Abstract
Natural populations of self-incompatible species often exhibit marked phenotypic variation among individuals in the strength of self-incompatibility (SI). In previous studies, we found that the strength of the SI response in Solanum carolinense, a weedy invasive with RNase-mediated SI, is a plastic trait. Selfing can be particularly important for weeds and other successional species that typically undergo repeated colonization and local extinction events and whose population sizes are often small. We applied a PCR-based protocol to identify the S-alleles present in 16 maternal genotypes and their offspring and performed a two-generation greenhouse study to determine whether variation in the strength of SI is due to the existence of weak and strong S-alleles differing in their ability to recognize and reject self-pollen. We found that allele S9 sets significantly more self seed than the other S-alleles in the population we sampled and that its ability to self is not dependent on interactions with other S-alleles. Our data suggest that the observed variations in self-fertility are likely due to factors that directly influence the expression of SI by altering the translation, turnover, or activity of the S-RNase. The variability in the strength of SI among individuals that we have observed in this and our previous studies raises the possibility that plasticity in the strength of SI in S. carolinense may play a role in the colonization and establishment of this weedy species.
IN many species of angiosperms, the occurrence of self-fertilization is discouraged by the presence of a self-incompatibility (SI) system, a genetic mechanism controlled by a multiallelic S-locus that disrupts the growth of self-pollen tubes before reaching the ovules. In the Solanaceae, the SI response involves specific ribonucleases (called S-RNases) that are produced by the pistil and enter the growing pollen tubes, where they degrade messenger and ribosomal RNA of pollen tubes that have an S-allele in common with the pistil. This generalized degradation eventually causes tube growth to arrest within the upper one-third of the style. Although S-RNases determine the specificity of the SI reaction, other genes are required for the rejection mechanism to operate (Cruz-Garcia et al. 2003; Kao and Tsukamoto 2004; Goldraij et al. 2006). Furthermore, it now seems that several unlinked genes are involved in the formation of a multi-protein complex that represents the active form of S-RNase (Cruz-Garcia et al. 2003).
Because of its genetic determination, SI has often been considered to be a qualitative trait of the breeding system of a species, i.e., species with a functional SI system are therefore obligate outcrossers and selfing is not possible. Natural populations, however, often exhibit marked phenotypic variation among individuals in the strength of SI (e.g., Levin 1996; Tsukamoto et al. 1999; Stephenson et al. 2000; Stone et al. 2006). The strength of SI is known to be influenced in some species by environmental conditions such as temperature, by internal stylar conditions such as the age of the flowers, by mutations that directly affect the strength of S-alleles (e.g., weak and strong S-alleles), by mutations that render a specific S-allele functionless, and by unlinked genetic modifiers that can affect the strength of S-alleles in the population (see Levin 1996; Stephenson et al. 2000; Good-Avila and Stephenson 2002; Tsukamoto et al. 2003a,b). In short, there appears to be multiple mechanisms through which the SI system of a species can be partially compromised and thereby permit the possibility of self-fertilization.
When a mutation that allows the possibility of self-fertilization arises in a population (or a genetic variant migrates into the population), its evolutionary fate (i.e., going extinct or becoming fixed) will be determined by the major forces favoring and opposing self-fertilization. There are some advantages associated with the ability to self-fertilize. First, there is an inherent genetic transmission advantage to selfing: a plant donates two haploid sets of chromosomes to each selfed seed and can still donate pollen to conspecifics (Fisher 1941). Second, selfing can be advantageous if it provides reproductive assurance (i.e., when pollinators are scarce or unreliable or when there are few S-alleles in the population) when seed set is limited by the availability of cross pollen (e.g., Stebbins 1957; Baker 1965; Lloyd 1992; Lloyd and Schoen 1992; Schoen et al. 1996). This is particularly important for weeds (Baker 1965) and other successional species (Burd 1994; Busch 2005), which typically undergo repeated colonization and local extinction events and whose population sizes are often small.
Solanum carolinense is a weed that inhabits disturbed habitats, including crop fields, pastures, and occasionally gardens. This species is listed as a noxious weed by the USDA and NRCS (2002) and the Seeds Act and Regulations of Canada (Basset and Munro 1986), and it is classified as an invasive weed in all of the 43 states in which it has been reported (USDA and NRCS 2002). As with other members of the Solanaceae, S. carolinense displays a typical RNase-mediated gametophytic self incompatibility (GSI) system. In previous studies, we have found that the SI response in S. carolinense is a plastic trait in that self-fertility increases with floral age and when no fruits are produced on the first several inflorescences (Stephenson et al. 2003; Travers et al. 2004). A series of controlled pollinations were carried out in the greenhouse using plants collected from a large population of horsenettle from Cumberland, Maryland, and we determined that the ability of the styles to arrest self-pollen tubes in the upper style changes with floral age and the presence of developing fruits (Stephenson et al. 2003). We further examined the effect of prior fruit set on the strength of SI (Travers et al. 2004). This study compared the self-seed production of plants that had a substantial fruit load due to outcrossing with plants bearing very few fruits. As expected for a species that is self-incompatible, outcross pollinations produced more fruits and fruits with more seeds than did self-pollinations. Most (80%) of the outcrossed flowers produced mature fruits with an average of 78 seeds per fruit. In contrast, only 4.5% of the self-pollinations produced fruits and each fruit had an average of only 8 seeds. However, the self-pollinations on the ramets with outcross fruit loads produced significantly fewer fruits (2.7% vs. 6.2%) and fewer seeds per self fruit (2.0 vs. 13.8) than did ramets with no outcross fruit load. There were also significant differences among genets in fruit and seed production from self-pollinations, indicating that there is broadsense heritable variation for this trait. Taken together, these studies suggest that the SI response in horsenettle may have a labile component: some genotypes of S. carolinense are more capable of producing self seed when cross pollen is scarce (older unpollinated flowers and low fruit production), even though the plants have a functional GSI system.
The strong genet effects (consistent among ramets) observed in these experiments indicate the presence of a genetic component in the ability to self-fertilize. The data from these studies, however, suggest that variations in self-fertility are not likely to be due to mutations that render specific S-alleles functionless because such mutations are unlikely to alter the strength of SI with floral age or prior fruit production (see Good-Avila and Stephenson 2002) and because seed set in selfed flowers with functionless S-alleles is likely to be greater than that observed in our earlier studies, especially following hand self-pollinations in which the stigmatic surface was saturated with self-pollen as occurred in those studies. The observed leakiness in the SI system is more likely to be due to factors that directly influence the expression of SI (e.g., by altering the transcription or translation of the S-RNase, its turnover, or its activity) or due to factors that indirectly influence the ability of self-pollen tubes to achieve fertilization (for instance, the nutritional or physical characteristics of the stylar transmitting tissue). Genetically, these factors could be the result of differences among S-haplotypes (weak and strong S-alleles) or unlinked modifiers. In this study, we performed a two-generation greenhouse study to determine whether variation in the strength of SI is due to the existence of weak and strong S-alleles differing in their ability to recognize and reject self-pollen.
MATERIALS AND METHODS
Plant material:
Horsenettle plants were collected from a large population located near State College, Pennsylvania. Rhizome cuttings were collected from 20 plants that were at least 5 m apart to decrease the possibility of sampling sprouts from the same rhizome. These cuttings were brought to the greenhouse, planted in 1-gallon pots, and allowed to resprout, grow, and flower. After flowering, we cut the stems off and moved the pots to a cold room set at 4° to vernalize for 6–8 weeks. After the cold treatment, the pots were returned to the greenhouse and allowed to acclimate for a week. We then created ramets from each of the 20 genets (plants) by dividing the rhizome into 5–6 pieces of similar size. Each rhizome cutting was replanted in a 1-gallon pot and allowed to resprout and grow. Four of the ramets were used in the controlled pollination experiment, and the remaining ramets were returned to the cold room. All of the ramets from 2 of the original 20 genets failed to resprout and therefore could not be used in this study.
Parental generation:
We divided the four ramets per genet into two groups. We performed only outcross pollinations on two ramets and only self-pollinations on the other two ramets. On both self-only ramets and both cross-only ramets per genet, we pollinated six flowers (three young flowers and three older flowers). The outcross pollinations were performed by collecting pollen from at least five different genets using a buzz-pollination device (a modified electric toothbrush) in a microcentrifuge tube, vibrating the tube to thoroughly mix the pollen and then touching the mixture to a stigma. Self-pollinations were made in the same manner except that pollen was collected from 2–3 flowers on the same plants as the flowers to be pollinated. After 48 hr, we collected the styles in 70% ethanol to stop any additional pollen tube growth. These styles were then digested in 500 μl of NaOH 1 m for 1 hr at 50° and then stained in 500 μl of 1% decolorized aniline blue for 24 hr (modified from Martin 1959). Each style was then examined under a UV-light microscope and the number of pollen tubes was counted at three points along the style: right below the stigmatic surface, at the transition zone (25% of the way down the style, where the pollen tubes enter the transmitting tissue of the style), and at the base of the style. On both self-only ramets and both cross-only ramets per genet, we then repeated the assigned (i.e., self or outcross) pollinations every 3–4 days on every flower that opened until a total of 40 flowers per ramet were pollinated. At maturity (∼6 weeks), the fruits were collected and the number of mature seeds produced per fruit was recorded; the seeds were air-dried for 1–2 days and then stored in plastic vials with some desiccant. Two of the 18 genets used in this experiment did not produce enough flowers to complete all pollinations and were therefore excluded from this study. All 16 remaining genets produced at least 20 selfed seeds.
We used two estimates of self-fertilization: (i) the number of pollen tubes reaching the base of the style after 48 hr, and (ii) the number of seeds per pollination to calculate the index of self-compatibility (ISC) using the formula  where nself is the count obtained after self-pollinations and noutcrossed is the count obtained after outcross pollinations; an ISC value of 1 indicates a completely self-compatible genet, whereas an ISC of 0 corresponds to a completely self-incompatible genet. A two-way ANOVA with one replication per treatment (proc GLM, SAS Institute 2002) was performed to compare the arc sine-transformed ISC values obtained from pollen tube growth and self-seed set to detect differences among genets for their ability to self-fertilize.
where nself is the count obtained after self-pollinations and noutcrossed is the count obtained after outcross pollinations; an ISC value of 1 indicates a completely self-compatible genet, whereas an ISC of 0 corresponds to a completely self-incompatible genet. A two-way ANOVA with one replication per treatment (proc GLM, SAS Institute 2002) was performed to compare the arc sine-transformed ISC values obtained from pollen tube growth and self-seed set to detect differences among genets for their ability to self-fertilize.
S-allele determination—parental generation:
To determine the S-genotype of the parental plants, we used a modification of the protocol used by Richman et al. (1995) to isolate and purify total RNA. We used RNA-based amplification in the parental generation because we had no prior knowledge as to which S-alleles were present in this population or whether it contained any S-alleles not previously described. We collected styles from fresh flowers, placed them immediately in RNAlater solution (Invitrogen Carlsbad, CA), and stored them at −20°. Five styles per genet (parental plant) were ground to a fine powder with liquid nitrogen; total RNA was extracted using RNAzol solution (Invitrogen). RT–PCR was performed to obtain cDNAs for the S-RNase-encoding mRNAs present in the styles using the RT–PCR one-step kit (Invitrogen). This cDNA was used in a 20-μl PCR reaction containing 10× PCR buffer, 0.2 mm of each dNTP, 25 ng of the degenerate primers 1 and 3 (Richman et al. 1995), and 1 unit of JumpStart Taq DNA polymerase (Sigma, St. Louis). PCR products were cloned using the TOPO-TA transformation kit (Invitrogen). Single copies of each S-allele were purified from positive transformants and sent to the Nucleic Acid Facility at Pennsylvania State University (University Park, PA) for sequencing. We performed three separate rounds of RNA extraction, cDNA synthesis, and cloning. Each cloning round included a triplicate cloning reaction and two plates per reaction. Ten positive colonies per plate were screened and sent to sequence. Clean, edited sequences were compared against the GenBank database using BLASTn at the NCBI website to assign their allelic identity. For all parental plants, we were able to find two S-alleles, as expected.
Progeny pollinations:
To determine the ISC for selfed and outcrossed progeny produced by each genet in the parental generation, we sowed 20 outcrossed and 20 selfed seeds for each of the 16 genets in plastic trays in the greenhouse. We recorded the number of days to germination and the total number of seeds that germinated. After the first true pair of leaves was produced, we randomly selected 6 outcrossed and 6 selfed seedlings per genet and transplanted them into 1-gallon pots. These pots were distributed on greenhouse benches in a randomized block design with one plant per cross (self or outcross) per genet in each block (for a total of six blocks). We performed self-pollinations on each of the 6 outcrossed and the 6 selfed plants every 3–4 days until 5–7 flowers were pollinated; these flowers were allowed to set fruits. Four weeks later, we performed outcross pollinations in the same fashion. (Our previous studies indicated that the presence of similarly aged outcross fruits often lead to the abortion of selfed fruits.) At maturity, we collected the fruits, counted the number of seeds in each fruit, and calculated the ISC for each of the six selfed and six outcrossed progeny from each genet in the parental generation.
S-allele determination—progeny generation:
To determine the S-genotype of the progeny plants, we applied a modified PCR-based screening protocol using allele-specific primers (Lu 2006). Young leaves were collected in the greenhouse in liquid nitrogen and stored at −80°. Total genomic DNA was extracted from leaf tissue using plant DNAzol extraction buffer (Invitrogen) and ribonuclease A (Invitrogen) and resuspended in 50 μl of DEPC-treated water. Each plant was screened simultaneously for all S-alleles present in the parental population (see Table 1) to ensure proper genotype determination and to reduce the possibility of false positive amplification; selected parental genets comprising all S-alleles present in the original population were amplified along with the progeny samples to serve as positive controls. The PCR amplification of S-alleles was carried out in a 20-μl volume reaction containing 20 ng of DNA, 10× PCR buffer, 10 mm of each dNTP, 10 ng of each forward and reverse allele-specific primers, and 1 unit of HotStart Taq DNA polymerase. The reaction was incubated at 95° for 3 min, followed by 30 cycles of 1 min at 95°, 1 min 30 sec at 60° and 1 min 30 sec at 72°, and a final extension step of 5 min at 72°. For the screening of allele S18 a touchdown protocol was used, with 6 cycles of 1 min at 95°, 1 min 30 sec at an initial annealing temperature of 60° with a 1° decrease per cycle and 1 min 30 sec at 72°, followed by 24 cycles of 1 min at 95°, 1 min 30 sec at 55° and 1 min 30 sec at 72°, and a final extension step of 5 min at 72°. We performed three separate rounds of genotype determination via PCR to further ensure that any observed homozygotes were not due to problems during the screening process. PCR products were run in a 1% agarose gel; to confirm that the amplified bands corresponded to the expected allele sequence, PCR products were cleaned with ExoSAP and sent for sequencing at the Nucleic Acid Facility at Pennsylvania State University. Clean sequences were compared against the NCBI database using BLASTn.
TABLE 1.
S-allele genotypes for 16 genets of S. carolinense and their selfed and outcrossed progeny based on PCR amplification of the S-RNase gene using allele-specific primers
| Selfed progeny
|
Outcrossed progeny
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parental genets | S1 | S2 | S3 | S4 | S5 | S6 | X1 | X2 | X3 | X4 | X5 | X6 | |
| A2 | S5S8 | S8S8 | S8S8 | S5S8 | S8S8 | S5S5 | S5S5 | S5S18 | S8S18 | S5S18 | S8S9 | S8S18 | S5S18 |
| A4 | S5S17 | S5S5 | S5S17 | S5S5 | S5S5 | — | S5S17 | S17S18 | S17S18 | S17S18 | S8S17 | S8S17 | S8S17 |
| A5 | S8S9 | S8S9 | S8S9 | S8S9 | — | S8S9 | S8S9 | S9S18 | S8S18 | S9S18 | S8S18 | S9S18 | S9S18 |
| A6 | S8S9 | S9S9 | S8S9 | S8S8 | S9S9 | S8S9 | S9S9 | S8S17 | S8S18 | S8S18 | S1S9 | S9S18 | S8S18 |
| A7 | S8S9 | S8S8 | S8S9 | S9S9 | S9S9 | S8S9 | S8S9 | S8S18 | S8S18 | S8S18 | S8S18 | S9S18 | S9S18 |
| A9 | S1S17 | — | — | S1S1 | S1S1 | S1S1 | S1S1 | S1S8 | S8S17 | S5S17 | S1S8 | S1S5 | S17S18 |
| B1 | S8S18 | S8S18 | S8S8 | S8S18 | S8S8 | S8S18 | S8S18 | S1S8 | S5S18 | S5S18 | S5S8 | S8S17 | S5S8 |
| B3 | S8S18 | S8S8 | S8S8 | S8S8 | S8S8 | S8S8 | S8S8 | S9S18 | S17S18 | S9S18 | S5S18 | S8S9 | S8S17 |
| B4 | S8S18 | — | S8S18 | S18S18 | S8S18 | — | S8S18 | S1S18 | S5S8 | S1S18 | S5S8 | S9S18 | S1S8 |
| B8 | S8S18 | S8S18 | S8S8 | S8S8 | S8S18 | S8S18 | S8S8 | S5S8 | S1S18 | S1S8 | S5S8 | S5S8 | S5S18 |
| B9 | S8S18 | S8S8 | S8S18 | S8S8 | S8S18 | S8S18 | S8S8 | S9S18 | S9S18 | S1S8 | S8S9 | S8S9 | S8S9 |
| B9b | S8S18 | S8S8 | S8S18 | S8S8 | S8S18 | S8S8 | S8S8 | S1S8 | S1S18 | S1S8 | S1S18 | S1S18 | S1S18 |
| B10 | S1S5 | S5S5 | S5S5 | S1S1 | S1S1 | S1S1 | S5S5 | S1S8 | S5S8 | S1S8 | S1S8 | S5S8 | S5S8 |
| B11 | S8S18 | — | S8S8 | S8S18 | S8S18 | S8S18 | S8S8 | S1S8 | S5S18 | S5S8 | S5S8 | S8S17 | S5S8 |
| C8 | S1S18 | S18S18 | S1S1 | S18S18 | — | — | S1S1 | S1S8 | S8S18 | S1S8 | S1S8 | S1S8 | S1S8 |
| C10 | S1S8 | S8S8 | S8S8 | S8S8 | S8S8 | S1S8 | S8S8 | S8S17 | S1S5 | S1S5 | S8S18 | S5S8 | S8S18 |
Segregation analysis of selfing ability:
To determine if there is an association between specific S-alleles and self-fertility, we compared the self-fertility (self-fruit set, self-seed set, and their ISC) from the outcrossed progeny on the basis of their S-allele composition. To do this, we scored all individuals on the basis of the presence (1) or absence (0) of each allele and treated the data for each allele as a separate random effect. For example, if a plant is S8S9, then this plant will be scored as “1” for both the S8 and S9 alleles and as “0” for all remaining alleles. For each allele, we performed a one-way random effects model ANOVA for the number of selfed seeds per pollination (proc GLM, SAS Institute 2002). Because the selfing ability of particular S-alleles might depend on their association with other S-alleles, we performed a second set of analyses, looking at the ability of the outcrossed progeny to set self seed by genotype. Finally, we analyzed the selfed progeny for their ability to self-fertilize (self-fruit set, self-seed set, and their ISC). We performed a one-way random effects model ANOVA to examine the relationship between the presence of each of five S-alleles with the ability of the selfed progeny to set self seed—allele S17 was excluded because only two selfed individuals were found to carry it (see Table 1).
To test for the effects of genet (i.e., maternal parentage) and cross (selfed or outcrossed progeny) on selfing ability, we performed a mixed model ANOVA (proc GLM, SAS Institute 2002) with cross as a fixed effect and block, genet, and the interaction between genet and cross as random effects, for the ratio of selfed and outcrossed fruits per pollination, the ratio of selfed and outcrossed seeds per fruit, the ratio of selfed and outcrossed seeds per pollination, and the ISC values from fruit and seed set. The block effect did not significantly affect the results and therefore was removed from the final analysis. All proportion variables were arcsine-square root transformed prior to analysis to meet the assumption of normality.
RESULTS
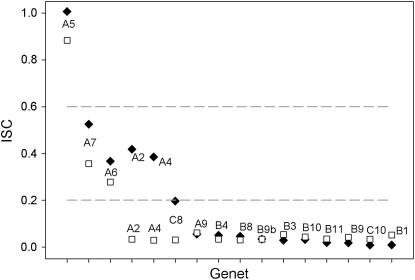
In this study, we performed a series of controlled pollinations in the greenhouse to determine the extent of the variation in the strength of SI among genets and identified the S-allele composition from two generations of plants to determine whether this variation is due to the existence of weak and strong S-alleles. Parental genets collected from a large population in Central Pennsylvania show a low diversity of S-alleles, with only six S-alleles present in the population. Alleles S2, S5, S8, S9, and S17 have been previously reported by Richman et al. (1995) and Lu (2006), while the allele S18 is a new allele (Mena-Alí 2006). Furthermore, these parental genets display a high level of variation for their ability to self (Figure 1). While most genets display very low ISC values, as expected for a fully functional SI system (values of ISC <0.1), some genets had greater pollen tube growth and self-fertility. Similar differences in ISC among genets are observed from both pollen tube growth and seed set, suggesting that the ability to self is indeed highly determined by genet identity (F15,15 = 13.04; P < 0.0001; see Figure 1). In two genets (A2 and A4) we found different estimates of ISC from pollen tube growth and seed set data, with a larger ISC obtained from seed set. While these genets set few self seeds, similar to the remaining 14 genets, both the A2 and A4 families also had a much lower seed set following outcross pollinations. This inflated the estimated ISC for seed set for A2 and A4, since outcross seed set was the denominator in our calculation of ISC for seed set. For the remaining 14 genets, the correspondence between ISC obtained from pollen tube growth and seed set is remarkable. In fact, the genet identity accounts for 91% of the variation observed in ISC values.
Figure 1.—
Variation in the ISC among 16 genets (maternal lines) of S. carolinense. The values of ISC were estimated using two methods: seed set counts (solid diamonds) and pollen tube growth (open squares). Three groups can be distinguished: most genets are strongly self-incompatible (ISC < 0.2). Genets A6 and A7 consistently show intermediate levels of self-compatibility (0.2 < ISC < 0.6); genet A5 seems to be highly self-compatible (ISC >> 0.6).
The ability to self is maintained in the selfed progeny. Overall, selfed offspring are able to set more selfed fruit and seed than the outcrossed progeny (Table 2), with a general twofold increase in their ability to self-fertilize when compared to the outcrossed progeny (Table 3). We did not detect a significant effect of maternal genet on the ability to self-fertilize in the progeny. However, in three of the five variables measured (self fruits per pollination, ISC from fruit set, and ISC from seed set) the genet-by-cross interaction was highly significant (Table 2). This genet by cross merits closer inspection; we will address the significance of this interaction below.
TABLE 2.
ANOVA for the effect of parental genet identity and level of breeding of the progeny (“cross”: selfed or outcrossed) for five different measures of self-compatibility
| F | d.f. | P | |
|---|---|---|---|
| Fruits per pollination | |||
| Genet | 1.27 | 15, 15 | 0.3261 |
| Cross | 6.98 | 1, 15 | 0.0183 |
| G × C | 1.94 | 15, 150 | 0.0230 |
| Seeds per fruit | |||
| Genet | 2.41 | 14, 14 | 0.0554 |
| Cross | 0.00 | 1, 20 | 0.9513 |
| G × C | 0.92 | 14, 60 | 0.5400 |
| Seeds per pollination | |||
| Genet | 1.57 | 15, 15 | 0.1953 |
| Cross | 1.44 | 1, 15 | 0.2492 |
| G × C | 1.50 | 15, 149 | 0.1109 |
| ISC fruit set | |||
| Genet | 1.33 | 15, 15 | 0.2941 |
| Cross | 4.03 | 1, 15 | 0.0628 |
| G × C | 2.01 | 15, 148 | 0.0182 |
| ISC seed set | |||
| Genet | 1.24 | 15, 15 | 0.3437 |
| Cross | 3.98 | 1, 15 | 0.0643 |
| G × C | 3.14 | 15, 148 | 0.0002 |
TABLE 3.
Mean ± SE for five variables of selfing ability from selfed and outcrossed progeny obtained from 16 genets of S. carolinense
| Breeding history | Fruits per pollination | Seeds per fruit | Seeds per pollination | ISC fruit | ISC seed |
|---|---|---|---|---|---|
| Selfed | 0.44 ± 0.04 | 20.2 ± 5.1 | 11.1 ± 2.1 | 0.40 ± 0.03 | 0.22 ± 0.02 |
| Outcrossed | 0.24 ± 0.04 | 20.6 ± 4.7 | 6.9 ± 2.0 | 0.27 ± 0.03 | 0.10 ± 0.02 |
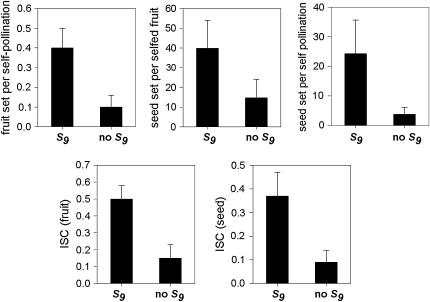
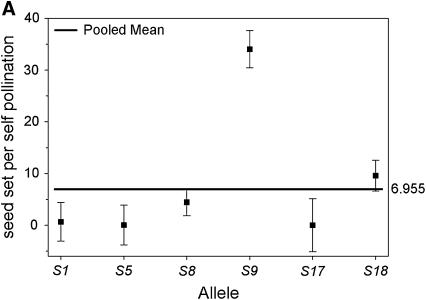
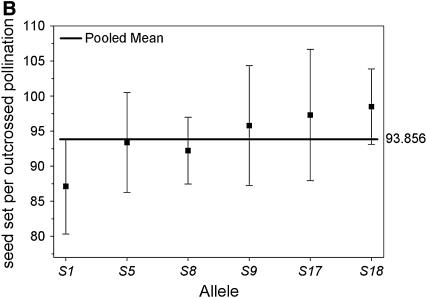
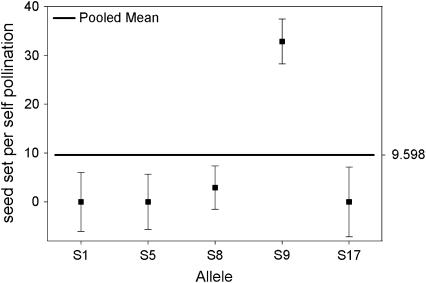
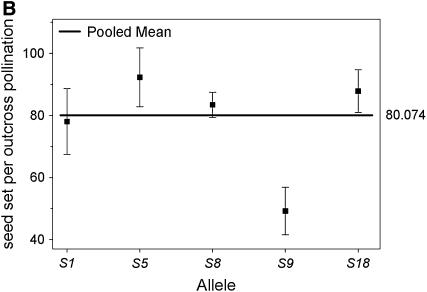
We compared the S-allele composition of all 16 parental genets (shown in Table 1) to the values of self-compatibility obtained for the parental plants. Only 3 of the 16 plants present in our population carry allele S9 (A5, A6, and A7). Interestingly, these three genets display the highest values of ISC obtained from pollen tube growth and self-seed set (Figure 1). When we compared the outcrossed progeny from these three genets, we found that their ability to set self seed segregates with the S9 allele in the progeny (Figure 2). Progeny carrying allele S9 set more self fruit and more self seeds than their siblings that do not carry S9. This difference is significant in four of the five variables compared (Table 4). These results suggest an association between the presence of allele S9 and the ability to set self seed. We further examined this possibility by comparing the number of seeds per self-pollination among all outcrossed progeny. The outcrossed progeny provides a larger sample size and also allows us to examine the presence of S9 in association with different S-alleles (due to the reshuffling of pollen carrying different alleles during our controlled pollinations). We performed a one-way random effects model ANOVA to examine the effects of each of the six S-alleles in our population (Table 1) on the ability of the plants to set self seed. Plants that carry allele S9 were able to set significantly more self seeds per pollination compared to plants carrying any other allele (Figure 3A). Plants carrying allele S18 were able to set intermediate numbers of self seed, although the mean values are only 28% of those carrying allele S9 (Figure 3A). The four remaining alleles did not show any significant ability to set self seed. In contrast, the ability to set seed following outcross pollinations is not significantly affected by the S-genotype of the plant (Figure 3B), suggesting that the differences in self seed set among S-alleles are not due to differences in the general vigor (or resource availability) of plants carrying the S9 allele.
Figure 2.—
Mean ± SE for five variables of self-fertility among outcrossed progeny of three genets (A5, A6 and A7) based on the presence or absence of allele S9. In four of the five variables, outcross offspring that carry S9 show a significantly higher ability to self-fertilization; see Table 4 for statistical analysis.
TABLE 4.
Comparison of the outcrossed progeny of three genets (A5, A6, and A7) of S. carolinense
| t | d.f. | P | |
|---|---|---|---|
| Fruits per pollination | 2.78 | 16 | 0.0135 |
| Seeds per fruit | 1.47 | 16 | 0.1619 |
| Seeds per pollination | 1.97 | 16 | 0.0665 |
| ISC fruit set | 3.00 | 16 | 0.0084 |
| ISC seed set | 2.63 | 16 | 0.0180 |
A t-test was used to compare these progeny for five measurements of selfing ability on the basis of the presence or absence of allele S9.
Figure 3.—
Mean number of seed set per self-pollination (A) and seed set per outcross pollination (B) by S-allele among the outcrossed progeny of 16 maternal genets. The values for each allele on the horizontal axis represent the mean of all genotypes in the outcrossed progeny that carried that allele (e.g., S1_, S5_, etc.). The bold horizontal line represents the overall mean for all plants. Plants carrying allele S9 set significantly more seeds following self-pollinations than plants carrying any other allele present in the population. There are no differences among alleles for the amount of seed set after outcross pollinations.
As mentioned above, the genet-by-cross interaction observed in the parental generation was highly significant (Table 2). Upon closer examination, we found that this interaction can be explained, at least in part, by the presence of allele S9. In genets A5, A6, and A7, the outcrossed progeny has a lower frequency of allele S9 compared to their selfed progeny such that the selfed progeny show a higher ability to self-fertilize compared to the outcrossed progeny. In contrast, other genets show a higher ability to self-fertilize in their outcrossed progeny compared to their selfed progeny. For instance, in genets A2, B3, and B9, some of the outcrossed progeny carry allele S9, while none of the selfed progeny have the S9 allele and therefore the outcross progeny for these genets show higher self-fertilization rates compared to the selfed progeny. Finally, some genets in which none of the progeny carried the S9 allele (e.g., A4, A9, and C8) show similarly low rates of self-fertilization in both selfed and outcrossed progeny.
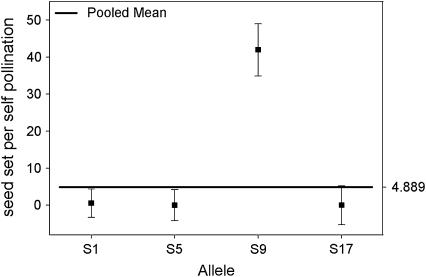
To further examine the link between allele S9 and the ability to self-fertilize, we looked at the interaction between alleles by examining different combinations of alleles. This analysis takes advantage of the high frequency of the S8 and S18 alleles in our population. We examined those plants carrying allele S18 and compared their selfing ability vs. the identity of the second allele in their genotype. If the moderate ability to self-fertilize of plants carrying allele S18 is due to the S18 allele (or some closely linked modifier gene), then we expect similar mean values of self-seed set per pollination for all SjS18, where Sj represent any allele other than S18. A one-way ANOVA for random effects for all plants with genotype SjS18 shows that individuals carrying S18 are able to set self seed only when found in association with allele S9 (F4,40 = 8.73; P < 0.0001). Plants carrying allele S18 in association with any allele other than S9 failed to set any seed following self-pollinations (Figure 4). We found a similar result when we compared all plants carrying allele S8: these plants were able to set self seed only when they carried allele S9 (F4,41 = 10.38; P < 0.0001) (Figure 5).
Figure 4.—
Mean number of seed set per self-pollination in outcrossed progeny with genotype SjS18. The bold horizontal line represents the overall mean for all plants. Plants carrying allele S9 set significantly more seed after self-pollinations than plants carrying any other allele. The presence of allele S18 does not seem to account for an increase in selfing ability.
Figure 5.—
Mean number of seed set per self-pollination in outcrossed progeny with genotype SjS8. The bold horizontal line represents the overall mean for all plants. Plants carrying allele S9 set significantly more seed after self-pollinations than plants carrying any other allele. The presence of allele S8 does not seem to account for an increase in selfing ability.
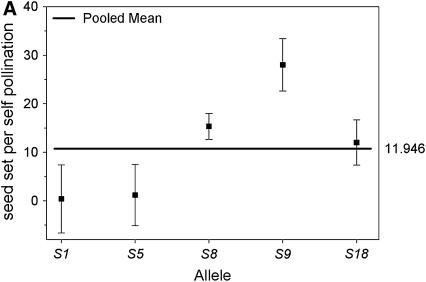
When we look at the association between specific S-alleles and their ability to self in the selfed progeny we find that, once again, plants that carry allele S9 were able to set more seeds per self-pollination compared to plants carrying any other allele (Figure 6A). Plants carrying allele S18 or allele S8 were also able to set some self seed, although the mean values are only 50% of those from allele S9 (Figure 6A). The two remaining alleles did not show any significant ability to set self seed. In contrast with the outcrossed progeny, in which the mean number of seed set after outcross pollination did not differ significantly among S-alleles, selfed progeny carrying allele S9 set significantly fewer outcrossed seeds than selfed individuals carrying any other S-allele (Figure 6B).
Figure 6.—
Mean number of seed set per self-pollination (A) and seed set per outcross pollination (B) by S-allele among selfed progeny of 16 maternal genets. The values for each allele on the horizontal axis represent the mean of all genotypes in the selfed progeny that carried that allele (e.g., S1_, S5_, etc.). The bold horizontal line represents the overall mean for all plants. Plants carrying allele S9 set significantly more seed per self-pollination and significantly less seed per outcross pollination than plants carrying any other allele present in the population.
Finally, 55 of the 87 selfed progeny that we were able to score were homozygous, which deviates significantly from the expected 1:2:1 ratio based on Mendelian segregation (χ2= 12.72; d.f. = 2; P < 0.0001). This deviation is caused by an excess rather than a deficit of homozygotes. Among the selfed progeny, heterozygous plants set significantly more seeds per self-pollination than homozygous plants (mean seed set per self-pollination for heterozygous plants, 20.7 ± 3.9; homozygous plants, 6.0 ± 2.7; F1,69 = 8.19; P < 0.01) and fewer seeds per outcross pollination (mean seed set per cross-pollination for heterozygous plants, 84.9 ± 6.7; homozygous plants, 77.1 ± 4.6) although this difference was not statistically significant (F1,69 = 0.77; P = 0.38).
DISCUSSION
In this study, we applied a PCR-based protocol to identify the S-alleles present in 16 maternal genotypes and their offspring and performed a series of greenhouse experiments to determine whether variation in the strength of SI in S. carolinense is due to the existence of weak and strong S-alleles differing in their ability to recognize and reject self-pollen. Our analysis of the parental genets showed that there is variation among individuals of S. carolinense for their strength of SI—most individuals have very low seed set following self-pollinations, but genets A5, A6, and A7 had intermediate to high levels of self-fertility (Figure 1). When compared against their S-allele composition, we found these three genets are the only individuals carrying the S9 allele, suggesting that S9 is a weak S-allele in this population of horsenettle. The outcrossed progeny from these three genets were found to have a greater self-fertility only when carrying allele S9 (Figure 2). This observation was further supported when we analyzed both the outcrossed (Figure 3) and selfed (Figure 6) progeny for their ability to set self seed. In addition, the ability of allele S9 to self-fertilize does not seem to be affected by interactions with other specific S-alleles. Plants with genotypes S9S18 and S8S9 showed similar levels of self-fertilization (see Figures 4 and 5), suggesting that the ability to self-fertilize is directly associated with the S9 haplotype. It should be noted that S9 is not a rare allele: it was first described by Richman et al. (1995), who found it present in two populations of S. carolinense from North Carolina and Tennessee. Lu (2006) examined 2–13 individuals from each of 13 populations distributed from Indiana, Illinois, and Arkansas in the west to Kentucky, Tennessee, Georgia, North and South Carolina, and New Jersey in the east and found the S9 allele in 23% of the populations. S9 is clearly a widespread allele; however, to date it is not known whether copies of S9 from the other populations behave similarly to the allele isolated from our Pennsylvania population.
The variation in the strength of SI within and between populations has been reported for a variety of plant species (see Good-Avila and Stephenson 2002; Stephenson et al. 2003; Stone et al. 2006). For instance, Ando et al. (1998) described mixed populations of Petunia axillaris composed of both self-incompatible and self-compatible individuals. In Laevenworthia alabamica, self-compatible individuals are found mostly at the periphery of the species range (Busch 2005). The mechanisms that result in this variability are diverse. In Campanula rapunculoides, analyses of 31 parental families suggested the presence of 3–5 unlinked, mostly recessive genetic modifiers affecting the strength of the SI response (Good-Avila and Stephenson 2002). In P. axillaris, the presence of self-compatible plants has been shown to be due to three different mechanisms: a linked gene that suppresses the expression of a specific S-RNase gene (Tsukamoto et al. 2003a), a loss of function on the pollen component of SI (Tsukamoto et al. 2003b), and the presence of unlinked modifiers (Tsukamoto et al. 1999). The variable strength of SI has been shown in most cases in species with small patchy populations such as weeds (Baker 1965), island colonizers (Bateman 1952; Burd 1994), and populations growing in peripheral habitats (Busch 2005); these populations typically undergo repeated events of colonization and local extinction and usually consist of a small number of individuals and are likely to experience reduced pollinator and mate availability.
Our findings are unlikely to be due to a functionless allele. All six alleles found in our population were successfully amplified from RNA extractions of fresh styles; therefore, at least at the transcription level, allele S9 is expressed and functional (Mena-Alí 2006). However, S9 could still be a weak allele if the mature protein shows reduced specificity or activity. In addition, if the breakdown of SI was due to mutations that render allele S9 functionless, then plants that carry this allele (for instance, genets A5, A6, and A7 in our parental population) should still be able to recognize and reject their other S-allele (S8, see Table 1) and should have not been able to set homozygotes for this second allele in the selfed progeny (Table 1). The presence of S8S8 progeny for these three genets suggests that any allele found in association with S9 may also be able to escape rejection and achieve fertilization.
Several studies have also shown that duplication of the S-locus can lead to breakdown of the GSI response (De Nettancourt 1977, 1997; Stone 2002), specifically the duplication of the pollen-expressed component of the GSI reaction of the Solanaceae (Tsukamoto et al. 1999). Our findings, however, are unlikely to be the result of a duplicated pollen determinant of SI because if the pollen component of the S-locus, S-locus F-box protein (Sijacic et al. 2004), is duplicated, then an allele carrying such duplication will be rendered fully self-compatible (Kao and Tsukamoto 2004) and such mutations are not likely to vary the strength of the SI response with floral age or prior fruit development (Good-Avila and Stephenson 2002) as we observed in our previous studies (Stephenson et al. 2003; Travers et al. 2004). A third possibility involves unlinked modifier genes that may affect the timing or magnitude of the S-RNase expression, its turnover, or the normal function of the SI biochemical pathways (Tsukamoto et al. 1999). Unlinked modifiers cannot explain the tight association of the breakdown phenotype with allele S9 in the outcrossed progeny (Figures 2, 4, and 5) because the unlinked modifiers would segregate in the progeny. In this case, some of the outcross progeny that do not possess the S9 allele would also be expected to be partially self-fertile. A modifier gene linked to allele S9 on the other hand may explain the association between the presence of this allele and its ability to self. If some of the molecular machinery involved in the SI reaction is defective and associated with S9, then even strong alleles should be able to escape the SI recognition and achieve fertilization when found in association with S9 (Figures 2–4). Several genes are known to be involved in SI response (Cruz-Garcia et al. 2003; Kao and Tsukamoto 2004; Goldraij et al. 2006), but none of them are found in close enough proximity to the S-locus to account for this tight association between S9 and the weakened SI response. One possible target may lie on the flanking regions surrounding different S-alleles. These sequences are known to be highly divergent (Coleman and Kao 1992); therefore, it could be possible for specific regulatory proteins to be either required for the appropriate transcription of different S-alleles or involved in the turnover of specific RNA transcripts (Tsukamoto et al. 2003a).
Although the variability in the strength of SI among individuals in a population may represent a transitional step in the evolution of self-compatibility from self-incompatible individuals, it may also be an evolutionarily stable state rather than a transient state between SI and full compatibility (Levin 1996; Stephenson et al. 2000; Vallejo-Marín and Uyenoyama 2004). In previous studies we have found that the ability of genotypes to set self seed increases with the age of unpollinated flowers (Stephenson et al. 2003) and under conditions of low fruit production (Travers et al. 2004). The presence of older unpollinated flowers on inflorescences and poor fruit set on prior inflorescences are conditions that are expected when pollinator activity is low and/or when outcross pollen sources are scarce (mates with suitable S-alleles). These studies have shown that most if not all S-genotypes become more self compatible with floral age and lack of fruit set; the results presented here indicate that this trend is more pronounced in the presence of allele S9.
Recently, Vallejo-Marín and O'Brien (2007) found an association between SI and clonal reproduction across the genus Solanum and suggest that clonality may provide reproductive assurance under conditions of pollen limitation. In S. carolinense, the existence of a weak S-allele and a generalized relaxation of SI with floral age and poor fruit set could allow the population to persist until new S-alleles arrive via migration, as well as providing some reproductive assurance. In both cases, selective pressures seem to be acting on small populations of self-incompatible species to favor strategies that increase the size and persistence of small patches. Recent theoretical studies have suggested that conditions of reduced S-allele diversity and limited mate availability favor the spread of self-compatible and “leaky” alleles (Vallejo-Marín and Uyenoyama 2004; Porcher and Lande 2005). Furthermore, Porcher and Lande's (2005) results indicate that a high level of inbreeding depression and the presence of sheltered load at the S-locus would allow the maintenance of SI. In our study, there is an indication that the selfed progeny suffer from inbreeding depression in that they produce fewer seeds following outcross pollinations than the outcross progeny (Figures 3B and 6B). Moreover, there is also an indication that selfed progeny that are S-homozygotes have greater inbreeding depression than selfed progeny that are S-heterozygotes (i.e., there appears to be sheltered load sensu; Uyenoyama 1997) in that S-homozygotes have reduced seed set following both self and outcross pollinations compared to self progeny that are S-heterozygotes (Uyenoyama 2003; Stone 2004). We are currently examining the extent of inbreeding depression and the presence of sheltered load in this population to elucidate the role of the variability in the SI system on the population dynamics of S. carolinense.
Acknowledgments
We thank T. Omeis and his staff at the Buckhout Greenhouse for assistance with the cultivation of the plants; L. Kessler, E. Drechsel, J. Thaller, R. Zwerin, and G. Stephenson for greenhouse and lab assistance; and T-h. Kao and S. Schaeffer for comments. This research was supported by a National Science Foundation grant DEB99-82086 to A.G.S. and a U. S. Department of Agriculture Cooperative State Research, Education, and Extension Service grant no. 2005-35320-15351 to A.G.S.
References
- Ando, T., T. Tsukamoto, N. Akiba, H. Kokubun, H. Watanabe et al., 1998. Differentiation in the degree of self-incompatibility in Petunia axillaris (Solanaceae) occurring in Uruguay. Acta Phytotax. Geobot. 49: 37–47. [Google Scholar]
- Baker, H. G., 1965. Characteristics and modes of origin of weeds, pp. 147–168 in The Genetics of Colonizing Species, edited by H. G. Baker and G. L. Stebbins. Academic Press, New York.
- Basset, I. J., and D. B. Munro, 1986. The biology of Canadian weeds. 78. Solanum carolinense L. and Solanum rostratum Dunal. Can. J. Plant Sci. 66: 977–991. [Google Scholar]
- Bateman, A. J., 1952. Self-incompatibility systems in angiosperms. I. Theory. Heredity 6: 285–310. [Google Scholar]
- Burd, M., 1994. Bateman's principle and plant reproduction: the role of pollen limitation in fruit and seed set. Bot. Rev. 60: 83–139. [Google Scholar]
- Busch, J. W., 2005. The evolution of self-compatibility in geographically peripheral populations of Leavenworthia alabamica (Brassicaceae). Am. J. Bot. 92: 1503–1512. [DOI] [PubMed] [Google Scholar]
- Coleman, C. E., and T.-H. Kao, 1992. The flanking regions of two Petunia inflata S-alleles are heterogeneous and contain repetitive sequences. Plant Mol. Biol. 18: 725–737. [DOI] [PubMed] [Google Scholar]
- Cruz-Garcia, F., C. N. Hancock and B. McClure, 2003. S-RNase complexes and pollen rejection. J. Exp. Bot. 54: 123–130. [DOI] [PubMed] [Google Scholar]
- de Nettancourt, D., 1977. Incompatibility in Angiosperms. Springer-Verlag, Berlin.
- de Nettancourt, D., 1997. Incompatibility in angiosperms. Sex. Plant Reprod. 10: 185–199. [Google Scholar]
- Fisher, R. A., 1941. Average excess and average affect of a gene substitution. Ann. Eugen. 11: 53–63. [Google Scholar]
- Goldraij, A., K. Kondo, C. B. Lee, C. N. Hancock, M. Sivaguru et al., 2006. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439: 805–810. [DOI] [PubMed] [Google Scholar]
- Good-Avila, S. V., and A. G. Stephenson, 2002. The inheritance of modifiers conferring self-fertility in the partially self-incompatible perennial, Campanula rapunculoides, L. (Campanulaceae). Evolution 56: 263–272. [DOI] [PubMed] [Google Scholar]
- Kao, T. -H., and T. Tsukamoto, 2004. The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16: S72–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. A., 1996. The evolutionary significance of pseudo self-fertility. Am. Nat. 148: 321–332. [Google Scholar]
- Lloyd, D. G., 1992. Self- and cross-fertilization in plants. II. The selection of self-fertilization. Int. J. Plant Sci. 153: 358–369. [Google Scholar]
- Lloyd, D. G., and D. J. Schoen, 1992. Self- and cross-fertilization in plants. I. Functional dimensions. Int. J. Plant Sci. 153: 341–357. [Google Scholar]
- Lu, Y., 2006. Historical events and allelic polymorphism at the gametophytic self-incompatibility locus in Solanaceae. Heredity 96: 22–28. [DOI] [PubMed] [Google Scholar]
- Martin, F. M., 1959. Staining and observing pollen tubes by means of fluorescence. Stain Technol. 34: 436–437. [DOI] [PubMed] [Google Scholar]
- Mena-Alí, J. I., 2006. Dynamics of the self-incompatibility alleles in populations of Solanum carolinense. Ph.D. Dissertation Pennsylvania State University, University Park, PA.
- Porcher, E., and R. Lande, 2005. Loss of gametophytic self-incompatibility with evolution of inbreeding depression. Evolution 59: 46–60. [PubMed] [Google Scholar]
- Richman, A. D., T. -H. Kao, S. W. Schaeffer and M. K. Uyenoyama, 1995. S-allele sequence diversity in natural populations of Solanum carolinense (Horsenettle). Heredity 75: 405–415. [DOI] [PubMed] [Google Scholar]
- SAS Institute, 2002. SAS User's Guide: Statistics. SAS Institute, Cary, NC.
- Schoen, D. J., M. T. Morgan and T. Bataillon, 1996. How does self-pollination evolve? Inferences from floral ecology and molecular genetic variation. Philos. Trans. R. Soc. Lond. Ser. B 351: 1281–1290. [Google Scholar]
- Sijacic, P., X. Wang, A. L. Skirpan, Y. Wang, P. E. Dowd et al., 2004. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429: 302–305. [DOI] [PubMed] [Google Scholar]
- Stebbins, G. L., 1957. Self-fertilization and population viability in the higher plants. Am. Nat. 91: 337–354. [Google Scholar]
- Stephenson, A. G., S. V. Good and D. W. Vogler, 2000. Interrelationships among inbreeding depression, plasticity in the self-incompatibility system, and the breeding system of Campanula rapunculoides L. (Campanulaceae). Ann. Bot. 85: 211–219. [Google Scholar]
- Stephenson, A. G., S. E. Travers, J. I. Mena-Alí and J. A. Winsor, 2003. Pollen performance before and during the autotrophic-heterotrophic transition of pollen tube growth. Philos. Trans. R. Soc. Lond. Ser. B 358: 1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, J. L., 2002. Molecular mechanisms underlying the breakdown of gametophytic self-incompatibility. Q. Rev. Biol. 77: 17–32. [DOI] [PubMed] [Google Scholar]
- Stone, J. L., 2004. Sheltered load associated with S-alleles in Solanum carolinense. Heredity 92: 335–342. [DOI] [PubMed] [Google Scholar]
- Stone, J. L., M. A. Sasuclark and C. P. Blomberg, 2006. Variation in the self-incompatibility response within and among populations of the tropical shrub Witheringia solanacea (Solanaceae). Am. J. Bot. 93: 592–598. [DOI] [PubMed] [Google Scholar]
- Travers, S. E., J. I. Mena-Alí and A. G. Stephenson, 2004. Plasticity in the self-incompatibility of Solanum carolinense. Plant Species Biol. 19: 127–135. [Google Scholar]
- Tsukamoto, T., T. Ando, H. Kokubun, H. Watanabe, M. Masada et al., 1999. Breakdown of self-incompatibility in a natural population of Petunia axillaris (Solanaceae) in Uruguay containing both self-incompatible and self-compatible plants. Sex. Plant Reprod. 12: 6–13. [Google Scholar]
- Tsukamoto, T., T. Ando, H. Kokubun, H. Watanabe, T. Sato et al., 2003. a Breakdown of self-incompatibility in a natural population of Petunia axillaris caused by a modifier locus that suppresses the expression of an S-RNase gene. Sex. Plant Reprod. 15: 255–263. [Google Scholar]
- Tsukamoto, T., T. Ando, K. Takahashi, T. Omori, H. Watanabe et al., 2003. b Breakdown of self-incompatibility in a natural population of Petunia axillaris caused by loss of pollen function. Plant Physiol. 131: 1903–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA, and NRCS, 2002. The PLANTS Database, Version 3.5. National Plant Data Center, Baton Rouge, LA.
- Uyenoyama, M. K., 1997. Genealogical structure among alleles regulating self-incompatibility in natural populations of flowering plants. Genetics 147: 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama, M. K., 2003. Genealogy-dependent variation in viability among self-incompatibility genotypes. Theor. Popul. Biol. 63: 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo-Marín, M., and H. E. O'Brien, 2007. Correlated evolution of self-incompatibility and clonal reproduction in Solanum (Solanaceae). New Phytol. 173: 415–421. [DOI] [PubMed] [Google Scholar]
- Vallejo-Marín, M., and M. K. Uyenoyama, 2004. On the evolutionary costs of self-incompatibility: incomplete reproductive compensation due to pollen limitation. Evolution 58: 1924–1935. [DOI] [PubMed] [Google Scholar]