Abstract
The conserved multi-subunit Ccr4-Not complex regulates gene expression in diverse ways. In this work, we characterize the suppression of temperature sensitivity associated with a mutation in the gene encoding the scaffold subunit of the Ccr4-Not complex, NOT1, by the deletion of SPT3. We determine that the deletion of SPT3, but not the deletion of genes encoding other subunits of the SAGA complex, globally suppresses transcriptional defects of not1-2. We find that transcriptional activation in not1-2 is associated with increased binding of TFIID and SAGA at promoters of upregulated genes, and this is suppressed by the deletion of SPT3. Interestingly, Spt3p-dependent activation of transcription occurs in not1-2 even if the SAGA complex is disrupted by the deletion of SPT7 that encodes a subunit of SAGA required for its integrity. Consistent with a SAGA-independent function of Spt3p, the deletion of SPT3 displays synthetic phenotypes when combined with a deletion of SPT7. Taken together, our results provide a new view of the Spt3 protein by identifying a SAGA-independent function of this protein that is functionally linked to the Ccr4-Not complex.
THE Ccr4-Not complex is a multi-protein complex consisting of at least nine subunits (Not1p–Not5p, Caf1p, Caf40p, Caf130p, and Ccr4p) conserved from yeast to human and required for the appropriate control of gene expression (for reviews see Collart 2003; Denis and Chen 2003; Collart and Timmers 2004). It exists in several forms >1 MDa and carries at least two enzymes: the Ccr4p-Caf1p deadenylase (Tucker et al. 2001) and the Not4p E3 ligase (Albert et al. 2002). The specificity of these enzymes has not yet been clearly established. Ccr4p contributes to the degradation of transcripts encoding ribosomal proteins and ribosome assembly factors (Grigull et al. 2004). Caf1p, endowed with nuclease activity in vitro (Daugeron et al. 2001), does not seem to contribute to deadenylation in vivo (Tucker et al. 2002; Viswanathan et al. 2004), but its recruitment to an mRNA is sufficient to decrease the half-life of this transcript (Finoux and Séraphin 2006). For Not4p, the only substrate described to date is the nascent polypeptide-associated (EGD/NAC) complex (Panasenko et al. 2006).
The function of the Ccr4-Not complex, however, extends beyond its known enzymatic activities. Indeed, many studies in the yeast Saccharomyces cerevisiae have demonstrated that the complex is involved in the positive and negative regulation of transcription in diverse ways (for reviews see (Collart 2003; Denis and Chen 2003; Collart and Timmers 2004). For instance, integrity of the Ccr4-Not complex is important for the appropriate distribution of TFIID across promoters (Lenssen et al. 2005) and Not5p associates with promoters in a Taf1p-dependent manner (Deluen et al. 2002). Other studies showed that transcriptional activation by Gcn4p leads to the recruitment of several subunits of the Ccr4-Not complex to the ARG1 promoter (Swanson et al. 2003) or that appropriate recruitment of the transcription machinery to RNR genes in response to DNA damage requires the Ccr4-Not complex (Mulder et al. 2005). The Ccr4-Not complex also controls the post-translational status of the Msn2p stress transcription factor via the Glc7 type I protein phosphatase, thereby inhibiting its transcriptional activity (Lenssen et al. 2005). Other genetic studies connect the Ccr4-Not complex to transcription, since, for instance, mutations in CCR4-NOT genes lead to the functional expression of his4-912δ, similar to mutations in SPT15 encoding the TATA-binding protein TBP (Badarinarayana et al. 2000).
One interesting genetic interaction linking the Ccr4-Not complex to transcription is the suppression of a temperature-sensitive mutation in the gene encoding the scaffold subunit of the Ccr4-Not complex, NOT1, by the deletion of SPT3, encoding a subunit of the SAGA complex (Collart 1996). SAGA is a conserved multi-protein complex that is required for the normal transcription of many genes (for review see Timmers and Tora 2005). It is modular in structure and a functional organization (Sterner et al. 1999; Wu et al. 2004). Genetic analyses of the nonessential subunits show that they fall into several classes by mutant phenotypes. One class consists of the Spt7p, Spt20p, and Ada1p subunits that are essential for the integrity of the complex, and deletion of these subunits can have severe growth defects (Sterner et al. 1999). A second class consists of Spt3p and Spt8p, which have very similar profiles of gene expression and synthetic genetic interactions (Ingvarsdotir et al. 2005). Their functions have generally been linked to interactions of SAGA with TBP. A third class consists of the Gcn5p, Ada2p, and Ada3p, which contribute to the histone acetyl-transferase (HAT) activity of the complex. Indeed, Gcn5p is the HAT enzyme and acetylates histones H2B and H3, whereas Ada2p and Ada3p are required for Gcn5p to acetylate nucleosomal histones (Grant et al. 1997; Sterner et al. 1999; Balasubramanian et al. 2002). Sgf11p and Ubp8p constitute a discrete, more recently identified functional module within SAGA that provides deubiquitination activity. Ubp8p is the catalytic subunit and Sgf11p is required for its association in the SAGA complex (Powell et al. 2004; Ingvarsdotir et al. 2005). Recently, Sus1p, thought to bridge transcription and mRNA export (Rodriguez-Navarro et al. 2004), and Chd1p, important for HAT activity (Pray-Grant et al. 2005), were identified as other SAGA subunits. The SAGA complex also has essential subunits such as Tra1p, thought to be involved in interactions with acidic activators (Grant et al. 1998) and a subset of TBP-associated factors and Tafp's (which are also subunits of the TFIID complex), required for nucleosome acetylation and transcriptional stimulation (Grant et al. 1998). At present, while all studies converge to determine that subunits of the SAGA complex play different roles, no clear picture emerges for the specific role of Spt3p. Substantial evidence has supported the idea that it functions through interactions with TBP (Eisenmann et al. 1989; Eisenmann et al. 1992; Madison and Winston 1997; Dudley et al. 1999; Bhaumik and Green 2001; Bhaumik and Green 2002), but this has recently been challenged (Sermwittayawong and Tan 2006). Furthermore, Spt3p is also a subunit of a different complex, SLIK/SALSA, which is very similar in composition to SAGA, but lacks Spt8p and the C terminus of Spt7p (Wu and Winston 2002).
In this work, we were interested in determining the specificity and nature of the genetic interaction between Spt3p and Not1p. Our study reveals that the not1-2 mutation leads mostly to upregulation of transcription at restrictive temperature, and this correlates with increased recruitment of TFIID and SAGA to upregulated promoters. We found a global suppression of not1-2 mutant phenotypes by deletion of SPT3 and determined that this global suppression is specific to the Spt3p subunit of the SAGA complex, with some effect of the Spt8p subunit. Finally, we could show that suppression of transcriptional activation in not1-2 by deletion of SPT3 occurs in cells lacking a SAGA complex. Taken together, our results reveal a SAGA-independent function of Spt3p that mediates transcriptional deregulation in a mutant of the Ccr4-Not complex.
MATERIALS AND METHODS
Strains, plasmids, and media:
The strains used in this study are listed in Table 1. All media were standard, either YPD for glucose-rich medium or synthetic complete media when the presence of a plasmid was selected for. Single-step deletions were performed by PCR as described by Longtine et al. (1998). All of the strains were checked with a PCR reaction performed on genomic DNA extracts using a primer localized in the marker gene and a primer localized at the 5′ noncoding sequence of the target gene. To make strains overexpressing histones H3 and H4, the HHT1 and HHF1 genes were amplified by PCR and cloned in pRS426 and pRS425, respectively. All primers used are available upon request.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| MY1 | MATaura3-52 trp1 leu2Δ∷PET56 gcn4 gal2 (KY803) | Hope and Struhl (1986) |
| MY2 | MATα ura3-52 trp1 leu2Δ∷PET56 gcn4 gal2 (KY804) | Hope and Struhl (1986) |
| MY3 | MATaura3-52 trp1 leu2Δ∷PET56 gcn4 gal2 his3∷TRP1 | Maillet et al. (2000) |
| MY8 | Isogenic to MY2 except not1-2 | Liu et al. (1998) |
| MY896 | Isogenic to MY1 except spt8∷LEU2 not1-2 | This study |
| MY1048 | Isogenic to MY2 except spt3∷TRP1 | This study |
| MY3667 | Isogenic to MY1 except gcn5∷TRP1 | This study |
| MY3712 | Isogenic to MY8 except spt3∷TRP1 | This study |
| MY4136 | Isogenic to MY3667 except MATα not1-2 | This study |
| MY4260 | Isogenic to MY1 except spt8∷TRP1 | This study |
| MY4058 | Isogenic to MY1 except spt7∷SPT7-13MYC-KANMX4 | This study |
| MY4294 | Isogenic to MY4058 except MATα not1-2 | This study |
| MY4299 | Isogenic to MY4058 except MATα spt3∷TRP1 | This study |
| MY4330 | Isogenic to MY4294 except spt3∷TRP1 | This study |
| MY4813 | Isogenic to MY1 except ubp8∷NATMX | This study |
| MY4814 | Isogenic to MY4813 except not1-2 | This study |
| MY4841 | Isogenic to MY1 except spt3∷TRP1 spt7∷LEU2 | This study |
| MY4914 | Isogenic to MY4841 except not1-2 | This study |
| LY102 | Isogenic to MY2 except not1-2 spt7∷LEU2 | This study |
| LY104 | Isogenic to MY3 except spt7∷LEU2 | This study |
Microarray experiments:
Cells were harvested and total RNA was extracted by the acid phenol method. For microarray experiments, we followed the standard Affymetrix procedure. Description of this procedure and description of the probe sets are presented on their website (http://www.affymetrix.com/index.affx). Briefly, cells were harvested under the appropriate conditions and total RNA was extracted by the acid phenol method. The quality of total cellular RNA was tested on a RNA 6000 Nano Chip (Agilent). Total cellular RNA was next purified with a RNeasy mini handbook kit (QIAGEN, Valencia, CA). Then the synthesis of cDNA, cRNA, the hybridization, and the scan of the chip were performed according to the technical manual of Affimetrix (GeneChip expression analysis). Oligo(dT) was added to 15 μg of purified total RNA. After 10 min of incubation at 70° in first-strand buffer with DTT (10 mm), dNTP (500 μm each), and reverse transcriptase [SuperScript II from Invitrogen (San Diego) at 200 units/μl] were added for the first-strand cDNA synthesis (1 hr at 42°). For the second strand synthesis, dNTP (200 μm each), Escherichia coli DNA ligase (10 units), E. coli DNA polymerase I (40 units), and E. coli RNaseH (2 units) were added to the second-strand buffer and incubated for 2 hr at 16°. Finally, T4 DNA polymerase (10 units) was added to the reaction. The cDNA was purified and, after ethanol precipitation, 15 μg of purified cDNA was incubated for 5 hr at 37° with high yield buffer (Enzo), biotine-coupled ribonucleotides, DTT, an RNase inhibitor mix, and T7 RNA polymerase. The cRNA was then purified in the same way as total RNA and 20 μg of cRNA were fragmented in 5× hybridizing buffer (200 mm Tris–acetate, pH 8.1, 500 mm KOAc, 150 mm MgOAc) and incubated for 35 min at 95°.
Hybridization of cRNA:
A total of 15 μg of cRNA was hybridized to the yeast genome S98 chip for 16 hr at 45° with 1× hybridizing buffer [100 mm 2-N-morpholino-ethane sulfonic acid (MES), 1 m (Na+), 20 mm EDTA, 0.01 Tween 20], oligonucleotide B2 (50 pmol), eukaryotic hybridization controls (20×), herring sperm DNA (0.1 mg/ml), and acetylated BSA (0.5 mg/ml). The chip was washed in the Fluidics station, first with buffer A (6× SSPE, 0.01% Tween 20) and then with buffer B [100 mm MES, 0.1 m (Na+), 0.01% Tween 20]. Antibodies were added next [normal goat IgG, 0.1 mg/ml; biotinylated antibodies, 0.5 mg/ml; acetylated BSA 2 mg/ml; 100 mm MES; 1 m (Na+); 0.05% Tween 20] followed by the SAPE solution [streptavidine–phycoerythrine 10 μg/ml, acetylated BSA 2 mg/ml, 100 mm MES, 1 m (Na+), 0.05% Tween 20]. Finally, the chip was scanned and the scan was analyzed by Affymetrix software (MicroArraySuite, MicroDB, and DataMiningTool).
Results were analyzed using the Iobion Array Assist software provided by Stratagene (La Jolla, CA) at http://www.stratagene.com/products/showProduct.aspx?pid=731. The stringent analysis consisted of using P = 0.05 according to CHP, one of the three proposed methods of analysis. This analysis considers only mRNAs expressed at similar levels in the duplicate samples of each strain, which are also significantly different in the strains that are compared. This analysis certainly underestimates the total number of target genes but nevertheless is useful to globally compare two strains, and the results are presented in supplemental Figure S1 at http://www.genetics.org/supplemental/.
S1 analysis:
Total cellular RNA extraction and S1 analyses were performed as described previously (Collart 1996). The sequences of the specific oligonucleotides are available upon request.
Western blot analyses:
Cells (2.5 OD600) were collected for protein extract preparation by rapid post-alkaline lysis. Briefly, the cell pellet was resuspended in 100 μl of H2O and then 100 μl of NaOH 0.2 n was added. After a 5-min incubation at room temperature (RT), the samples were spun at 14,000 rpm for 5 min and the supernatant was discarded. The pellet was resuspended in 50 μl of post-alkaline loading buffer (0.06 m Tris, pH 6.8, 5% glycerol, 2% SDS, 4% β-mercapto-ethanol, bromophenol blue). The samples were loaded on a 15% SDS–PAGE gel for Western blot analysis. After transfer of separated proteins to nitrocellulose membranes, these were incubated overnight at 4° with polyclonal antibodies (anti-H3 ab1791 and anti-H4 ab10158-100 from Abcam). The secondary antibody was a polyclonal goat anti-rabbit antibody conjugated either to alkaline phosphatase or to horseradish peroxidase.
Chromatin immunoprecipitation experiments:
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously (Lenssen et al. 2005). Briefly, wild-type and mutant cells grown appropriately were fixed with 1% formaldehyde for 20 min at RT, and glycine was added to a final concentration of 125 mm to stop the reaction. The cells were washed three times (20 mm Tris–HCl, pH 7.5, 200 mm NaCl) and broken in FA–lysis buffer (50 mm HEPES–KOH, pH 7.5, 140 mm NaCl, 1 mm EDTA, pH 8, 1% Triton, 0.1% sodium deoxycholate, 1 mm PMSF) for 30 min and sonicated. A fraction of the extracts (input) was incubated overnight at 4° with specific antibodies (anti-Taf5p, our polyclonal antibody; anti-MYC, monoclonal 9E10; anti-H3-K4-Met3, polyclonal ab8580 from Abcam) and protein G sepharose. The sepharose beads were collected and washed once with FA–lysis buffer, followed by a wash with FA–lysis buffer plus 350 mm NaCl, then buffer III (10 mm Tris–HCl, pH 8, 1 mm EDTA, pH 8, 250 mm LiCl, 1% Igepal, 1% sodium deoxycholate), and finally twice with TE. Immunoprecipitated chromatin was eluted in 1% SDS, 50 mm Tris–HCl, pH 7.5, 10 mm EDTA, pH 8. Then 10 mg/ml of proteinase K was added to the precipitates and to a fraction of the input for 5 hr at 65° to reverse crosslinks. DNA was extracted and precipitated. Specific DNA was measured by real-time PCR using SYBR Green and the amount in the precipitates was expressed relative to the input DNA. The amount of a specific promoter DNA was normalized to the amount of an intergenic sequence. The sequences of specific oligonucleotides are available upon request.
RESULTS
Suppression of not1-2 is specific to the deletion of the Spt3p SAGA subunit:
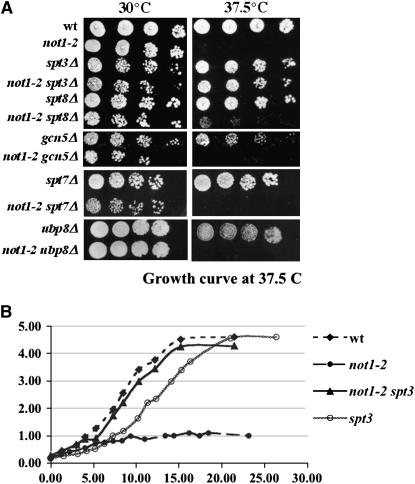
To address the specificity of the suppression of not1-2 temperature sensitivity by the deletion of the Spt3p subunit of the SAGA complex (Collart 1996), we tested whether the deletion of SAGA subunits in addition to Spt3p might suppress not1-2. We deleted SPT8, SPT7, GCN5, and UBP8 in our genetic background and created the not1-2 double-mutant strains (Table 1). Cells from the single- and double-mutant strains were grown to exponential phase, diluted to an equivalent OD600 of 1.0, serially diluted fivefold, and spotted on glucose-rich plates (YPD) that were placed at 30° or 37.5° (Figure 1). A suppression of not1-2 temperature sensitivity was obtained only with the deletion of SPT3 and, to a lesser extent, with the deletion of SPT8, but not with the deletion of other subunits of the SAGA complex. In particular, no suppression was observed upon deletion of SPT7 that disrupts the SAGA complex. Instead, a slight synthetic growth phenotype was observed (Figure 1A).
Figure 1.—
not1-2 temperature sensitivity is suppressed specifically by the deletion of SPT3. The indicated strains were grown exponentially in glucose-rich medium for 24 hr and then (A) serially diluted and plated on glucose-rich plates that were placed at 30° or 37.5° for 3 days or (B) were shifted to 37.5° after reaching an OD600 of 0.2. Growth of the cells was measured over time.
Genomewide profiling suggests that suppression of transcriptional deregulation in not1-2 by deletion of SPT3 is global:
Suppression of not1-2 temperature sensitivity by the deletion of SPT3 could be due to the restoration of appropriate expression of one or a select few genes required for growth at high temperature. Alternatively, deletion of SPT3 might generally restore appropriate gene expression in cells mutant for Not1p. To address this issue, we performed microarray experiments. We grew wild-type, not1-2, spt3Δ, and not1-2 spt3Δ cells for 24 hr exponentially in liquid glucose-rich medium, and then transferred cells at an OD600 of 0.2 to 37.5° (Figure 1B). As expected, growth of not1-2 cells stopped after a couple of divisions at restrictive temperature, whereas double-mutant cells grew like wild-type cells. spt3Δ cells grew to the same final density as wild-type cells, but displayed a slight delay in growth after transfer to high temperature (Figure 1B). Total cellular RNA was extracted for all strains after the shift to restrictive temperature, when the cells were at an OD600 of 0.8. Labeled cRNA was prepared for each sample and was hybridized to Affymetrix whole-genome arrays. For each strain, the experiment was performed twice independently, and the results were analyzed using the Array Assist software provided by Iobion (see materials and methods). We compared each single mutant and double mutant to the wild type to define genes with altered expression, and we also compared the not1-2 mutant to the not1-2 spt3Δ double mutant. Using an analysis that takes into account genes only for which expression is reproducible in the duplicates of the mutant and the wild-type cells, and which display at least a 2-fold difference in expression between the mutant and the wild type, there were 119 genes that displayed increased expression (from 2- to 40-fold) and 33 genes that displayed decreased expression (in general, 2- to 3-fold) in the not1-2 mutant relative to the wild-type cells.
Interestingly, 70% of 119 genes that displayed increased expression in not1-2 were totally (19%) or partially (51%) restored in the double mutant, and 58% of the 33 genes that displayed decreased expression in not1-2 were totally (18%) or partially (40%) suppressed in the double mutant. In contrast, 49% of the 37 genes that were increased in the spt3Δ single mutant were similarly increased in the double mutant, and 40% of the 65 genes with decreased expression in the spt3Δ single mutant were similarly decreased in the double mutant (Table 2). The other genes deregulated in the spt3Δ single mutant were slightly less deregulated in the double mutant, indicating a low level of suppression of spt3Δ by the not1-2 mutation.
TABLE 2.
Suppression of not1-2 by spt3Δ is global
| Mutant | No. of target genes | Expression phenotype | Expression in not1-2 spt3Δ |
|---|---|---|---|
| not1-2 | 119 | Increased | 23 totally suppressed |
| 61 partially suppressed | |||
| not1-2 | 33 | Decreased | 6 totally suppressed |
| 13 partially suppressed | |||
| spt3Δ | 37 | Increased | 18 no suppression |
| 17 partial suppression | |||
| spt3Δ | 65 | Decreased | 26 no suppression |
| 35 partial suppression |
Thus, mutation of NOT1 leads mostly to an increase in gene expression under the conditions where the growth arrests at high temperature, and the deletion of SPT3 has a global suppressive effect.
mRNA analysis confirms suppression of not1-2-increased gene expression by spt3Δ:
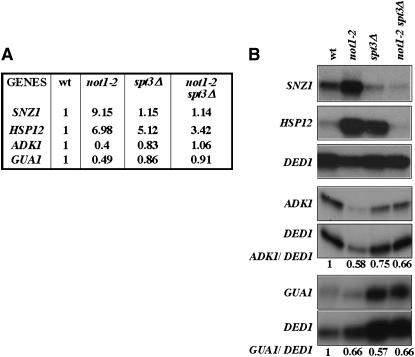
To obtain confirmation of the results found by the microarray analyses, we analyzed by S1 digestion the mRNA levels of genes induced in not1-2 and suppressed in the double mutant, such as SNZ1 and HSP12, or decreased in not1-2 and suppressed in the double mutant, such as GUA1 and ADK1 (Figure 2A). We grew cells as above, collected total cellular RNA, and analyzed the levels of specific mRNAs by S1 analysis. Consistently with the microarrays, HSP12 and SNZ1 were highly increased and GUA1 and ADK1 weakly decreased in not1-2 (Figure 2B). The deletion of SPT3 strongly suppressed the increased expression of HSP12 and SNZ1, but only partially suppressed the decreased expression of ADK1 and not at all that of GUA1 (Figure 2B). Thus, the transcriptional defects in not1-2 were most evident for genes upregulated in the mutant, and these defects were well suppressed by the deletion of SPT3. In contrast, downregulation of mRNAs in not1-2 was weak, consistent with the microarray results, but a suppressive effect by deletion of SPT3 for these mRNA reductions was not very convincing. Hence we focused further analyses on genes activated in not1-2.
Figure 2.—
mRNA analysis confirms suppression of transcriptional deregulation in not1-2 by the deletion of SPT3. (A) The results of the microarray experiments for the four indicated transcripts are given. (B) Cells were grown, as in Figure 1, to an OD600 of 0.8, and 50 μg of total cellular RNA was analyzed by S1 digestion for the levels of the indicated transcripts. DED1 was always measured in the same reaction as the other transcripts as an internal control, since the microarray experiments indicated that it was not affected by not1-2. The RNA was sampled and analyzed the same day for SNZ1 and HSP12, and the results for DED1 were the same and are shown only once. The levels of ADK1 and GUA1 relative to DED1 were quantified using phosphoimager analysis and are indicated below the radiographs.
Total restoration of appropriate transcription in not1-2 is specific to the deletion of the Spt3p subunit of the SAGA complex:
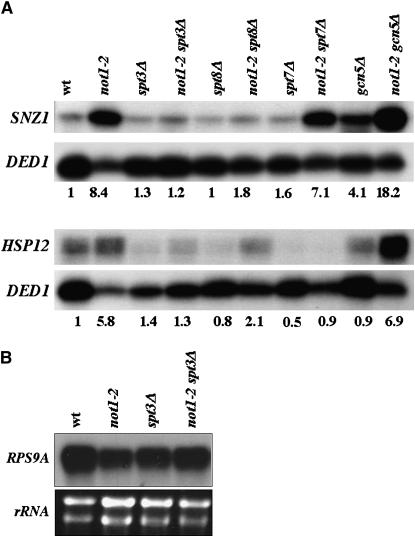
To investigate the nature of the suppression, we chose to concentrate on the HSP12 and SNZ1 genes. We first tested whether the deletion of other subunits of the SAGA complex that do not suppress the temperature sensitivity of not1-2 affect expression of HSP12 and SNZ1 in not1-2. For this, we grew, as above, not1-2, spt3Δ, spt8Δ, spt7Δ, and gcn5Δ single mutants and not1-2 double mutants and analyzed equivalent amounts of total cellular RNA for the levels of HSP12 and SNZ1 mRNAs. Deletion of GCN5 or SPT7 did not reduce SNZ1 mRNA levels in not1-2 cells, and, furthermore, deletion of GCN5 in itself increased SNZ1 expression. In contrast, the deletion of SPT3, and to a lesser extent that of SPT8, reduced the level of SNZ1 mRNA in not1-2 mutant cells (Figure 3A). Deletion of SPT3, SPT7, and, to a lesser extent, SPT8, but not GCN5, reduced the expression of HSP12 mRNA in not1-2 (Figure 3A).
Figure 3.—
The deletion of SPT3 specifically reduces increased HSP12 and SNZ1 mRNA in not1-2. (A) The indicated strains were grown exponentially for 24 hr and then shifted at an OD600 of 0.2 to 37.5°. Total cellular RNA was prepared when cells reached an OD600 of 0.8 as in Figure 2. The levels of HSP12 and SNZ1 mRNAs were measured simultaneously to that of DED1 in 50 μg of total cellular RNA by S1 analysis. Because of the unequal loading of some lanes, the results were quantified by phosphoimager analysis. (B) The levels of RPS9A in 50 μg of total RNA were measured for the indicated strains, and the levels of rRNA in 5 μg of total cellular RNA are indicated as a control.
Thus, in good correlation with suppression of temperature sensitivity, total restoration of wild-type transcript levels in not1-2 requires the deletion of SPT3 specifically, and the deletion of SPT8 also partially restores wild-type mRNA levels.
The trimethylation mark of histone H3 follows transcription in not1-2:
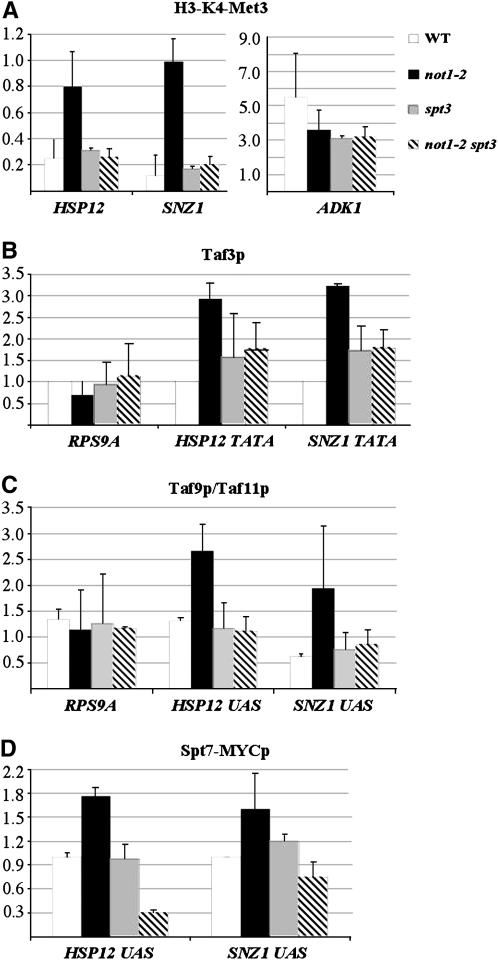
It has been previously described that activation of transcription correlates with increased trimethylation of histone H3 (H3-K4-Me3) at the core promoter (Santos-Rosa et al. 2002; Pokholok et al. 2005). To determine whether changes in mRNA levels in not1-2 are due to changes in transcription initiation, we investigated the presence of this histone modification mark at promoters of genes with altered expression in not1-2 by ChIP experiments. Wild-type, not1-2, spt3Δ, and not1-2 spt3Δ cells were grown as above, cells were crosslinked, and total protein extracts were incubated with antibodies against H3-K4-Me3. For each immunoprecipitation, we first expressed the amount of each DNA in the immunoprecipitate relative to its amount in the total input DNA. Then we normalized the amount of specific DNA immunoprecipitated with the antibody to the amount of an intergenic DNA that was immunoprecipitated. The level of H3-K4-Me3 at the HSP12 and SNZ1 promoters was strikingly increased in not1-2, and this was suppressed by the deletion of SPT3, which by itself had no effect on the levels of H3-K4-Me3 compared to wild-type cells (Figure 4A). In contrast, the presence of H3-K4-Me3 on the ADK1 promoter was decreased in not1-2 cells when compared to wild-type cells (Figure 4A), in good correlation with decreased expression from this promoter in not1-2 (see Figure 2). Thus, the levels of H3-K4-Me3 found at promoters in wild-type and not1-2 or not1-2 spt3Δ cells correlate well with the levels of transcripts measured in these cells, providing support that changes in transcription levels are the basis for the genetic interaction between Spt3p and Not1p.
Figure 4.—
The presence of Taf3p at HSP12 and SNZ1 promoters increases in not1-2 and this is suppressed by the deletion of SPT3. The indicated strains were grown as in Figure 3 to an OD600 of 0.8 at 37.5°, and the cells were crosslinked prior to total extract preparation and immunoprecpitation with the indicated antibodies. The amount of indicated promoter DNAs in the immunoprecipitates was evaluated by real-time PCR, expressed relative to input DNA, and then normalized to the amount of an intergenic unexpressed DNA. (A) The antibodies were against H3-K4-Me3 and the results presented are the average of two experiments performed with independent cultures. (B) Antibodies were against Taf3p and the experiment was repeated three times with independent cultures. (C) Antibodies against Taf9p and Taf11p were used and the relative amount of DNA immunoprecipitated with Taf9p to that immunoprecipiated with Taf11p was calculated (Taf9p/Taf11p). The experiment was repeated two times with independent cultures. (D) The indicated strains expressed MYC-tagged Spt7p and antibodies against MYC were used. The experiment was repeated three times with independent cultures.
Deletion of SPT3 suppresses increased association of Taf3p with promoters upregulated in not1-2:
Previous studies have shown that the recruitment of TFIID to promoters was increased or decreased in a promoter-specific manner upon mutation of the Ccr4-Not complex (Deluen et al. 2002). To determine whether the not1-2 mutation had such a phenotype, we investigated by ChIP experiments whether the recruitment of Taf3p, a specific subunit of the TFIID complex, was altered in not1-2 cells at restrictive temperature on induced promoters such as HSP12 and SNZ1. Wild-type, not1-2, spt3Δ, and not1-2 spt3Δ cells were grown as before, cells were crosslinked, and total protein extracts were incubated with antibodies against Taf3p. The presence of the HSP12 or SNZ1 promoters in the immunoprecipitates was investigated by real-time PCR as before. As a control, we looked at the presence of the RPS9A promoter in the immunoprecipitates, since RPS9A mRNA is roughly similar in all four strains, but slightly decreased in not1-2 cells relative to wild-type cells, rather than increased as HSP12 and SNZ1 (Figure 3B). Taf3p association with the HSP12 and SNZ1 promoters was significantly higher in not1-2 than in wild-type cells, and this was suppressed by the deletion of SPT3 (Figure 4B). In contrast, there was no increase in the association of Taf3p with the RPS9A promoter in not1-2 relative to wild-type cells, but instead there was a slight decrease that was suppressed by the deletion of SPT3. The deletion of SPT3 in wild-type cells did not appear to significantly alter the association of Taf3p with any of the promoters (Figure 4B).
Thus, from our analysis of two relevant promoters, we conclude that recruitment of TFIID is increased at promoters that are upregulated in not1-2 and this is suppressed by the deletion of SPT3.
Spt3p mediates recruitment of SAGA to the UAS of genes activated in not1-2:
To understand how Spt3p might affect recruitment of TFIID in not1-2, we investigated the association of SAGA with genes upregulated in not1-2 by comparing the recruitment of Taf9p, a subunit of both SAGA and TFIID, and of Taf11p, a subunit of TFIID only, to the upstream activating sequence (UAS) of HSP12 and SNZ1. We grew cells as above, crosslinked them, and prepared total protein extracts that we incubated with antibodies against Taf9p or Taf11p. We evaluated, as before, by real-time PCR the presence of the RPS9A core promoter and the HSP12 or SNZ1 UAS DNAs in the immunoprecipitates. We expressed the amount of each DNA that was immunoprecipitated with Taf9p relative to Taf11p as a tool to measure the contribution of SAGA vs. TFIID for each DNA. The association of Taf9p vs. Taf11p to the RPS9A core promoter was very similar in all strains (ratio of ∼1.0), compatible with TFIID associated with this DNA sequence in all cases (Figure 4C). In contrast, there was a significant increase in the association of Taf9p relative to Taf11p with the UAS sequences of SNZ1 and HSP12 in not1-2 relative to wild-type cells, suggesting an increase of SAGA to this DNA sequence in not1-2. Interestingly, this increase was suppressed by the deletion of SPT3 that had no effect in wild-type cells (Figure 4C).
To confirm these results, we created strains expressing C-terminally MYC-tagged Spt7p (see Table 1) and investigated the association of Spt7p to the UAS of HSP12 and SNZ1 by ChIP experiments as before, using antibodies against MYC. In total agreement with the previous experiments, we found that the association of Spt7p was increased at the UAS of both HSP12 and SNZ1 in not1-2 cells compared to wild-type cells (Figure 4D). The deletion of SPT3 did not have a major impact on the association of Spt7p with these UAS sequences in wild-type cells, but it suppressed the increased association of Spt7p with these sequences in not1-2 cells (Figure 4D).
Taken together, these results reveal an Spt3p-dependent increase of SAGA association with UAS sequences of genes upregulated in not1-2, but they suggest no role of Spt3p for the basal level of SAGA associated with such sequences.
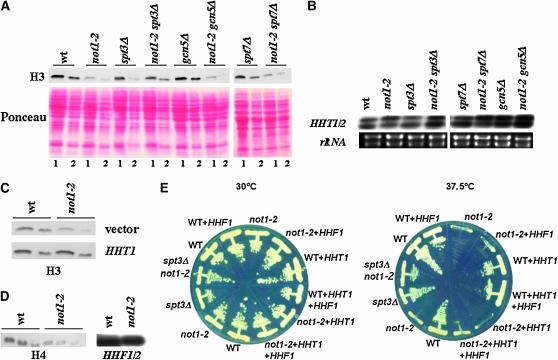
Deletion of SPT3 suppresses post-transcriptional loss of histone H3 in not1-2:
Surprisingly, during the course of the experiments presented above, we observed that the level of total H3-K4-Me3 in extracts from not1-2 cells was reduced when compared to wild-type cells at restrictive temperature (data not shown). We then discovered that the decreased levels of H3-K4-Me3 in not1-2 correlated with decreased levels of total histone H3 (Figure 5A). Astonishingly, this loss of histone H3 was suppressed by the deletion of SPT3 but not by the deletion of GCN5 or SPT7 (Figure 5A). Since deletion of SPT3 suppresses transcriptional deregulation in not1-2, we analyzed the levels of the mRNAs encoding histone H3 in wild-type, not1-2, spt3Δ, spt8Δ, spt7Δ gcn5Δ, and not1-2 double mutants by S1 digestion. However, the levels of histone H3 mRNA were roughly similar in all strains (Figure 5B), suggesting that the differences in total levels of histone H3 protein in the different strains described above are due to post-transcriptional control of histone H3 protein.
Figure 5.—
The level of total histone H3 and H4 protein but not mRNA is decreased in not1-2 and this is suppressed by deletion of SPT3. The indicated strains were grown as in Figure 3 to an OD600 of 0.8 at 37.5°. (A) Total protein extracts were prepared by post-alkaline lysis. Proteins were separated on 15% SDS–PAGE and analyzed by Western blot for the levels of histone H3. Lane 1, 14 μl of post-alkaline lysis corresponding to 0.7 OD600; lane 2, 7 μl of post-alkaline lysis corresponding to 0.35 OD600. (B) Total cellular RNA was extracted and 50 μg were analyzed by S1 digestion for the levels of HHT1/2 mRNAs. The probe cannot distinguish between the two different mRNAs that encode histone H3. The amount of rRNA is shown as the loading control. (C) Total protein extracts were prepared by post-alkaline lysis, as indicated in A, but also from wild-type or not1-2 cells carrying either an empty vector or a multicopy plasmid expressing histone H3 as indicated. (D) Total protein extracts were prepared by post-alkaline lysis. Proteins were separated on 15% SDS–PAGE and analyzed by Western blot for the levels of histone H4, using 28, 14 or 7 μl of post-alkaline lysis for each strain (left), total cellular RNA was extracted, and 50 μg was analyzed by S1 digestion for the levels of HHF1/2 mRNAs (right). (E) The indicated strains overexpressing or not overexpressing histone H3 and/or H4 as indicated were streaked on glucose-rich plates and placed at 30° or 37.5° as indicated.
To determine whether not1-2 mutant phenotypes might then result from reduced histone H3 levels, we increased histone H3 protein levels in not1-2 to a level similar to that in the wild type by transforming not1-2 mutant cells with a multicopy plasmid carrying the HHT1 gene encoding histone H3 (Figure 5C). Nevertheless, the transformants could not grow at high temperature (Figure 5E), suggesting that restoring histone H3 protein levels alone was not sufficient to suppress not1-2. An obvious explanation could be that a decrease of histone H4 might also occur, and indeed, we found a decrease also of histone H4 protein but not mRNA in not1-2 (Figure 5D). We then created a multicopy plasmid carrying the HHF1 gene encoding histone H4 that we transformed into wild-type and not1-2 cells carrying or not carrying the multicopy plasmid expressing histone H3. However, not1-2 cells carrying the multicopy plasmids expressing histones H3 and H4 still did not grow at high temperature (Figure 5E). Thus, decreased levels of histones H3 and H4 are unlikely to be the cause of temperature sensitivity in not1-2.
Evidence for SAGA-independent functions of Spt3p:
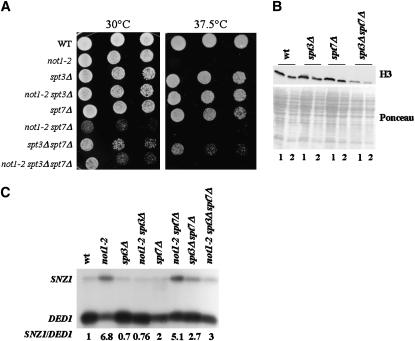
Since our experiments so far show that suppression of not1-2 is obtained specifically by deleting the Spt3p subunit of the SAGA complex, we investigated whether Spt3p might have a function outside of the SAGA complex. We created an spt3Δ spt7Δ double-mutant strain by crossing the single mutants, sporulating the diploids, and dissecting the tetrads. The double spt3Δ spt7Δ grew slower than either single mutant at both 30° and 37.5°, indicating a synthetic phenotype between the two deletions. The deletion of SPT7 in not1-2 also led to a synthetic slow-growth phenotype as mentioned above (Figure 1A), but interestingly, the triple not1-2 spt3Δ spt7Δ grew better than the not1-2 spt7Δ double mutant, indicating that the deletion of SPT3 suppressed not1-2 in the absence of SPT7. Suppression of not1-2 by the deletion of SPT3 nevertheless did not restore growth at high temperature in the context of spt7Δ (Figure 6A). The synthetic phenotype between spt3Δ and spt7Δ and the suppression of not1-2 in spt7Δ suggested a function for Spt3p in the absence of a SAGA complex. This idea was confirmed when we analyzed total histone H3 levels by Western blot analysis of total protein extracts. Indeed, the spt3Δ spt7Δ double mutant, but neither single mutant, displayed reduced levels of histone H3 protein when compared to the wild type, indicating once again a synthetic phenotype (Figure 6B).
Figure 6.—
spt3Δ and spt7Δ display a synthetic phenotype. The indicated strains were grown exponentially for 24 hr and then (A) serially diluted and plated on glucose-rich plates that were placed at 30° or 37.5° as indicated for 3 days. (B) Strains shifted to high temperature, as in Figure 1, to an OD600 of 0.8. Total extracts were made by post-alkaline lysis and analyzed by Western blot for the levels of histone H3. The Ponceau staining is shown for loading control. Lane 1, 14 μl of post-alkaline lysis corresponding to 0.7 OD600; lane 2, 7 μl of post-alkaline lysis corresponding to 0.35 OD600. (C) Strains shifted to high temperature, as in Figure 1, to an OD600 of 0.8. Total cellular RNA was extracted and 50 μg were analyzed by S1 digestion for the levels of SNZ1 mRNAs and DED1 as an internal control. Results were quantified by phosphoimager analysis.
To determine whether the SAGA-independent function of Spt3p suggested by the experiments presented above might be responsible for mutant phenotypes in not1-2, we looked at the expression of SNZ1 mRNA. Indeed, our first experiments (see Figure 3A) revealed that SNZ1 mRNA was upregulated by not1-2 in the absence of SAGA, namely in spt7Δ. We grew wild type, not1-2, spt3Δ, spt7Δ, spt3Δ spt7Δ, and the not1-2 double or triple mutants as before and prepared total cellular RNA that we analyzed by S1 digestion for the levels of SNZ1 mRNA. The levels of SNZ1 mRNA were increased by the not1-2 mutation in spt7Δ mutant cells, but not in spt3Δ cells or in spt7Δ spt3Δ double-mutant cells (Figure 6C). In other words, in the absence of SPT7, the deletion of SPT3 sup-pressed the not1-2-dependent induction of SNZ1 mRNA.
Taken together, these results show that Spt3p has a function outside of the SAGA complex and that the not1-2 mutation leads to an Spt3p-dependent increase in SNZ1 mRNA even in the absence of a SAGA complex.
Spt3 mediates transcription activation in not1-2 at permissive temperature:
The experiments presented so far demonstrate Spt3p-dependent transcriptional activation in not1-2 at restrictive temperature. To make sure that the observed effects might not be indirectly due to loss of viability or growth arrest at high temperature, we analyzed the levels of SNZ1 and HSP12 mRNAs in wild type, not1-2, spt3Δ, and not1-2 spt3Δ at permissive temperature. Both mRNAs were induced in not1-2, and this was suppressed by the deletion of SPT3 (Figure 7A). Thus, Spt3p-dependent activation of genes in not1-2 occurs at permissive temperature already.
Figure 7.—
Spt3p-dependent activation in not1-2 occurs at permissive temperature. The indicated strains were grown in glucose-rich medium to exponential phase and (A and B) total cellular RNA was extracted for analysis by S1 digestion of the indicated mRNAs. DED1 mRNA and rRNAs are shown as the loading control. (C) The cells were crosslinked prior to total extract preparation and immunoprecpitation with Taf9p polyclonal antibodies. The amount of indicated promoter DNAs in the immunoprecipitates was evaluated by real-time PCR, expressed relative to input DNA, and then normalized to the amount of an intergenic unexpressed DNA.
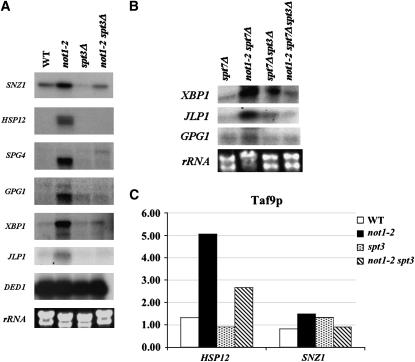
To extend our findings, we analyzed at permissive temperature the expression of other genes identified in the microarrays as targets for Spt3-dependent transcriptional activation in not1-2 at restrictive temperature, namely SPG4, GPG1, XBP1, and JLP1 (see supplemental Figure S1 at http://www.genetics.org/supplemental/). All four mRNAs were induced in not1-2 at permissive temperature, and this was suppressed by the deletion of SPT3 (Figure 7A).
Next, we wanted to determine whether Spt3p-dependent, SAGA-independent transcriptional activation in not1-2 might be relevant to more genes than just SNZ1, and whether this was also visible at permissive temperature. Indeed, we found that XBP1, JLP1, and GPG1 mRNAs were induced in not1-2 at permissive temperature in the absence of SPT7 and that this was Spt3p dependent (Figure 7B).
Finally, for HSP12 and SNZ1, we investigated whether Spt3p-dependent mRNA induction in not1-2 at permissive temperature correlated with an Spt3p-dependent increased presence of SAGA and/or TFIID at the promoters, as observed before at restrictive temperature. We performed ChIP experiments as above, but grew the cells at permissive temperature to exponential phase and used antibodies to Taf9p, a subunit of both TFIID and SAGA. We observed that, indeed, even at permissive temperature, the presence of Taf9p at the promoters of HSP12 and SNZ1 was increased in not1-2 and this was suppressed by the deletion of SPT3 (Figure 7C).
DISCUSSION
Spt3p mediates mutant phenotypes in not1-2:
In this work, we have used genetic and molecular tools to investigate the interaction between Not1p, the essential and scaffold subunit of the Ccr4-Not complex, and Spt3p, a subunit of one functional module of the SAGA complex. Indeed, a previous study demonstrated that the thermo-sensitivity associated with a not1-2 mutation could be suppressed by deletion of SPT3 (Collart 1996). not1-2 is a nonsense mutation in which a C-terminally truncated form of Not1p accumulates and only low levels of full-length protein containing the essential C-terminal domain are expressed. The mutant phenotypes of not1-2 most certainly result from reduced levels of Not1p, and thus of the Ccr4-Not complex, as the overexpression of the N-terminal portion of Not1p has no observable mutant phenotype (our unpublished observations) and all Not1p protein in the cell is associated in Ccr4-Not complexes (Maillet et al. 2000). We found that the mutant phenotypes associated with not1-2 were entirely mediated by Spt3p. Indeed, microarray analyses revealed that transcriptional deregulation in not1-2 was generally dependent upon Spt3p and consisted mostly of increased gene expression. Furthermore, by ChIP experiments we could show that the Spt3p-dependent increased transcription in not1-2 correlated with Spt3p-dependent increased recruitment of SAGA and TFIID to upregulated promoters. Finally, we observed that transcriptional deregulation and temperature sensitivity in not1-2 correlated with an Spt3p-dependent post-transcriptional decrease of total histone H3 and H4 protein levels in the cell. Taken together, our results suggest that the Ccr4-Not complex regulates an Spt3p-dependent function to maintain growth at high temperature, as well as appropriate histone H3 and H4 protein levels and transcriptional repression at this temperature.
We were able to show that Spt3p-dependent transcriptional deregulation in not1-2 already occurs at permissive temperature when not1-2 cells grow as well as wild-type cells. At this temperature, in contrast, histone H3 and H4 levels are not affected in not1-2 compared to the wild type (see supplemental Figure S2 at http://www.genetics.org/supplemental/). Furthermore, overexpression of histones H3 and H4 in not1-2 leading to histone levels similar to the wild type does not restore growth at high temperature. This would suggest that decreased histone levels are not the cause of Spt3p-dependent transcriptional upregulation in not1-2, nor a cause for growth arrest at restrictive temperature, but instead might be a consequence, for instance, of growth arrest at high temperature.
A function of Spt3p independent of SAGA is controlled by Not1p:
Spt3p is a subunit of the SAGA complex and of its very similar SLIK/SALSA complex. Its precise function in these complexes has not been clearly established, and no SAGA- or SLIK/SALSA-independent role for Spt3 has been described. Generally, studies of Spt3p have been global, and they have shown that it plays roles in both activation and repression of transcription. While original experiments suggested that Spt3p functions through interactions with TBP (Eisenmann et al. 1989, 1992), it has not been demonstrated that TBP co-immunoprecipitates with Spt3p, and in fact interaction of SAGA with TBP clearly requires Spt8p but not Spt3p (Sermwittayawong and Tan 2006). Interactions between Spt3p and Spt8p might explain the confusion about the interaction between Spt3p and TBP. Indeed, genetic studies suggest that the functions of Spt3p and Spt8p are very similar (Ingvarsdotir et al. 2005). However, Spt3p, but not Spt8p, is a subunit of the SLIK/SALSA complex, suggesting that the functions of the two proteins are certainly not identical. Our study shows that deletion of SAGA subunits other than Spt3p does not suppress the not1-2 mutation, except for the deletion of SPT8 that also suppresses not1-2 but to a lesser extent than the deletion of SPT3. These results are in agreement with all previous studies showing that the function of Spt3p is closest to that of Spt8p within the SAGA complex, but is different from that of the other SAGA subunits. The results also show that the roles of Spt3p and Spt8p are not exactly identical. The partial suppression of not1-2 obtained by deletion of SPT8 might suggest that Spt8p contributes toward a function(s) of Spt3p that is independent of the SAGA complex.
We were able to determine that the deletion of SPT3 suppresses transcriptional induction in not1-2 cells lacking Spt7p, and thus SAGA or SLIK/SALSA complexes. These observations suggest that Spt3p has a function(s) outside of the SAGA complex and that this function(s) of Spt3p mediates transcriptional deregulation in not1-2. Compatible with this idea is the observation that the deletion of SPT3 displays a synthetic phenotype when combined with a deletion of SPT7. Indeed, this synthetic phenotype supports a function for Spt3p in the absence of a SAGA complex. Furthermore, the deletion of SPT3 was observed to suppress the synthetic growth phenotype obtained by combining a deletion of SPT7 that results in the loss of SAGA and not1-2. Again, such an observation is compatible with a function of Spt3p outside of SAGA.
The Ccr4-Not complex regulates the capacity of Spt3p to contribute to recruit SAGA and TFIID to certain promoters:
The not1-2 mutant leads to increases in transcription initiation, since increases in mRNA levels correlate with increased association of TFIID and SAGA, as well as with increased presence of the H3-K4-Me3 mark, at promoters of upregulated genes in not1-2. In the not1-2 mutant cells, Spt3p mediates the recruitment and/or stabilization of SAGA and TFIID to promoters of upregulated genes, but Spt3p is not required for the basal levels of either TFIID or SAGA associated with these same promoters in wild-type cells. Thus, in wild-type cells the capacity of Spt3p to recruit and/or stabilize SAGA and TFIID to these promoters is under the control of the Ccr4-Not complex.
So far, contradictory results concerning the role of Spt3p in the recruitment of SAGA to promoters have been published. It was generally accepted that Spt3p is not required for the recruitment of the SAGA complex at SAGA-dependent promoters (Larschan and Winston 2001), but a recent study (Qiu et al. 2005) showed that high-level recruitment of Tra1p and other SAGA subunits requires Spt3p. Our results suggest that if the Ccr4-Not complex is not completely wild type in some strains, this could explain the different results obtained in different studies.
Our studies also raise the possibility that TFIID and SAGA co-occupy promoters, since Spt3p mediates recruitment and/or stabilization of both complexes to the same promoters of genes upregulated in not1-2. This finding is in agreement with a previous study that has suggested that Spt3p, like Gcn5p, might help to recruit Taf1p (Van Oevelen et al. 2005).
Spt3p mediates activation of transcription in not1-2 even in the absence of a SAGA complex, suggesting that it can lead to the recruitment of TFIID in the absence of a SAGA complex. Thus, Spt3p is likely to be capable of recruiting and/or stabilizing TFIID at the core promoter from within the SAGA complex at the UAS or without being associated with the SAGA complex.
How might Spt3p mediate recruitment of SAGA and TFIID in not1-2? Spt3p might be able to provide a contact surface for transcriptional activators and connect them to TFIID. Alternatively, it could be that Spt3p interacts with complexes such as mediator that can have a stimulatory effect on SAGA recruitment as suggested previously (Qiu et al. 2005). Spt3p has also been shown to be important for nucleosome remodeling by cooperating with Mot1p (Madison and Winston 1997; Topalidou et al. 2004; Van Oevelen et al. 2005). A possible connection between Spt3p and chromatin is also suggested by our work since we observe an Spt3p-dependent decrease of cellular histone H3 and H4 levels in not1-2 that correlates with an Spt3p-dependent increased recruitment and/or stabilization of SAGA and TFIID to promoters of genes upregulated in not1-2. In other studies, evidence connecting Spt3p and Spt8p to TFIIA has been provided. Indeed, overexpression of TFIIA suppresses spt3 mutations (Madison and Winston 1997) and a recent study has shown that Spt8p reduces affinity of SAGA for the other general transcription factors because of competition with TFIIA (Warfield et al. 2004). However, overexpression of TFIIA does not suppress not1-2 or alter suppression of not1-2 by the deletion of SPT3 (data not shown), suggesting that interactions between TFIIA and Spt3p are not involved in the not1-2 mutant phenotypes.
Mechanism of transcriptional deregulation in not1-2:
The identification of Spt3p as the protein that specifically mediates mutant phenotypes in not1-2 leads to the important question of how the Ccr4-Not complex controls Spt3p-dependent transcriptional activation. A relevant question in this regard is whether the Ccr4-Not complex controls Spt3p directly or whether Spt3p happens to be required for transcriptional activation of all the genes induced in not1-2. We were not able to show any co-immunoprecipitation of SAGA subunits with Ccr4-Not subunits (data not shown), yet it is possible that the Ccr4-Not complex regulates post-translational modification of SAGA subunits. Indeed, we have preliminary data suggesting subtle changes in the association of Spt3p with Spt7p when compared to that of Taf5p with Spt7p in not1-2 (data not shown). These findings are consistent with some direct effect of the Ccr4-Not complex on Spt3p and/or the SAGA complex. Alternatively, the Ccr4-Not complex might regulate Spt3p-dependent recruitment of SAGA to promoters by regulating the 19S proteasomal subunits that contribute to SAGA recruitment as recently shown (Lee et al. 2005) or by regulating transcription factors that recruit SAGA to activate transcription. We addressed a possible role of the proteasome in not1-2 by analyzing induction of HSP12 and SNZ1 mRNAs in not1-2 cells carrying the temperature-sensitive proteasomal mutation sug1-25. However, induction of both genes was similar in not1-2 or not1-2 sug1-25 mutants at restrictive temperature (data not shown), suggesting that the proteasome is not required for upregulation of these genes in not1-2. In contrast, we already know that the Ccr4-Not complex controls the activity of at least one transcription factor, Msn2p, through control of its post-translational modification (Lenssen et al. 2005). Thus, one could imagine that the Ccr4-Not complex controls a subset of transcription factors that act to recruit SAGA to genes through the Spt3p subunit. We previously determined that the alterations in TFIID distribution to promoters in mutants of the Ccr4-Not complex were independent of Msn2p (Lenssen et al. 2005). Nevertheless, the Ccr4-Not complex may control more than one transcription factor such that the deletion of one individual transcription factor would indeed not act as a general suppressor. In contrast, the deletion of a protein that connects all of these transcription factors to the general transcription machinery should act as a general suppressor. Spt3p could be such a connecting protein. If this latter model is correct—namely if the primary target of the Ccr4-Not complex is a number of Spt3p-dependent transcription factors—it is at present unclear why the not1-2 mutation would lead to an Spt3p-dependent global reduction of histone H3 and H4 levels. It could be that this latter phenotype is completely independent or, as mentioned above, is really only a consequence of growth arrest at high temperature. We are currently undertaking a biochemical study of Spt3p and SAGA in wild-type and not1-2 cells with the hope of providing answers to the remaining questions.
Acknowledgments
We thank Gregory Prelich for a critical reading of the manuscript. This work was supported by a grant (3100AO-100793) from the National Science Foundation and an Improving Human Potential/Network European grant (HPRN-CTG-2002-00261) supported by the Office Fédéral de l'Education et de la Science (NJ 02.0017) to M.A.C. Finally, this work was also supported by a grant from the Leenards foundation awarded to M.A.C.
References
- Albert, T., H. Hanzawa, Y. I. A. Legtenberg, M. J. deRuwe, F. A. J. van der Heuvel et al., 2002. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 21: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badarinarayana, V., Y.-C. Chiang and C. L. Denis, 2000. Functional interaction of CCR4-NOT proteins with TATAAA-binding protein (TBP) and its associated factors in yeast. Genetics 155: 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant and S. Tan, 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277: 7989–7995. [DOI] [PubMed] [Google Scholar]
- Bhaumik, S. R., and M. R. Green, 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15: 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik, S. R., and M. R. Green, 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. 22: 7365–7371. [DOI] [PMC free article] [PubMed]
- Collart, M. A., 1996. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol. Cell. Biol. 16: 6668–6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart, M. A., 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313: 1–16. [DOI] [PubMed] [Google Scholar]
- Collart, M. A., and M. H. T. Timmers, 2004. The eukaryotic Ccr4-Not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways. Prog. Nucleic Acid. Res. Mol. Biol. 77: 289–322. [DOI] [PubMed] [Google Scholar]
- Daugeron, M.-C., F. Mauxion and B. Séraphin, 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 29: 2448–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluen, C., N. James, L. Maillet, M. Molinete, G. Theiler et al., 2002. The Ccr4-Not complex and yTAF1 (yTaf(II)130p/yTaf(II)145p) present physical and functional interactions. Mol. Cell. Biol. 22: 6735–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis, C. L., and J. Chen, 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73: 221–250. [DOI] [PubMed] [Google Scholar]
- Dudley, A. M., C. Rougeulle and F. Winston, 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13: 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann, D. M., C. Dollard and F. Winston, 1989. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell 58: 1183–1191. [DOI] [PubMed] [Google Scholar]
- Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney and F. Winston, 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6: 1319–1331. [DOI] [PubMed] [Google Scholar]
- Finoux, A.-L., and B. Séraphin, 2006. In vivo targeting of the yeast Pop2 deadenylase subunit to reporter transcripts induces their rapid degradation and generates new decay intermediates. J. Biol. Chem. 281: 25940–25947. [DOI] [PubMed] [Google Scholar]
- Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell et al., 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11: 1640–1650. [DOI] [PubMed] [Google Scholar]
- Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese et al., 1998. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94: 45–53. [DOI] [PubMed] [Google Scholar]
- Grigull, J., S. Mnaimneh, J. Pootoolal, M. D. Robinson and T. R. Hughes, 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 24: 5534–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope, I., and K. Struhl, 1986. Functional dissection of a eukaryotic transcriptional activator protein. Cell 46: 885–894. [DOI] [PubMed] [Google Scholar]
- Ingvarsdotir, K., N. J. Krogan, N. C. T. Emre, A. Wyce, N. J. Thompson et al., 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitue a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 25: 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan, E., and F. Winston, 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15: 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D., E. Ezhkova, B. Li, S. G. Pattenden, W. P. Tansey et al., 2005. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123: 423–436. [DOI] [PubMed] [Google Scholar]
- Lenssen, E., N. James, I. Pedruzzi, F. Dubouloz, E. Cameroni et al., 2005. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol. Cell. Biol. 25: 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.-Y., V. Badarinarayana, D. C. Audino, J. Rappsilber, M. Mann et al., 1998. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17: 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Madison, J. M., and F. Winston, 1997. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet, L., C. Tu, Y. K. Hong, E. O. Shuster and M. A. Collart, 2000. The essential function of NOT1 lies within the CCR4-NOT complex. J. Mol. Biol. 303: 131–143. [DOI] [PubMed] [Google Scholar]
- Mulder, K. W., G. S. Winkler and M. H. T. Timmers, 2005. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. Nucleic Acids Res. 33: 6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko, O., E. Landrieux, M. Feuermann, A. Finka, N. Paquet et al., 2006. The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J. Biol. Chem. 281: 31389–31398. [DOI] [PubMed] [Google Scholar]
- Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett et al., 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527. [DOI] [PubMed] [Google Scholar]
- Powell, D. W., C. M. Weaver, J. L. Jennings, K. J. McAfee, Y. He et al., 2004. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell. Biol. 24: 7249–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates, III and P. A. Grant, 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433: 434–437. [DOI] [PubMed] [Google Scholar]
- Qiu, H., C. Hu, G. J. Zhang, M. J. Swanson, C. Boonchird et al., 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 25: 3461–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro, S., T. Fischer, M.-J. Luo, A. Oreto, S. Brettschneider et al., 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein et al., 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411. [DOI] [PubMed] [Google Scholar]
- Sermwittayawong, D., and S. Tan, 2006. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO J. 25: 3791–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya et al., 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19: 86–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, M. J., Q. Hongfang, L. Sumibcay, A. Krueger, S.-J. Kim et al., 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23: 2800–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers, H. T., and L. Tora, 2005. SAGA unveiled. Trends Biochem. Sci. 30: 7–10. [DOI] [PubMed] [Google Scholar]
- Topalidou, I., M. Papamichos-Chronakis, G. Thireos and D. Tzamarias, 2004. Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment. EMBO J. 23: 1943–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis et al., 2001. The transcription factor associated proteins, Ccr4p and Caf1p, are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386. [DOI] [PubMed] [Google Scholar]
- Tucker, M., R. R. Staples, M. A. Valencia-Sanchez, D. Muhlrad and R. Parker, 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21: 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oevelen, C. J. C., H. A. A. M. Van Teeffelen and M. H. T. Timmers, 2005. Differential requirement of SAGA subunits for Mot1p and Taf1p recruitment in gene activation. Mol. Cell. Biol. 25: 4863–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan, P., T. Ohn, Y.-C. Chiang, J. Chen and C. D. Denis, 2004. Mouse CAF1 can function as a processive deadenylase/3′-5′ exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 279: 23988–23995. [DOI] [PubMed] [Google Scholar]
- Warfield, L., J. A. Ranish and S. Hahn, 2004. Positive and negative functions of the SAGA complex mediated through interaction of Spt8p with TBP and the N-terminal domain of TFIIA. Genes Dev. 18: 1022–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P.-Y. J., and F. Winston, 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22: 5367–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, P.-Y. J., C. Ruhlmann, F. Winston and P. Schultz, 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15: 199–208. [DOI] [PubMed] [Google Scholar]