Abstract
In Saccharomyces cerevisiae exit from mitosis requires the Cdc14 phosphatase to reverse CDK-mediated phosphorylation. Cdc14 is released from the nucleolus by the Cdc14 early anaphase release (FEAR) and mitotic exit network (MEN) pathways. In meiosis, the FEAR pathway is essential for exit from anaphase I. The MEN component Cdc15 is required for the formation of mature spores. To analyze the role of Cdc15 during sporulation, a conditional mutant in which CDC15 expression was controlled by the CLB2 promoter was used. Cdc15-depleted cells proceeded normally through the meiotic divisions but were unable to properly disassemble meiosis II spindles. The morphology of the prospore membrane was aberrant and failed to capture the nuclear lobes. Cdc15 was not required for Cdc14 release from the nucleoli, but it was essential to maintain Cdc14 released and for its nucleo-cytoplasmic transport. However, cells carrying a CDC14 allele with defects in nuclear export (Cdc14-ΔNES) were able to disassemble the spindle and to complete spore formation, suggesting that the Cdc14 nuclear export defect was not the cause of the phenotypes observed in cdc15 mutants.
MEIOSIS is a specialized cell division that produces the haploid cells needed for sexual reproduction, and the completion of meiosis is normally coupled to differentiation programs that produce gametes. In Saccharomyces cerevisiae, sporulation is initiated when diploid cells are grown in the absence of nitrogen and the presence of a nonfermentable carbon source such as acetate (Esposito and Klapholtz 1981). When triggered to enter the sporulation program, cells exit the mitotic cycle from the G1 phase. This is followed by one round of S phase and two meiotic divisions that generate four haploid nuclei, which are packaged into individual spores. As a consequence of this process, four haploid gametes (spores) are formed in the cytoplasm of the mother cell (ascus) (reviewed by Neiman 2005). At the onset of meiosis II, the spindle pole body (SPB) is modified by the incorporation of several meiosis-specific proteins, which together make up the meiosis II outer plaque (Knop and Strasser 2000; Bajgier et al. 2001; Nickas et al. 2003). This change in composition converts the function of the cytoplasmic face of the SPB from microtubule nucleation to membrane nucleation. The process of spore construction requires the de novo assembly of two cellular structures, the prospore membrane (PSM) that is generated around the daughter nuclei to create prospores and a protective spore wall that surrounds the prospores. Once the meiosis II outer plaque of the SPB has been assembled, it becomes a site for the docking and coalescence of secretory vesicles to form flattened double-membrane sheets termed PSMs (Moens 1971; Neiman 1998). These nascent PSMs expand during meiosis II, and at the time of nuclear division each PSM completely engulfs the nuclear lobe to which it is anchored via the SPB. At the completion of meiosis II, each PSM fuses with itself so that each daughter nucleus (and associated cytoplasm) is captured inside the PSM, creating immature spores. The prospores mature into spores through the synthesis of a specific spore wall, which initially occurs in the lumen between the two PSMs (reviewed by Neiman 2005).
During the mitotic cycle, nuclear division is tightly linked to cytokinesis, the final event of the cell cycle that gives rise to two daughter cells. Exit from mitosis is brought about by the inactivation of mitotic cyclin-dependent kinase (CDK) activity. In S. cerevisiae, the Cdc14 protein is a temporally and spatially regulated phosphatase that mediates mitotic exit by promoting CDK inactivation and by dephosphorylating CDK substrates (reviewed by Bardin and Amon 2001; Surana et al. 2002). Cdc14 is sequestered in the nucleolus throughout most of the cell cycle (Shou et al. 1999; Visintin et al. 1999). The Cdc14 early anaphase release network (FEAR) and mitotic exit network (MEN) have been shown to act in a coordinated fashion to trigger exit from mitosis by mediating the release of Cdc14 from the nucleolus. Cdc14 is transiently released in early anaphase by the actions of the FEAR pathway, and the MEN allows levels of nuclear Cdc14 to remain high until mitotic exit is complete (reviewed by Bardin and Amon 2001; Mccollum and Gould 2001; D'amours and Amon 2004). In addition to mediating Cdc14 release at different times, both pathways have been shown to affect different processes. The FEAR pathway is critical for nuclear positioning in anaphase while the MEN is essential for mitotic exit. The MEN is a signal transduction pathway composed of the GTP-binding protein Tem1 and its putative guanine nucleotide exchange factor Lte1, the protein kinases Cdc5, Cdc15 and Dbf2/Dbf20, the Dbf2-associated factor Mob1, and the SPB protein Nud1 (reviewed by Bardin and Amon 2001). Conditional lethal temperature-sensitive (ts) mutations in any of the MEN genes cause cells to arrest in late mitosis with elevated mitotic CDK activity when shifted to the restrictive temperature.
Recent studies have suggested that the roles of Cdc14 and the FEAR network during meiosis I are similar to their roles in mitosis. Cdc14 release by the FEAR pathway is essential for the inactivation of Clb-CDKs and exit from meiosis I (Buonomo et al. 2003; Marston et al. 2003). FEAR mutants (such as slk19Δ and spo12Δ) are unable to disassemble the spindle at anaphase I. The FEAR pathway performs its meiotic role by regulating Cdc14 release from the nucleolus. Conversely, MEN genes are not required for exit from meiosis I (Kamieniecki et al. 2005). Cdc15 or Tem1-depleted cells are able to complete the two meiotic divisions with similar efficiencies to wild-type cells. Interestingly, Cdc15-depleted cells—but not Tem1-depleted cells or lte1 mutants—fail to form mature spores, suggesting that Cdc15, but not Tem1 and Lte1, plays a role in linking exit from meiosis II to spore differentiation.
Here we analyze the role of Cdc15 during meiosis and spore morphogenesis. Cdc15-depleted cells underwent meiotic divisions with kinetics and efficiency similar to those of wild-type cells, but failed to form mature spores. Several defects were observed in these mutants: the inability to disassemble anaphase II spindles, morphological defects in PSM assembly resulting in failure to capture the nuclear lobes, and the persistence of Spo21 at the SPB at the end of meiosis II. Cdc15 was not required for Cdc14 release from the nucleolus at the end of anaphase II, but it was necessary to maintain Cdc14 in its released state and for its transport from the nucleus to the cytoplasm. The phenotypes of Cdc14-ΔNES mutants suggest that Cdc15 could be required to coordinate the meiotic cell cycle with the morphogenetic program that results in the formation of mature spores.
MATERIALS AND METHODS
Yeast strains and methods:
The S. cerevisiae strains used in this study were constructed in the fast-sporulating SK1 background. The wild-type strain AN120 (MATa/MATα ura3/ura3 leu2/leu2 trp1∷hisG/trp1∷hisG ARG4/arg4-NspI his3ΔSK/his3ΔSK hoΔ∷LYS2/hoΔ∷LYS2 lys2/lys2 RME1/rme1Δ∷LEU2) has been described previously (Neiman et al. 2000). HI50 is isogenic to AN120, except that it contains CDC15 under the control of the CLB2 promoter, which is repressed during sporulation. HI50 was constructed by PCR-mediated integration of the CLB2 promoter using pRK69 (Kamieniecki et al. 2005) as template into the haploids AN117-4B and AN117-16D, followed by mating to produce the diploid HI50. Strain SEP41 [his3ΔSK/his3ΔSK trp1∷hisG/trp1∷hisG ura3/ura3 leu2/leu2 lys2/lys2 arg4-NspI/arg4-NspI hoΔ∷LYS2/hoΔ∷LYS2 rme1Δ∷LEU2/rme1Δ∷LEU2 cdc14∷kanMX4/cdc14∷kanMX4 pEP36 (CDC14-ΔNES)] contains the Cdc14 nuclear export allele (Cdc14-ΔNES; with residues 359–367 deleted) (Bembenek et al. 2005), and it was constructed by deleting the wild-type CDC14 gene in two spores of opposite mating type derived from AN120 that carried plasmid pEP36 and then mating the transformants to generate the diploid SEP41.
For sporulation, overnight liquid cultures were grown in YEPD or SC media (Sherman et al. 1986) to saturation. Cultures were then diluted 1:300 in YPAcetate (1% yeast extract, 2% peptone, 2% potassium acetate), grown to midlog phase, and transferred to 2% potassium acetate at a final concentration of 3 × 107 cells/ml (Neiman 1998). Aliquots of the cultures were removed at different times. Yeast strains were transformed by the lithium acetate protocol (Gietz and Woods 2002).
Plasmids:
Plasmid pC763 carries a XhoI–BamHI fragment containing the CDC14 gene obtained from plasmid pUG121 (Gruneberg et al. 2000), cloned into the same sites of pRS426. pC774 carries SIC1 under the control of the SPO20 promoter cloned in pRS424. To construct it, a PCR fragment carrying nucleotides −731 to −1 of the SPO20 promoter was used to replace the TEF1 promoter in plasmid pRS424-TEF to generate pRS424-Spo20pr. The SIC1 coding region was then isolated from plasmid YIpPGK-SIC1 (Toyn et al. 1997) by digestion with EcoRI and cloned into the similarly digested pRS424-SPO20pr plasmid. Plasmid pEP36 contains the CDC14-ΔNES allele (Bembenek et al. 2005) cloned into vector pRS426, and it was constructed by PCR amplification of a DNA fragment in which amino acids 359–367 were deleted, and then replacing the EcoRI–XhoI from plasmid pEP17 (containing the wild-type CDC14 gene) with the amplified fragment.
Microscopy techniques:
Analysis of meiotic progression and visualization of in vivo fluorescent proteins and indirect immunofluorescence were accomplished using a Leica DMXRA microscope equipped for Nomarski optics and epifluorescence. Images were captured using a Photometrics Sensys CCD camera. For visualization of GFP fluorescence, samples were fixed with formaldehyde 4% for 10 min, washed once in PBS, and placed in mounting media (Vectashield; Vector Laboratories, Burlingame, CA) containing DAPI (4′,6′-diamino-2-phenylindole). For some figures, an Axioplan2 microscope (Carl Zeiss, Thornwood, NY) was used. Images were then acquired using a Zeiss mRM Axiocam and deconvoluted using Zeiss Axiovision 3.1 software. Transmission electron microscopy was performed as previously described (Coluccio et al. 2004); images were captured on a JEOL 1200 EX microscope at 80 kV.
Indirect immunofluorescence was performed as previously described (Salah and Nasmyth 2000). Primary antibodies were used at 1:100 dilution and incubated for 1 hr. Samples were washed five times with PBS-BSA 1% and incubated with secondary antibodies (1:200) for 1 hr. After incubation, antibodies were removed by washing thoroughly with PBS buffer. Samples were mounted with DAPI mounting media. The primary antibodies used were anti-Tubulin (Sigma 75168, St. Louis); anti-Cdc14 (YE17 sc12045, Santa Cruz Biotechnology, Santa Cruz, CA); anti-Cdc11 (Y415 sc7170, Santa Cruz Biotechnology); or anti-GFP (Living colors 8371, Becton Dickinson). Different secondary antibodies coupled to Alexa Fluor 488, 594, or 633 (Invitrogen Molecular Probes, Invitrogen, Carlsbad, CA) were used.
Protein methods:
Protein extracts were prepared using TCA precipitation as described (Klein et al. 1999). Samples were separated in NUPAGE Novex 10–12% Bis-Tris gels (Invitrogen) according to the recommendations of the supplier. For Western blot, 25 μg of protein extracts were transferred to P-Hybond membranes (GE Healthcare) using an X-Cell Sure Lock mini-cell system (Invitrogen). Anti-Cdc14 antibody (YE-17 sc12045, Santa Cruz Biotechnology; 1:500) or anti-Pgk1 antibody (mouse monoclonal 22C5, A6457 Invitrogen Molecular Probes, 1:3000) were used. Horseradish peroxidase-conjugated anti-goat antibody (R21459, Invitrogen Molecular Probes) or anti-mouse antibody (NA931V GE Healthcare) were used as secondary antibodies. Detection of proteins was performed using the ECL kit (GE Healthcare). For detection of Cdc15-HA, protein extracts from cells incubated in sporulation medium for different times were prepared in B70 buffer (Zachariae et al. 1998), adding protease inhibitors (Complete X, Roche). Cdc15-HA was immunoprecipitated from 1 mg of protein extract using the μMACS-HA isolation kit (Miltenyi Biotec) according to supplier recommendations. Immunoprecipitated material was separated by SDS–PAGE and incubated anti-HA antibody (Covance, 1:1000) at 4° and developed with horseradish peroxidase-conjugated anti-mouse antibody as secondary antibody.
RESULTS
Cdc15 function is required at the end of meiosis II:
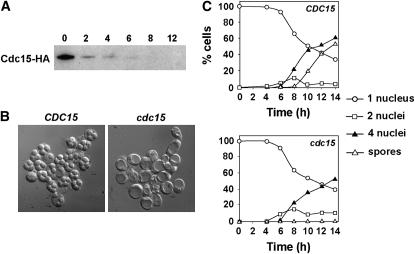
To analyze the role of Cdc15 during meiosis, we used a conditional mutant in which the expression of CDC15 was under the control of the CLB2 promoter (PCLB2-CDC15), which is expressed during vegetative growth but is repressed during sporulation. This allele significantly reduced Cdc15 protein after 4 hr of transfer to sporulation medium, and the protein was undetectable by Western blot after 6 hr of incubation (Figure 1A). For this reason, this strain behaved as a conditional mutant, which was viable during vegetative growth but was unable to form spores. This construct was introduced into the fast-sporulating SK-1 background, and we observed that mature spores were never seen in this strain, even after prolonged incubation times in sporulation medium (Figure 1B). This suggested that Cdc15 is essential for completion of sporogenesis.
Figure 1.—
CDC15 is not required for DNA replication or meiotic segregation. (A) Levels of Cdc15 protein in cells with the PCLB2-CDC15 construct. Cdc15 was immunoprecipitated from 1 mg of protein extracts before Western blot analysis. Antibodies against the N-terminal HA epitope tags were used for detection and immunoprecipitation. Samples were collected at the indicated number of hours after transfer to sporulation medium (0, 2, 4, 6, 8, and 12 hr). (B) Depletion of CDC15 during sporulation blocks the formation of spores. DIC images of wild type (CDC15) or PCLB2-CDC15 mutant (cdc15) incubated for 24 hr in sporulation medium. (C) Time course of appearance of binucleate cells (squares) or tetranucleate cells (triangles) in wild type (CDC15) or PCLB2-CDC15 mutant (cdc15). Strains were incubated in sporulation medium for the indicated times (hours) before aliquots were collected and the cells were stained with DAPI. At least 200 cells were counted for each time point.
To determine the moment at which Cdc15 exerts its function during meiosis, the kinetics of the appearance of bi- and tetranucleate cells was measured in wild-type and cdc15-depleted cells incubated in sporulation medium for different time intervals. Microscopic inspection of the DAPI-stained cells indicated that bi- and tetranucleate cells were present in the PCLB2-CDC15 mutant. When progression through meiosis was quantified, the results indicated that the kinetics of appearance of bi- and tretanucleate cells in PCLB2-CDC15 mutant was slightly delayed and they showed a reduction in the percentage of cells with four nuclei after 14 hr of incubation compared to the wild-type strain (Figure 1C). Mature spores started to appear after 10 hr of incubation and accounted for ∼80% of the culture by 24 hr of incubation in the wild-type strain. However, PCLB2-CDC15 mutants arrested at the end of meiosis after the four nuclei had separated, and mature spores were never observed in the mutant. No differences in the timing of DNA replication as assayed by FACS analysis were observed between the wild-type and cdc15 mutant strain (data not shown). Together, these results indicate that CDC15 is not required for DNA duplication or meiotic segregation, but that it does function at the end of meiosis II.
Cdc15 is required for postanaphase II spindle disassembly:
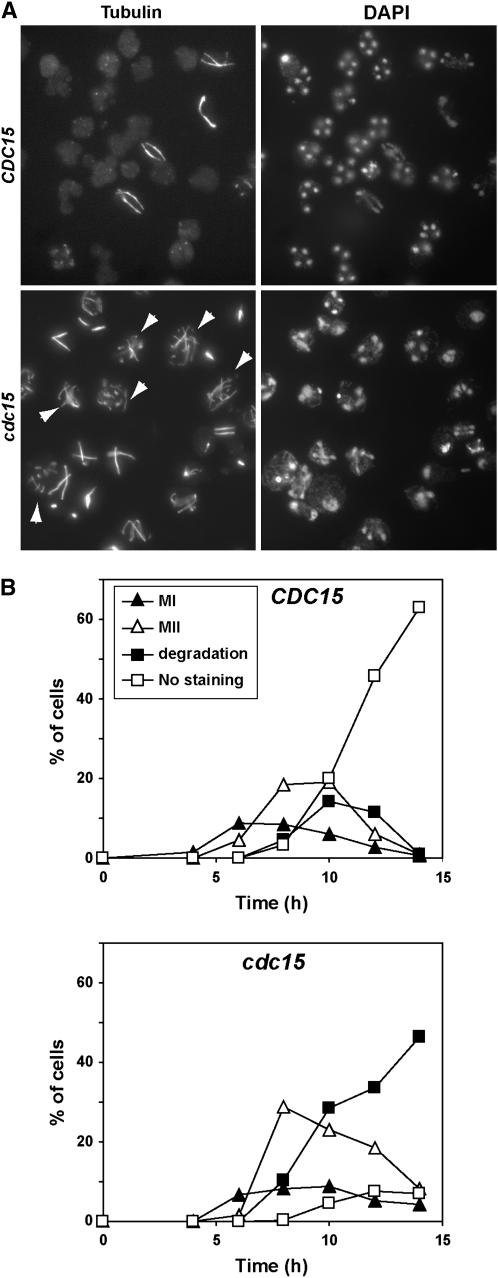
To analyze the nature of the defect of PCLB2-CDC15 mutants in more detail, we monitored the kinetics of spindle assembly and disassembly during the different phases of meiosis using indirect immunofluorescence. The appearance of meiosis I spindles and their morphology were similar in wild-type and PCLB2-CDC15 mutants, suggesting that Cdc15 function is not essential during the first meiotic division. The assembly of meiosis II spindles was also similar in both strains, but the PCLB2-CDC15 mutant was delayed in meiosis II spindle disassembly as judged by the persistence of cells with anaphase II spindles (Figure 2A). At the end of meiosis II, wild-type cells disassembled the spindle and initiated the morphogenetic program that led to the formation of mature spores. In contrast, the PCLB2-CDC15 mutant failed to normally disassemble the anaphase II spindles, and in many cells these collapsed or broke down into several fragments (arrowheads in Figure 2A).
Figure 2.—
Cdc15-depleted cells have defects in meiotic spindle disassembly. (A) Nuclear and spindle morphology in wild-type and cdc15 mutants. Wild type (CDC15) and PCLB2-CDC15 mutant (cdc15) were incubated in sporulation medium; samples were collected at different time intervals and stained with anti-tubulin antibody (left) and DAPI (right). The image shows an example of cells that had been incubated in sporulation medium for 10 hr. (B) Quantification of the different spindle morphologies during the sporulation process in wild-type (CDC15) and PCLB2-CDC15 (cdc15) cells. Cells from the meiotic time course shown in Figure 1C were stained with anti-tubulin antibody and classified according to the spindle morphology. Meiosis I includes cells in prophase, metaphase I, and anaphase I, whereas cells with metaphase II or anaphase II spindles were classified as meiosis II. Postmeiotic cells that had already disassembled the meiotic spindle and were negative for tubulin staining were classified as “no staining.” Cells in meiosis II with broken or collapsed spindles were classified as “degradation.”
Quantitation of the number of cells with different types of spindles confirmed these results. No apparent differences were observed for meiosis I spindles (Figure 2B), which disassembled with similar kinetics in both strains. Assembly of the meiosis II spindle in the PCLB2-CDC15 strain took place at approximately the same time as the wild-type strain. However, there was a slight delay in meiosis II progression in Cdc15-depleted cells, as judged by the accumulation of meiosis II spindles (Figure 2B). The percentage of cells with meiosis II spindles was higher in the PCLB2-CDC15 mutant than in the wild type, suggesting either a defect in spindle disassembly or a delay in progression through meiosis II. However, the most significant differences were observed after meiosis II, since most of the wild-type cells had disassembled the spindle and showed no tubulin staining after 10–12 hr of incubation in sporulation medium (70% of the cells), while only 8% of the cdc15 depleted cells showed no staining. In contrast, ∼50% of the PCLB2-CDC15 mutant cells showed abnormal tubulin staining. Thus, these results indicate that Cdc15 is required for proper spindle disassembly after meiosis II.
PSM formation is aberrant in cdc15 mutants:
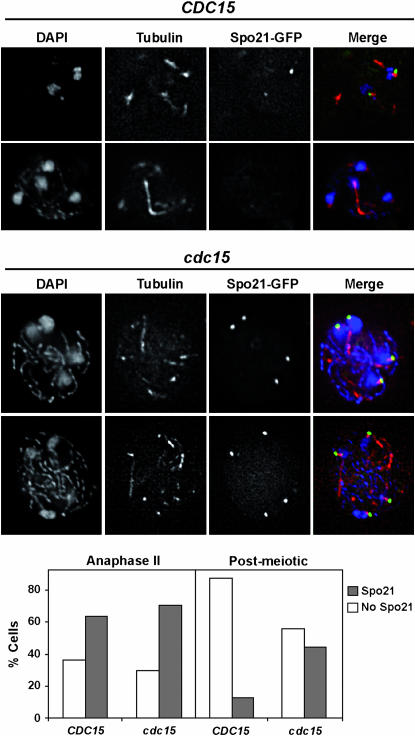
Since the depletion of Cdc15 results in defects at the end of meiosis, we analyzed whether the morphogenetic program leading to spore morphogenesis was affected in these cells. During the second meiotic division, the outer plaque of the SPB acts as a nucleation center for the assembly of the PSM. Spo21 is a sporulation-specific component of the outer plaque, and it localizes to the meiotic SPB (Knop and Strasser 2000; Bajgier et al. 2001). As meiosis II progresses, the PSM engulfs the adjacent nuclear lobe and later fuses with itself, creating immature spores. To test whether spore morphogenesis was altered in Cdc15-depleted cells, we first checked if SPB outer plaque assembly occurred normally in mutant cells using a functional Spo21-GFP fusion (Bajgier et al. 2001). We found that Spo21 localized to the four SPBs of anaphase II cells, and no significant differences in the percentage of cells displaying Spo21 staining were detected between wild-type and PCLB2-CDC15 mutants (Figure 3). However, the Spo21-GFP fluorescence disappeared at the time of spindle disassembly in wild-type cells, while in cdc15 mutants it persisted and remained associated with the four SPBs even after some of the microtubules had started to fragment. The percentage of cdc15 postmeiotic cells showing Spo21 at the SPBs when the spindle collapsed was ∼45%.
Figure 3.—
cdc15 mutants have defects in SPB modification. Spo21 localizes to the SPB in Cdc15-depleted cells during sporulation. Wild type (CDC15, top) or PCLB2-CDC15 mutant (cdc15, bottom) expressing SPO21-GFP (from plasmid pSPO21) were transferred to sporulation medium. After 8 hr of incubation, cells were collected and stained with anti-tubulin antibody and DAPI. Images of Spo21-GFP (green), tubulin (red), and DAPI (blue) and the merging of the three channels are shown. The graph shows the percentage of wild-type (CDC15) or cdc15 mutant cells with Spo21 staining (open rectangles) or without staining (shaded rectangles) in anaphase II or postmeiotic cells. At least 100 cells were counted for each category.
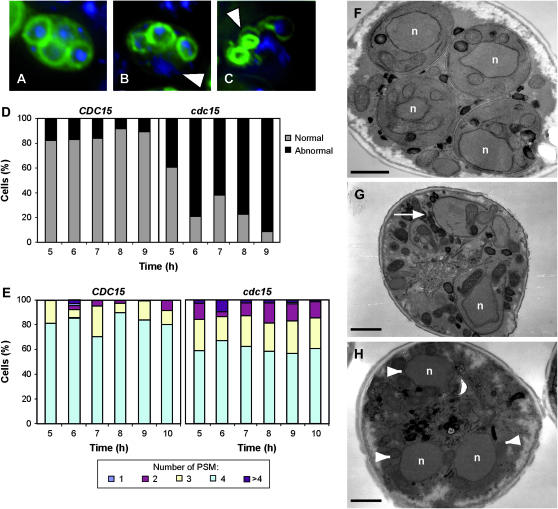
The formation of PSM was monitored using a Spo20-GFP fusion protein (Nakanishi et al. 2004). Spo20p is a sporulation-specific SNAP-25 homolog that is required for the developmentally regulated branch of the secretory pathway that leads to spore morphogenesis (Neiman 1998). In wild-type cells, each nucleus was usually surrounded by a PSM, which displayed a regular shape, and the four compartments in each cell were similar in size (Figure 4A). However, in PCLB2-CDC15 mutants there was considerable variation in the number, size, and shape of the PSMs. In particular, cells often displayed fewer than four PSMs, and the membranes that did form were frequently misshapen and failed to capture nuclei (Figure 4, B and C). The defects in PSM formation and nuclear capture were quantified to gain an idea of the extent of the phenotype. The results indicated that the morphology of the PSM was aberrant in a large proportion of the cells and that the defect increased over time, ∼90% of abnormal PSMs being observed at 9 hr (Figure 4D). Also, the number of PSMs per cell varied more than in wild-type cells, and the percentage of cells with four PSMs was lower than in wild-type cells (Figure 4E). Finally, nuclear capture was also deficient in PCLB2-CDC15 cells, since only 45% of the PSMs were able to correctly surround a nucleus in mutant cells as compared to the wild type (∼83%). The failure to capture nuclei was also evident in ultrastructural studies of the PCLB2-CDC15 cells, and examples of PSMs without a nucleus and uncaptured nuclei were often seen (Figure 4, G and H). This was in contrast to wild-type cells, in which the nuclei were almost always seen to be within PSMs (Figure 4F). Interestingly, the electron microscopy analysis also revealed that prospores that formed in the PCLB2-CDC15 strain failed to initiate spore wall assembly, as indicated by the lack of spore wall material in the lumen of the PSM. Taken together, these results clearly suggest that CDC15 function is required for normal PSM formation and nuclear capture after the assembly of the membrane-nucleating center on the SPB.
Figure 4.—
PCLB2-CDC15 cells display PSM defects. Fluorescence micrographs of sporulating wild-type (A) and PCLB2-CDC15 cells (B and C). PSMs are visualized in green using GFP-Spo2051−91, and nuclei are shown in blue using DAPI. Arrowheads in B indicate unpackaged nuclei in postmeiotic cells and abnormal PSMs in C. (D) Quantification of the PSM morphology during sporulation in wild type (CDC15) and PCLB2-CDC15 mutant (cdc15). At least 200 cells in which the PSM was visible were counted for each strain. The graph shows the percentage of cells with normal morphology (shaded bars) or aberrant shapes (solid bars) in cells with visible PSMs. Cells with at least one abnormal PSM were counted as aberrant. (E) Quantification of the number of PSMs during sporulation in wild type (CDC15) and PCLB2-CDC15 mutant (cdc15). At least 200 cells in which the PSM was visible were counted for each strain. (F–H) Electron micrographs of sporulating wild type (F) and PCLB2-CDC15 cells (G and H). White arrow in G indicates a growing PSM that is not encapsulating a nucleus (indicated with n in all images). Arrowheads in H indicate multiple, unpackaged nuclei in a postmeiotic cell. Bars, 1 μm.
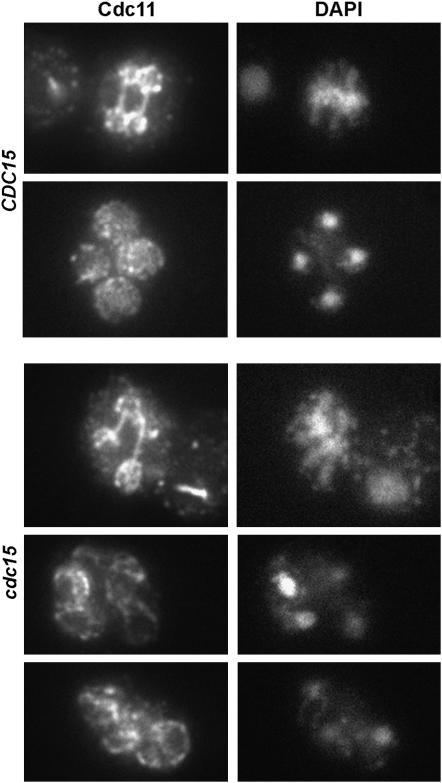
Septin organization is normal in Cdc15-depleted cells:
Since the defect produced by the depletion of Cdc15 resulted in a block in the late steps of the sporulation process, we also analyzed the localization of septins in wild-type and cdc15 mutants. During meiosis, septins are assembled as sheets that line the PSM (De Virgilio et al. 1996; Fares et al. 1996; Tachikawa et al. 2001). The localization of Cdc11 was analyzed by indirect immunofluorescence using anti-Cdc11 antibodies. In wild-type cells, Cdc11 was found to form rounded structures that surrounded the nuclear lobes after anaphase II, later becoming localized in a pattern that closely resembled the PSM (Figure 5). No apparent differences were found in the PCLB2-CDC15 mutant at any stage of the sporulation process, and the localization was very similar to that observed for wild-type cells (Figure 5). Similar results were obtained with a GFP fusion to the sporulation-specific septin Spr28 (De Virgilio et al. 1996) (data not shown). Thus, these results suggest that Cdc15 is required for late steps of spore morphogenesis, but that it is not necessary for the assembly of the septin structure that underlies the PSM.
Figure 5.—
Septins are normally assembled in cdc15 mutants. Localization of Cdc11 in wild type and PCLB2-CDC15 mutant. Cells were incubated in sporulation medium. Samples were collected at different times and stained with anti-Cdc11 antibody and DAPI and inspected for septin morphology. Images of Cdc11 (left) and DAPI (right) are shown.
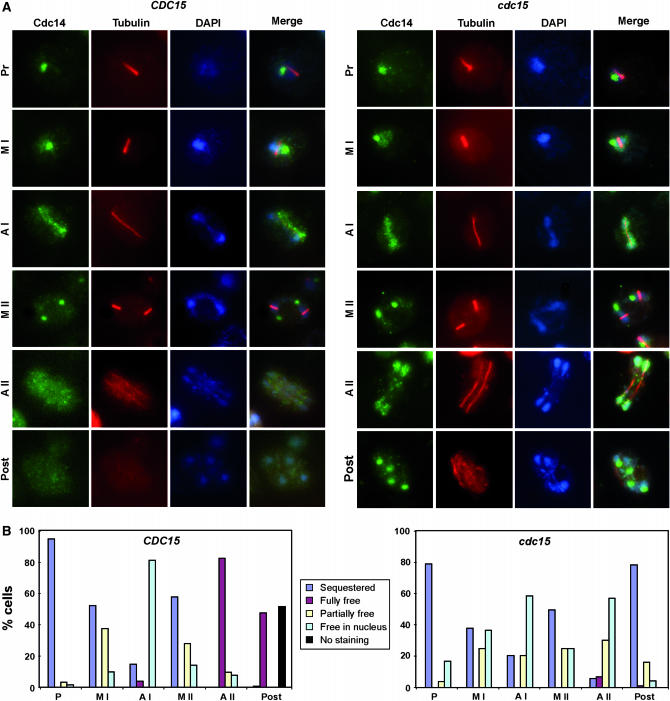
Cdc15 is not required for Cdc14 release from the nucleolus but is involved in its nucleo-cytoplasmic transport:
During vegetative growth, Cdc15 is required for exit from mitosis. This kinase is an upstream component of the MEN, a signaling pathway whose activation results in the release of the protein phosphatase Cdc14 from the nucleolus. Cdc14 remains sequestered in the nucleolus during most of the cell cycle (Shou et al. 1999; Visintin et al. 1999). To test whether Cdc15 might perform a similar function during meiosis, we analyzed Cdc14 localization as the cells progressed through meiosis by indirect immunofluorescence. We found that in wild-type cells, Cdc14 exhibited a localization pattern similar to that described previously (Buonomo et al. 2003; Marston et al. 2003); that is, Cdc14 resided in the nucleolus throughout meiotic prophase I and metaphase I (Figure 6, A and B), and it was released from the nucleolus to the nucleus upon entry into anaphase I. During metaphase II, Cdc14 was resequestered into the nucleoli before being finally released uniformly into the nucleus and the cytoplasm upon anaphase II entry (Cdc14 was released in >80% of the wild-type cells). Finally, as sporogenesis progressed Cdc14 staining was observed in the cytoplasm (40% of the cells), but it also disappeared from a large proportion of the cells (40%).
Figure 6.—
Localization of Cdc14 during sporulation. (A) Wild type (CDC15, left) or PCLB2-CDC15 mutant (cdc15, right) were transferred to sporulation medium. At different times after the induction of sporulation, cells were collected and stained with anti-Cdc14 and anti-tubulin antibodies and DAPI. Anti-goat Alexa Fluor 488 and anti-mouse Alexa Fluor 633 were used as secondary antibodies. Cdc14 (green), tubulin (red), and DAPI (blue) images are shown, as well as the merged image. Representative images of the different meiotic phases are shown for each strain. (B) Localization of Cdc14 during the different stages of the sporulation in wild-type (left) or PCLB2-CDC15 (right) cells. Localization of Cdc14 in each phase of the sporulation process was classified as sequestered in the nucleolus (blue), fully (maroon) or partially (yellow) released from the nucleolus, free in the nucleus (light blue), or cells showing no staining (black). For wild-type cells, the different localization is presented as a percentage of metaphase I (MI; n = 109), anaphase I (AI; n = 92), metaphase II (MII; n = 149), anaphase II (AII; n = 155), or spores (n = 113) with visible Cdc14. For PCLB2-CDC15 cells, a similar percentage for metaphase I (MI; n = 112), anaphase I (AI; n = 74), metaphase II (MII; n = 128), anaphase II (AII; n = 132), or postmeiotic cells (n = 64) is shown.
In PCLB2-CDC15 mutants, Cdc14 localization during meiosis I and metaphase II was similar to that observed in the wild-type strain (Figure 6, A and B). However, during anaphase II important differences between the wild-type and mutant were detected, since in most of the mutant cells Cdc14 was released from the nucleoli, as happens in the wild-type strain, but failed to reach the cytoplasm and accumulated in the nucleus of the cells (∼80% of the cells presented Cdc14 nuclear staining). In addition, at later steps Cdc14 was resequestered into four intense foci in postmeiotic cells (∼80%) instead of being exported. These results therefore indicate that Cdc15 is not required for Cdc14 release from the nucleoli during meiosis, but it is necessary to maintain Cdc14 in its released state and for its transport from the nucleus to the cytoplasm of the cells in anaphase II.
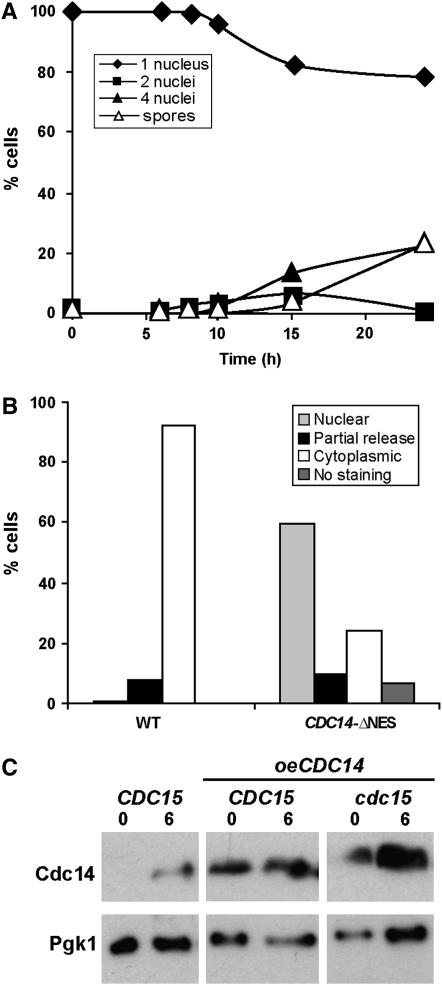
A Cdc14 allele with a nuclear export defect affects entry into meiosis but not spore formation:
To examine if Cdc14 nuclear export is important for sporogenesis, we examined the sporulation in strains containing a Cdc14 allele in which the nuclear export signal (NES) had been deleted. The Crm1-dependent nuclear export of Cdc14 is important for the coordination of mitotic exit and cytokinesis during vegetative growth, since cells carrying a Cdc14-ΔNES allele have an abnormal morphology and defects in cytokinesis (Bembenek et al. 2005). We therefore constructed a homozygous diploid strain carrying the CDC14-ΔNES allele in the SK1 background. This strain was induced to sporulate, and progression through meiosis was monitored by DAPI staining and microscopic inspection of spores. The results indicated that the Cdc14-ΔNES allele produced a marked delay in the onset of sporulation and a reduction in the percentage of mature spores in comparison with the wild-type strain, in which mature spore production was never >20% (Figure 7A). This delay could be due to the defects in cytokinesis described for this mutant allele (Bembenek et al. 2005) and the consequent defects in exiting the mitotic cycle from the G1 phase to enter the meiotic program. However, cells that started meiosis were able to form mature spores, as judged by the fact that the percentages of tetranucleate cells and mature spores were similar.
Figure 7.—
Importance of Cdc14 nuclear export in sporulation. Cdc14-ΔNES strains have defects in sporulation. Time course of appearance of binucleate (squares) and tetranucleate cells (triangles) and spores (white triangles) in cells carrying Cdc14-ΔNES (SEP41). Cells were incubated in sporulation medium for the indicated times (hours) before aliquots were collected and stained with DAPI. At least 200 cells were counted for each time point. (B) Localization of Cdc14 and Cdc14-ΔNES in anaphase II cells. Cells were collected and stained with anti-Cdc14 and anti-tubulin antibodies and DAPI. Localization of Cdc14 in the fraction of cells that had entered meiosis was classified as nuclear (lightly shaded bar), cytoplasmic (open bars), partially released (solid bars), or without staining (darkly shaded bar). At least 100 cells in anaphase II were counted. (C) Analysis of Cdc14 protein levels in the wild-type strain (CDC15) and in the wild-type and cdc15 mutant strains overexpressing CDC14 (CDC15 oeCDC14 and cdc15 oeCDC14, respectively). Samples were collected at 0 and 6 hr after transfer to sporulation medium to prepare protein extracts. Western blot was performed using anti-Cdc14 antibody. Anti-Pgk1 was used as loading control.
To test whether the Cdc14-ΔNES also produced a defect in nuclear export during sporulation, samples were stained with anti-Cdc14 antibody for microscopic inspection. In Cdc14-ΔNES cells that had entered the meiotic cycle, Cdc14 localization during meiosis I and metaphase II was similar to that observed in the wild type. However, in anaphase II cells important differences between the wild type and mutant were detected. From those cells that had entered the meiotic program, only ∼25% of them showed a clear cytoplasmic staining for Cdc14, while in the rest the Cdc14 nuclear export was impaired (Figure 7B). Furthermore, staining with anti-tubulin antibody failed to indicate defects in meiosis II spindle disassembly in Cdc14-ΔNES cells (data not shown), even though Cdc14 was retained in the nucleus in a large number of the cells. These observations suggest that the absence of complete Cdc14 nuclear export is not the basis for the cdc15 sporulation defect.
Overexpression of MEN components does not complement the absence of Cdc15:
The components of the MEN pathway show multiple genetic interactions with one another during vegetative growth, including high-copy suppression of the conditional lethal growth defect of some mutations (Jaspersen et al. 1998). For example, overexpression of CDC14 can rescue the phenotype of cdc15-2 mutants (Grandin et al. 1998). In addition, Sic1 is a specific inhibitor of cyclin-Cdc28 complexes whose overexpression can also complement a cdc15 mutation (Mendenhall 1993; Schwob et al. 1994; Jaspersen et al. 1998). We tested whether overexpression of Cdc14 phosphatase was enough to bypass the Cdc15 requirement during sporulation. Since CDC14 expression increases during sporulation (Chu et al. 1998), we used a plasmid containing CDC14 cloned under the control of its own promoter. Increase in Cdc14 protein levels during sporulation in these strains was assessed by Western blot (Figure 7C). Even though Cdc14 protein levels were higher in the strains overexpressing CDC14, no complementation of the spore formation defect or the spindle disassembly defect of PCLB2-CDC15 mutants was observed (data not shown). Additionally, we tested whether overexpression of the SIC1 inhibitor had any effect on PCLB2-CDC15 mutants during meiosis. To increase the expression level of this gene during sporulation, SIC1 was placed under the control of the SPO20 sporulation-specific promoter, which is induced midway through sporulation (Neiman 1998). We found that SIC1 overexpression was also unable to rescue the sporulation or spindle disassembly phenotypes of PCLB2-CDC15 mutants (data not shown). Taken together with the results of the CDC14-NES experiments described above, these data suggest that the critical function of CDC15 in sporulation is not mediated through Cdc14 and the downregulation of Cdc28 activity.
DISCUSSION
In mitotically dividing cells, exit from mitosis requires the inactivation of mitotic CDKs. Cells lacking Cdc14 function arrest in late anaphase with high CDK activity (Visintin et al. 1998). For most of the cell cycle, Cdc14 resides in the nucleolus owing to its association with the Cfi1/Net1 inhibitor (Shou et al. 1999; Visintin et al. 1999). The dissociation of Cdc14 from the nucleolus during anaphase is controlled by the FEAR and MEN networks (reviewed by D'amours and Amon 2004; Stegmeier and Amon 2004). At the onset of anaphase, the FEAR network initiates the release of Cdc14, and the phosphatase spreads throughout the nucleus. During later stages of anaphase, the MEN promotes further release of Cdc14 and maintains the phosphatase in its released state, and during telophase the phosphatase is also found in the cytoplasm. Cdc14 activated by the MEN is mainly responsible for promoting exit from mitosis, because this function does not occur in the absence of MEN function (Visintin et al. 1997; Jaspersen et al. 1998).
The functions of Cdc14, FEAR, and MEN during meiosis are less well known. The transition from meiosis I to meiosis II requires that meiotic CDK activity should be sufficiently lowered to allow the disassembly of the meiosis I spindle but not so much as to permit the assembly of prereplicative complexes onto the DNA, which would result in a new S phase. It has been shown that exit from meiosis I requires only the FEAR pathway, since mutants in this pathway are impaired in meiosis I spindle disassembly, whereas MEN inactivation does not affect exit from the first meiotic division (Kamieniecki et al. 2000; Buonomo et al. 2003; Marston et al. 2003; Kamieniecki et al. 2005). Notably, even though chromosome segregation during meiosis I is not efficient in FEAR network mutants, they are able to progress through meiosis II and form two spores (dyads) instead of four (tetrads) (Sharon and Simchen 1990; Marston et al. 2003). As for the MEN, previous work has shown that cells bearing the PCLB2-CDC15 or the PCLB2-TEM1 alleles or a deletion of LTE1 all proceed through the meiotic nuclear divisions, but only PCLB2-CDC15 cells cannot form spores (Kamieniecki et al. 2005). Our more detailed characterization of the PCLB2-CDC15 phenotype indicates that despite the normal kinetics of chromosome behavior, Cdc15 is required for proper exit from meiosis II. In particular, the mutant segregates the four nuclei with similar kinetics to that of the wild type, but fails to properly break down the spindle at the end of meiosis II and fails to export Cdc14 from the nucleus to the cytoplasm. These results suggest Cdc15 has an important function during exit from meiosis II. Similar to its role during exit from mitosis, Cdc15 was required for full Cdc14 release and cytoplasmic accumulation at the end of meiosis II. No differences in Cdc14 localization were observed during meiosis I between the wild type and the cdc15 cells, suggesting that MEN function is not necessary for Cdc14 release at the end of the first meiotic division, as previously described (Kamieniecki et al. 2005). In contrast, the main differences were found at the end of meiosis II, where Cdc14 was partially released to the nucleus in cdc15 cells but was never transported to the cytoplasm. Interestingly, Cdc14 concentrated in four intense foci at the end of meiosis II in cdc15 cells, similar to what has been described for the cdc15-2 mutant during mitosis (Stegmeier et al. 2002; Yoshida et al. 2002). These observations suggest that Cdc14 release from the nucleoli at the end of the second meiotic division is independent of the MEN. It is very likely that the FEAR network is required to initiate Cdc14 release from the nucleoli during anaphase II, but that this function has not been detected before because of its essential function at the end of meiosis I. However, Cdc15 is necessary to maintain Cdc14 in a released state until the end of meiosis II and for its nucleo-cytoplasmic transport. It is interesting that two screens aimed at identifying reduction-of-function mutants that bypass the mitotic exit defect of MEN mutants identified proteins involved in nucleo-cytoplasmic transport, such as the karyopherins Kap104 and Mtr10 (Asakawa and Toh-E 2002; Shou and Deshaies 2002). Furthermore, Crm1-dependent nuclear export of Cdc14 is necessary to coordinate mitotic exit and cytokinesis (Maurer et al. 2001; Bembenek et al. 2005). This suggests that Cdc15-mediated control of Cdc14 export is important for mitotic exit and cytokinesis. Our unpublished observations support the idea that Cdc15 is also involved in nucleo-cytoplasmic transport during sporulation. A screening for high-copy suppressors of the sporulation defect of cdc15 mutants led to the isolation of YRB2. Yrb2 is a Ran-binding GTPase involved in nuclear protein export transport during mitosis (Taura et al. 1997) and interacts with the Crm1/Xpo1 exportin. Overexpression of YRB2 or CRM1 weakly suppressed the phenotypes of PCLB2-CDC15 cells. Although progression through the chromosomal events of mitosis and meiosis displays different requirements for the FEAR and MEN networks (Stegmeier et al. 2002; Buonomo et al. 2003; Marston et al. 2003; Kamieniecki et al. 2005), the data presented here reveal a common requirement for the MEN kinase Cdc15 for promoting Cdc14 export at the end of the nuclear division.
Unlike mitotic cells, however, the principle functions of Cdc15 in sporulation appear not to be tied to Cdc14 localization. The observation that Cdc14-ΔNES cells are able to complete spore formation in the absence of Cdc14 nuclear export (Cdc14-ΔNES) suggests that Cdc15-mediated Cdc14 export is not essential for spore morphogenesis. Although we cannot rule out that a small fraction of Cdc14 not detected by immunofluorescence could be present in the cell cytoplasm in Cdc14-ΔNES cells, the observation that overexpression of the wild-type Cdc14 does not rescue Cdc15 depletion also suggests that the function of Cdc15 in spore formation is independent of Cdc14. Furthermore, cells carrying the ts cdc14-3 allele have defects in chromosome segregation, but are able to complete the sporulation program and form dyads (Sharon and Simchen 1990). Interestingly, studies of several cdc15 mutant alleles have suggested that Cdc15 might play some additional functions that are independent of its effects on Cdc14 activation during mitosis (Shirayama et al. 1996; Jiménez et al. 1998; Jaspersen and Morgan 2000). In particular, Cdc15 function is critical for the formation of the division septum and might also be involved in the direct control of cytokinesis. During sporulation, the main morphogenetic events are PSM formation and spore wall assembly, and the cytokinetic process in meiosis is the closure of the PSM (reviewed by Neiman 2005). In this way, PCLB2-CDC15 cells display abnormally shaped and reduced numbers of PSMs, suggesting a defect in membrane assembly. It is also noteworthy that the prospores that are formed in the PCLB2-CDC15 cells do not appear to initiate spore wall formation. This phenotype may indicate a defect in PSM closure (Tachikawa et al. 2001). Thus, in both vegetative and sporulating cells Cdc15 could be required to complete a cytokinesis event.
It is unclear whether the sporulation morphogenetic defects seen in PCLB2-CDC15 cells would be linked to the meiosis II exit phenotypes or whether they would represent a second function for Cdc15. Several observations suggest that Cdc15 might perform an additional function in spore morphogenesis. First, PSM defects cannot be a consequence of the cell cycle block at the end of meiosis II because PSM synthesis begins shortly after the onset of meiosis II (Neiman 2005). The prospore defects in Cdc15-depleted cells are already evident early in meiosis II, suggesting that Cdc15 might have a function in this stage. Second, perturbation of the cell cycle machinery does not always result in PSM defects or spore wall defects. This can be seen in several cases: the Swm1 subunit of the anaphase promoting complex/cyclosome (APC/C) is required for the efficient turnover of specific APC substrates and for the degradation of Clb2 at the end of mitosis (Ufano et al. 2004; Page et al. 2005). swm1Δ mutants are able to complete PSM closure and arrest at a later stage in spore development (Ufano et al. 1999). Also, mutants in components of the FEAR network (such as cdc14-3, spo12Δ, or slk19Δ) have defects in exit from meiosis I but are able to complete the morphogenesis program and form asci with two spores (dyads) (Sharon and Simchen 1990; Marston et al. 2003). Finally, the Spo13 protein plays a key role in preventing sister centromere separation in meiosis I (Lee et al. 2002; Shonn et al. 2002). Even though spo13Δ mutants have defects in nuclear segregation, they are able to produce two viable diploid spores (Klapholz and Esposito 1980a; Klapholz and Esposito 1980b).
One important question still to be resolved is how MEN is activated at the end of meiosis II and whether the entire MEN functions in exit from meiosis or whether this is a unique function for Cdc15. As noted, mutation of the MEN components TEM1 and LTE1 does not cause sporulation defects (Kamieniecki et al. 2005), although the function of the downstream kinases Dbf2 and Dbf20 during meiosis remains to be tested. In addition, unlike the situation with cdc15-ts mutations in mitotic cells (Grandin et al. 1998), overexpression of Cdc14 failed to rescue the PCLB2-CDC15 phenotype. These results suggest that the sporulation function of CDC15 might be independent of the MEN pathway. Alternatively, it could be speculated that two upstream activators Tem1 and Lte1 might be dispensable during meiosis due to the intrinsic differences between mitotic and meiotic division. Although we have been unable to localize Cdc15 during sporulation, either using a Cdc15-GFP or by indirect immunofluorescence, this kinase localizes to the SPB in mitosis (Cenamor et al. 1999; Xu et al. 2000; Menssen et al. 2001; Visintin and Amon 2001). Thus, it is tempting to surmise that Cdc15 would also reside in the SPBs during meiosis, and that its activation could be related to some specific modification of the outer plaque of the SPB that occurs at the beginning of meiosis II. In fact, Cdc15-depleted cells have defects in the disassembly of the meiotic outer plaque at the end of anaphase II.
In S. pombe, the analog of the MEN is referred to as the septation initiation network (SIN) (for reviews, see Bardin and Amon 2001; Krapp et al. 2004). The SIN has recently been shown to be required for sporulation in S. pombe (Krapp et al. 2006). Interestingly, mutants lacking SIN function proceed through meiosis with normal kinetics but display abnormal forespore membranes, the S. pombe equivalent of the PSM, which fail to package nuclei and form mature spores. No unusual organization of microtubules at late time points was noted in this study, but it is tempting to speculate that the MEN and the SIN might play conserved roles in meiotic exit in the two yeasts. One SIN component characterized in this work was the SPB component cdc11, which serves as the organizing center for the SIN. NUD1, the S. cerevisiae homolog of cdc11, plays an analogous role for the MEN, and it too has been reported to have a sporulation defect (Gordon et al. 2006). Strains harboring a ts allele of NUD1 proceed normally through meiosis but failed to form spores efficiently at the restrictive temperature. Although this is a less severe phenotype than PCLB2-CDC15 or cdc11-ts mutants in S. pombe, it is conceivable that the nud1 sporulation defect might result from a failure to adequately activate Cdc15. Taken together, these different results suggest that the phenotypes seen in the PCLB2-CDC15 strain may indicate a general requirement for the MEN in meiosis II exit and a specific Cdc15 function in spore development.
Acknowledgments
We thank Javier Jiménez, Jaime Correa, and Pedro San Segundo for helpful comments and discussions of the manuscript; Carmen Castro for help and advice with microscopy; Javier Jiménez and Alberto González-Novo for antibodies; Wolfgang Zachariae's lab members for technical advice, suggestions, and discussion, and Nick Skinner for language revision. This research was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (BFU2004-00778) to C.R.V and F.R.I and by National Institutes of Health grant GM62184 to A.M.N. M.E. Pablo is recipient of a fellowship from the Ministerio de Educación y Ciencia (Spain).
References
- Asakawa, K., and A. Toh-e, 2002. A defect of Kap104 alleviates the requirement of mitotic exit network gene functions in Saccharomyces cerevisiae. Genetics 162: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajgier, B. K., M. Malzone, M. Nickas and A. M. Neiman, 2001. SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol. Biol. Cell 12: 1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin, A. J., and A. Amon, 2001. MEN and SIN: What's the difference? Nat. Rev. Mol. Cell. Biol. 2: 815–826. [DOI] [PubMed] [Google Scholar]
- Bembenek, J., J. Kang, C. Kurischko, B. Li, J. R. Raab et al., 2005. Crm1-mediated nuclear export of Cdc14 is required for the completion of cytokinesis in budding yeast. Cell Cycle 4: 961–971. [DOI] [PubMed] [Google Scholar]
- Buonomo, S. B., K. P. Rabitsch, J. Fuchs, S. Gruber, M. Sullivan et al., 2003. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev. Cell. 4: 727–739. [DOI] [PubMed] [Google Scholar]
- Cenamor, R., J. Jimenez, V. J. Cid, C. Nombela and M. Sanchez, 1999. The budding yeast Cdc15 localizes to the spindle pole body in a cell-cycle-dependent manner. Mol. Cell Biol. Res. Commun. 2: 178–184. [DOI] [PubMed] [Google Scholar]
- Coluccio, A., M. Malzone and A. M. Neiman, 2004. Genetic evidence of a role for membrane lipid composition in the regulation of soluble NEM-sensitive factor receptor function in Saccharomyces cerevisiae. Genetics 166: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, S., J. DeRisi, J. Mulholland, D. Botstein, P. O. Brown et al., 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., and A. Amon, 2004. At the interface between signaling and executing anaphase–Cdc14 and the FEAR network. Genes Dev. 18: 2581–2595. [DOI] [PubMed] [Google Scholar]
- De Virgilio, C., D. J. Demarini and J. R. Pringle, 1996. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology 142: 2897–2905. [DOI] [PubMed] [Google Scholar]
- Esposito, R. E., and S. Klapholtz, 1981. Meiosis and ascospore development, pp. 211–287 in The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae, edited by J. N. Strathern, E. W. Jones and J. R. Broach. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Fares, H., L. Goetsch and L. Pringle, 1996. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol. 132: 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., and R. A. Woods, 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Gordon, O., C. Taxis, P. J. Keller, A. Benjak, E. H. Stelzer et al., 2006. Nud1p, the yeast homolog of Centriolin, regulates spindle pole body inheritance in meiosis. EMBO J. 25: 3856–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin, N., A. de Almeida and M. Charbonneau, 1998. The Cdc14 phosphatase is functionally associated with the Dbf2 protein kinase in Saccharomyces cerevisiae. Mol. Gen. Genet. 258: 104–116. [DOI] [PubMed] [Google Scholar]
- Gruneberg, U., K. Campbell, C. Simpson, J. Grindlay and E. Schiebel, 2000. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 19: 6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S. L., J. F. Charles, R. L. Tinker-Kulberg and D. O. Morgan, 1998. A late mitotic regulatory network controling cyclin destruction in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 2803–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S. L., and D. O. Morgan, 2000. Cdc14 activates Cdc15 to promote mitotic exit in budding yeast. Curr. Biol. 10: 615–618. [DOI] [PubMed] [Google Scholar]
- Jiménez, J., V. J. Cid, R. Cenamor, M. Yuste-Rojas, G. Molero et al., 1998. Morphogenesis beyond cytokinetic arrest in Saccharomyces cerevisiae. J. Cell Biol. 143: 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamieniecki, R. J., L. Liu and D. S. Dawson, 2005. FEAR but not MEN genes are required for exit from meiosis I. Cell Cycle 4: e34–e39. [PubMed] [Google Scholar]
- Kamieniecki, R. J., R. M. Shanks and D. S. Dawson, 2000. Slk19p is necessary to prevent separation of sister chromatids in meiosis I. Curr. Biol. 10: 1182–1190. [DOI] [PubMed] [Google Scholar]
- Klapholz, S., and R. E. Esposito, 1980. a Isolation of SPO12–1 and SPO13–1 from a natural variant of yeast that undergoes a single meiotic division. Genetics 96: 567–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz, S., and R. E. Esposito, 1980. b Recombination and chromosome segregation during the single division meiosis in SPO12-1 and SPO13-1 diploids. Genetics 96: 589–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, F., P. Mahr, M. Galova, S. B. Buonomo, C. Michaelis et al., 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103. [DOI] [PubMed] [Google Scholar]
- Knop, M., and K. Strasser, 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19: 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp, A., P. Collin, A. Cokoja, S. Dischinger, E. Cano et al., 2006. The Schizosaccharomyces pombe septation initiation network (SIN) is required for spore formation in meiosis. J. Cell Sci. 119: 2882–2891. [DOI] [PubMed] [Google Scholar]
- Krapp, A., M. P. Gulli and V. Simanis, 2004. SIN and the art of splitting the fission yeast cell. Curr. Biol. 14: R722–R730. [DOI] [PubMed] [Google Scholar]
- Lee, B. H., A. Amon and S. Prinz, 2002. Spo13 regulates cohesin cleavage. Genes Dev. 16: 1672–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston, A. L., B. H. Lee and A. Amon, 2003. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev. Cell. 4: 711–726. [DOI] [PubMed] [Google Scholar]
- Maurer, P., M. Redd, J. Solsbacher, F. R. Bischoff, M. Greiner et al., 2001. The nuclear export receptor Xpo1p forms distinct complexes with NES transport substrates and the yeast Ran binding protein 1 (Yrb1p). Mol. Biol. Cell 12: 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum, D., and K. L. Gould, 2001. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell. Biol. 11: 89–95. [DOI] [PubMed] [Google Scholar]
- Mendenhall, M. D., 1993. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science 259: 216–219. [DOI] [PubMed] [Google Scholar]
- Menssen, R., A. Neutzner and W. Seufert, 2001. Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr. Biol. 11: 345–350. [DOI] [PubMed] [Google Scholar]
- Moens, P. B., 1971. Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. Can. J. Microbiol. 17: 507–510. [DOI] [PubMed] [Google Scholar]
- Nakanishi, H., P. de los Santos and A. M. Neiman, 2004. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 15: 1802–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, A. M., 2005. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 565–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, A. M., L. Katz and P. J. Brennwald, 2000. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 155: 1643–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, A. N., 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas, M. E., C. Schwartz and A. M. Neiman, 2003. Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot. Cell 2: 431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, A. M., V. Aneliunas, J. R. Lamb and P. Hieter, 2005. In vivo characterization of the nonessential budding yeast anaphase-promoting complex/cyclosome components Swm1p, Mnd2p and Apc9p. Genetics 170: 1045–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah, S. M., and K. Nasmyth, 2000. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma 109: 27–34. [DOI] [PubMed] [Google Scholar]
- Schwob, E., T. Bohm, M. D. Mendenhall and K. Nasmyth, 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244. [DOI] [PubMed] [Google Scholar]
- Sharon, G., and G. Simchen, 1990. Mixed segregation of chromosomes during single-division meiosis of Saccharomyces cerevisiae. Genetics 125: 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1986. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shirayama, M., Y. Matsui and A. Toh-e, 1996. Dominant mutant alleles of yeast protein kinase gene CDC15 suppress the lte1 defect in termination of M phase and genetically interact with CDC14. Mol. Gen. Genet. 251: 176–185. [DOI] [PubMed] [Google Scholar]
- Shonn, M. A., R. McCarroll and A. W. Murray, 2002. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev. 16: 1659–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, W., and R. J. Deshaies, 2002. Multiple telophase arrest bypassed (tab) mutants alleviate the essential requirement for Cdc15 in exit from mitosis in S. cerevisiae. BMC Genet. 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed et al., 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233–244. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., and A. Amon, 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38: 203–232. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., R. Visintin and A. Amon, 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108: 207–220. [DOI] [PubMed] [Google Scholar]
- Surana, U., F. M. Yeong and H. H. Lim, 2002. MEN, destruction and separation: mechanistic links between mitotic exit and cytokinesis in budding yeast. BioEssays 24: 659–666. [DOI] [PubMed] [Google Scholar]
- Tachikawa, H., A. Bloecher, K. Tatchell and A. M. Neiman, 2001. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155: 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura, T., G. Schlenstedt and P. A. Silver, 1997. Yrb2p is a nuclear protein that interacts with Prp20p, a yeast Rcc1 homologue. J. Biol. Chem. 272: 31877–31884. [DOI] [PubMed] [Google Scholar]
- Toyn, J. H., A. L. Johnson, J. D. Donovan, W. M. Toone and L. H. Johnston, 1997. The Swi5 transcription factor of Saccharomyces cerevisiae has a role in exit from mitosis through induction of the cdk-inhibitor Sic1 in telophase. Genetics 145: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufano, S., M. E. Pablo, A. Calzada, F. del Rey and C. R. Vázquez de Aldana, 2004. The Swm1p subunit of the APC/Cyclosome is required for activation of the daughter-specific gene expression program mediated by Ace2p during growth at high temperature in Saccharomyces cerevisiae. J. Cell Sci. 117: 545–557. [DOI] [PubMed] [Google Scholar]
- Ufano, S., P. San Segundo, F. del Rey and C. R. Vázquez de Aldana, 1999. SWM1, a developmentally regulated gene is required for spore wall assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 2118–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., and A. Amon, 2001. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell 12: 2961–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers et al., 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk- dependent phosphorylation. Mol. Cell 2: 709–718. [DOI] [PubMed] [Google Scholar]
- Visintin, R., E. S. Hwang and A. Amon, 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398: 818–823. [DOI] [PubMed] [Google Scholar]
- Visintin, R., S. Prinz and A. Amon, 1997. CDC20 and CDH1: a family of substrate-specific activators of APC- dependent proteolysis. Science 278: 460–463. [DOI] [PubMed] [Google Scholar]
- Xu, S., H. K. Huang, P. Kaiser, M. Latterich and T. Hunter, 2000. Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr. Biol. 10: 329–332. [DOI] [PubMed] [Google Scholar]
- Yoshida, S., K. Asakawa and A. Toh-e, 2002. Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr. Biol. 12: 944–950. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., A. Shevchenko, P. D. Andrews, R. Ciosk, M. Galova et al., 1998. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science 279: 1216–1219. [DOI] [PubMed] [Google Scholar]