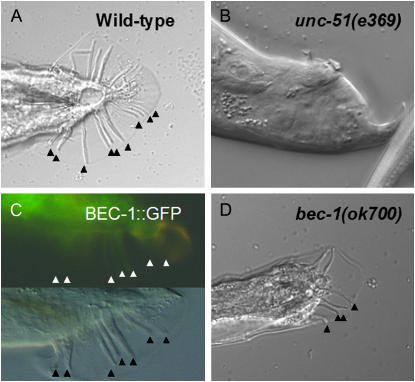
In C. elegans, several components of the TGF-β signaling pathway are involved in male tail ray pattern formation (Suzuki et al. 1999; Morita et al. 1999, 2002). To demonstrate whether UNC-51 and BEC-1 are also interacting with TGF-β signaling to affect male tail development, we assayed patterning of male tail structures in unc-51(−) mutants and bec-1(−); Ex[bec-1(+)] animals. unc-51 mutant males showed severe tail abnormalities, including the complete loss of the sensory rays and fan (Figure 4B). Moreover, BEC-1 was expressed in all structures of the adult male tail, and in bec-1 mosaic males the neighboring rays are often fused with each other (Figure 4). Together, our results indicate that unc-51 and bec-1 influence male tail patterning, possibly by interacting with the TGF-β system. To determine where these genes may act in the TGF pathway, we analyzed the expression of an integrated plgg-1∷GFP∷LGG-1 reporter (Tóth et al. 2007), which is supposed to label autophagosomal structures in hypodermal seam cells (Meléndez et al. 2003). Wild-type animals and sma-6(e1482) mutants carrying a transgene that expressed GFP∷LGG-1 had mainly a diffuse cytoplasmic staining pattern (Figure 5), whereas the number of GFP∷LGG-1-positive foci increased markedly in the lon-1(e185) background. This suggests that autophagy genes, at least some of them, may act downstream of and are inhibited by LON-1 in body size regulation.
Figure 4.—
UNC-51 and BEC-1 are required for male tail development. (A) Tail structure of wild-type adult male. (B) Tail phenotype of an unc-51(e369) male. In this animal the rays are completely missing. (C) pbec-1∷BEC-1∷GFP is expressed in wild-type male. Corresponding fluorescence (top) and Nomarski (bottom) images. (D) Male tail structure in a bec-1(ok691); Ex[pbec-1∷BEC-1∷GFP] adult. Arrowheads indicate the rays on one side. bec-1 mutants with affected tail always lost GFP expression in the tail.
Figure 5.—
Intracellular accumulation of LGG-1 in hypodermal seam cells is affected by TGF-β signaling. (A) Expression of GFP∷LGG-1 in the seam cells of wild-type animal. GFP foci are supposed to label autophagosomal structures. (B) Expression of the GFP∷LGG-1 reporter in the seam cells of sma-6(e1482) mutant L3 larva. (C) GFP∷LGG-1 accumulates in punctate areas in the seam cells of lon-1(e185) mutant L3 larva. (D) Quantification of GFP∷LGG-1-positive foci in individual seam cells of wild-type, sma-6(e1482), and lon-1(e185) mutant animals. The number of punctate area per seam cell is shown.
In summary, our data indicate that the insulin/IGF-1 and TGF-β signaling pathways may interact with the UNC-51 and BEC-1 autophagy genes to control cell size in C. elegans. The autophagy protein Atg5 has also been shown to influence cell size in mice; in Atg5-deficient animals cell size is not reduced in response to food withdrawal (Hosokawa et al. 2006). Furthermore, autophagy genes are also implicated in TOR (target of rapamycin) kinase-mediated cell growth control in Drosophila (Scott et al. 2004). Together, these data point to autophagy, an evolutionarily conserved cellular degradative pathway, as a possible mechanism to take part in size regulation of cells and organs in divergent animal phyla (Figure 6). This is in good accordance with the pivotal role of autophagy in regulated turnover of subcellular constituents (cytoplasmic macromolecules and organelles) (Klionsky and Emr 2001).
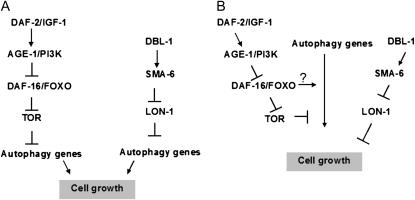
Figure 6.—
Two alternative models for how autophagy affects cell growth in C. elegans. (A) If autophagy genes mediate the effect of both insulin/IGF-1 and TGF-β signaling to control cell growth, i.e., insulin/IGF-1 and TGF-β signaling converge on autophagy to regulate cell size, unc-51 and bec-1 should function downstream of these two hormonal systems. (B) Because mutations in bec-1 and unc-51 do not completely suppress the Lon phenotype of daf-2 and certain TGF-β mutant strains, it is also possible that autophagy genes act in parallel to insulin/IGF-1 and TGF-β signaling in cell growth control. The question marks indicate whether the effect of the insulin/IGF-1 signaling pathway on body size is solely the result of changes in autophagy. Arrows indicate positive regulatory interactions, bars represent inhibitions. DAF-2, IGF-1 receptor; AGE-1, phosphatidylinositol-3-OH kinase; DAF-16, FOXO forkhead transcription factor; SMA-6, type I TGF-β receptor; DBL-1, TGF-β ligand for SMA-6; LON-1, cytoplasmic transducer of TGF-β signaling.
Further studies are needed to determine whether other C. elegans autophagy genes are also involved in cell growth control. However, at present only a very limited number of autophagy genes are available as mutant alleles and, what we did not show here, their RNAi-mediated silencing is often ineffective or leads to weak reactions (see also Kovács et al. 2004).
The characterization of the autophagic process itself in C. elegans is still in a very preliminary stage (Meléndez et al. 2003; Kovács et al. 2004). Feeding defective mutant worms also have a shorter body length and are proposed to have increased autophagy (Morck and Pilon 2006). This suggests that, similar to its possible dual role in neuronal cell survival and loss (Takács-Vellai et al. 2006), both deregulation and hyperactivation of autophagy genes cause reduction in cell size. Thus, fine tuning of autophagy gene activity is critical for maintaining normal cell size. The regulation of cell growth and proliferation are tightly integrated. Our study suggests that identifying autophagy genes as key modulators of cell size will be essential for understanding how uncontrolled cell growth leads to cancer.
Acknowledgments
We are grateful to Theresa Stiernagle and the C. elegans Genetic Center, founded by the National Institutes of Health, for providing strains. We thank Sara Simon and Emese Karácsony for excellent technical help and two anonymous referees for their valuable comments on the manuscript. This work was supported by grants from the Ministry of Health (no. 167/2006) and the National Office for Research and Technology (NKFP no. 1A/007/2004) to T. V. and the Hungarian Scientific Research Found (OTKA no. T047241) to A.L.K. T.V. is a grantee of the János Bolyai scholarship.
References
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielpour, D., and K. Song, 2005. Cross-talk between IGF-1 and TGF-beta signaling pathways. Cytokine Growth Factor Rev. 17: 59–74. [DOI] [PubMed] [Google Scholar]
- Derynck, R., and Y. E. Zhang, 2003. Smad-dependent and Smad-independent pathways in TGF-beta family signaling. Nature 425: 577–584. [DOI] [PubMed] [Google Scholar]
- Estevez, M., L. Attisano, J. L. Wrana, P. S. Albert, J. Massague et al., 1993. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature 365: 644–649. [DOI] [PubMed] [Google Scholar]
- Gunther, C. V., L. L. Georgi and D. L. Riddle, 2000. A Caenorhabditis elegans type I TGF-beta receptor can function in the absence of type II kinase to promote larval development. Development 127: 3337–3347. [DOI] [PubMed] [Google Scholar]
- Hirose, T., Y. Nakano, Y. Nagamatsu, T. Misumi, T. Ohta et al., 2003. Cyclic GMP-dependent protein kinase EGL-4 controls body size and lifespan in C. elegans. Development 130: 1089–1099. [DOI] [PubMed] [Google Scholar]
- Hosokawa, N., Y. Hara and N. Mizushima, 2006. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 580: 2623–2629. [DOI] [PubMed] [Google Scholar]
- Kammenga, J. E., A. Doroszuk, J. A. Riksen, E. Hazendonk, L. Spiridon et al., 2007. A Caenorhabditis elegans wild-type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet. 3: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., and S. D. Emr, 2001. Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., 2005. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács, A. L., T. Vellai and F. Müller, 2004. Autophagy in Caenorhabditis elegans, pp. 219–225 in Autophagy, edited by D. J. Klionsky. Landes Biosciences, Georgetown, TX.
- Krishna, S., L. L. Maduzia and R. W. Padgett, 1999. Specificity of TGF-beta signaling is conferred by distinct type I receptor and their associated SMAD proteins in Caenorhabditis elegans. Development 126: 251–260. [DOI] [PubMed] [Google Scholar]
- Leevers, S. J., and H. McNeill, 2005. Controlling the size of organs and organisms. Curr. Opin. Cell Biol. 17: 604–609. [DOI] [PubMed] [Google Scholar]
- McCulloch, D., and D. Gems, 2003. Body size, insulin/IGF-1 signaling and aging in the nematode Caenorhabditis elegans. Exp. Gerontol. 38: 129–136. [DOI] [PubMed] [Google Scholar]
- Meléndez, A., Z. Talloczy, M. Seaman, E. L. Eskelinen, D. H. Hall et al., 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301: 1387–1391. [DOI] [PubMed] [Google Scholar]
- Mohler, W. A., J. S. Simske, E. M. Williams-Masson, J. D. Hardin and J. G. White, 1998. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr. Biol. 8: 1087–1090. [DOI] [PubMed] [Google Scholar]
- Morck, C., and M. Pilon, 2006. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev. Biol. 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, K., K. L. Chow and N. Ueno, 1999. Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-beta family. Development 126: 1337–1347. [DOI] [PubMed] [Google Scholar]
- Morita, K., A. J. Flemming, Y. Sugihara, M. Mochii, Y. Suzuki et al., 2002. A Caenorhabditis elegans TGF-beta, DBL-1, controls the expression of LON-1, a PR-related protein, that regulates polyploidization and body length. EMBO J. 21: 1063–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura, K., C. Wicky, L. Magnenat, H. Tobler, F. Müller et al., 1994. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 8: 2389–2400. [DOI] [PubMed] [Google Scholar]
- Oldham, S., and E. Hafen, 2003. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13: 79–85. [DOI] [PubMed] [Google Scholar]
- Riddle, D. L., and P. S. Albert, 1997. Genetic and environmental regulation of dauer larva development, pp. 739–768 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. Priess. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Saltiel, A. R., and R. C. Kahn, 2001. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414: 799–806. [DOI] [PubMed] [Google Scholar]
- Saucedo, L. J., and B. A. Edgar, 2002. Why size matters: altering cell size. Curr. Opin. Genet. Dev. 12: 565–571. [DOI] [PubMed] [Google Scholar]
- Scott, R. C., O. Schuldiner and T. P. Neufeld, 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7: 167–178. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., M. D. Yandell, P. J. Roy, S. Krishna, C. Savage-Dunn et al., 1999. A BMP-homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126: 241–250. [DOI] [PubMed] [Google Scholar]
- Takács-Vellai, K., T. Vellai, A. Puoti, M. Passannante, A. Streit et al., 2005. Inactivation of the autophagy gene bec-1 trigger apoptotic cell death in C. elegans. Curr. Biol. 15: 1513–1517. [DOI] [PubMed] [Google Scholar]
- Takács-Vellai, K., A. Bayci and T. Vellai, 2006. Autophagy in neuronal cell loss: a road to death. BioEssays 28: 1126–1131. [DOI] [PubMed] [Google Scholar]
- Tóth, M. L., P. Simon, A. L. Kovács and T. Vellai, 2007. Influence of autophagy genes on ion-channel dependent neuronal degeneration in Caenorhabditis elegans. J. Cell Sci. 120: 1134–1141. [DOI] [PubMed] [Google Scholar]
- Wang, J., R. Tokarz and C. Savage-Dunn, 2002. The expression of TGFb signal transducers in the hypodermis regulates body size in C. elegans. Development 129: 4989–4998. [DOI] [PubMed] [Google Scholar]