Abstract
We describe a candidate gene approach for associating SNPs with variation in flowering time and water-soluble carbohydrate (WSC) content and other quality traits in the temperate forage grass species Lolium perenne. Three analysis methods were used, which took the significant population structure into account. First, a linear mixed model was used enabling a structured association analysis to be incorporated with the nine populations identified in the structure analysis as random variables. Second, a within-population analysis of variance was performed. Third, a tree-scanning method was used, in which haplotype trees were associated with phenotypes on the basis of inferred haplotypes. Analysis of variance within populations identified several associations between WSC, nitrogen (N), and dry matter digestibility with allelic variants within an alkaline invertase candidate gene LpcAI. These associations were only detected in material harvested in one of the two years. By contrast, consistent associations between the L. perenne homolog (LpHD1) of the rice photoperiod control gene HD1 and flowering time were identified. One SNP, in the immediate upstream region of the LpHD1 coding sequence (C-4443-A), was significant in the linear mixed model. Within-population analysis of variance and tree-scanning analysis confirmed and extended this result to the 2118 polymorphisms in some of the populations. The merits of the tree-scanning method are compared to the single SNP analysis. The potential usefulness of the 4443 SNP in marker-assisted selection is currently being evaluated in test crosses of genotypes from this work with turf-grass varieties.
ASSOCIATION or linkage disequilibrium (LD) mapping in crop plant species has received increasing attention in recent years owing to its potential for fine mapping of traits and the prospects for identifying functional markers (Nordborg and Tavare 2002; Nordborg et al. 2002; Flint-Garcia et al. 2003; Rafalski and Morgante 2004; Flint-Garcia et al. 2005; Gupta et al. 2005; Yu and Buckler 2006; Breseghello and Sorrells 2006a). By using populations of unknown pedigree, the recombination events that have occurred over many generations are exploited for more refined mapping than is possible in conventional F2 or backcross mapping families (Flint-Garcia et al. 2003). The method thus has the potential to provide useful markers for marker-assisted selection (MAS) in genetic improvement programs. It was first used as a candidate gene approach in plants by Thornsberry et al. (2001), who demonstrated association between allelic variants and flowering time in the Dwarf8 gene in maize. It has been followed by other analyses in maize (Wilson et al. 2004; Szalma et al. 2005; Yu et al. 2006), rice (Bao et al. 2006a, 2006b), Arabidopsis thaliana (Olsen et al. 2004; Aranzana et al. 2005), barley (Ivandic et al. 2002; Kraakman et al. 2006), and wheat (Breseghello and Sorrells 2006b). The method is dependent upon LD (the nonrandom occurrence of alleles at different loci) between marker and phenotype, and this is affected by recombination. The effective recombination rate in turn is influenced by the breeding system. In inbreeding species effective recombination is lower, whereas in self-incompatible species the opposite is the case. In species where LD has been studied, it has in general extended further in self-compatible species, than in those that are out-breeding (Flint-Garcia et al. 2003; Rafalski and Morgante 2004). The potential for higher resolution mapping would therefore be expected in the latter species.
The important temperate forage and amenity grass Lolium perenne is an obligate out-breeding species (Cornish et al. 1979). One would thus expect LD to decay to insignificant levels over short distances. The only data on LD in L. perenne come from an AFLP marker analysis of populations, in which the resolution was limited by the 2–3-cM resolution of the F2 mapping family onto which the markers were mapped (Skøt et al. 2005), and from preliminary data on the alkaline invertase being analyzed further in this work (Humphreys et al. 2006). In the absence of more extensive information of the extent of LD within genes and given the obligate out-breeding nature of the species, it seemed reasonable to assume that LD decays rapidly. A candidate gene approach to association mapping thus appeared most likely to be successful, since a genome-wide approach would require an excessive number of molecular markers to be certain of identifying markers in LD with any given QTL allele. A careful selection of target genes with a probable role in controlling a phenotype is more likely to lead to identification of useful markers associated with a trait.
Population structure also has a major effect on LD. It is influenced by population genetic forces such as drift, selection, population admixture, and gene flow (Gaut and Long 2003). The selection of populations for association mapping is therefore an important issue in terms of capturing the maximum variation in the trait of interest, while minimizing effects of population structure.
Here, we are focusing on two traits in L. perenne: flowering time or heading date (HD) and content of water-soluble carbohydrates (WSCs). Both traits are of fundamental importance for plant growth and development and affect traits of practical and economic significance in forage and turf grass breeding. In particular, HD has an impact on biomass production, persistency, and quality including WSC (Wilkins and Humphreys 2003). Both traits have a high degree of heritability, and high sugar grass varieties have been bred (Wilkins and Humphreys 2003). However, there is still a need for further improvement in this trait, and there are a number of genes that could be targeted as candidates, particularly those involved in fructan biosynthesis or breakdown and enzymes involved in sugar metabolism such as invertases (Gallagher et al. 2004; Chalmers et al. 2005). Here we use a cytosolic neutral/alkaline invertase (Gallagher and Pollock 1998), which has been mapped to a QTL on chromosome 6 for glucose and fructose content in L. perenne (Turner et al. 2006). This LpcAI gene encodes an enzyme that hydrolyses sucrose to produce glucose and fructose, the substrates for respiration and biosynthesis of primary and secondary compounds as well as regulation of gene expression by sugars (Gallagher and Pollock 1998; Gallagher et al. 2004). Expression of this neutral/alkaline invertase gene is more or less constant in response to a number of perturbations including variation in sucrose substrate concentration, light, and position in the leaf i.e., age of tissue (Gallagher and Pollock 1998).
The genetic control of flowering in L. perenne is mainly determined by day length and temperature. Short days and low temperatures (vernalization) are required as a primary induction, followed by longer days and higher temperatures. There is however, a large degree of genetic variability for this trait within L. perenne. Orthologous genes to some of those involved in the photoperiod-controlled flowering induction in the model species A. thaliana have been identified in rice (Yano et al. 2000) and forage grasses (Armstead et al. 2004; Armstead et al. 2005). In particular, the HD1 homolog of the CONSTANS gene in A. thaliana (Putterill et al. 1995) is located on chromosome 7 in L. perenne within a major QTL for flowering time (Jones et al. 2002; Armstead et al. 2004). Its expression is upregulated in response to long days, and it is capable of complementing a mutant CONSTANS line in A. thaliana (Martin et al. 2004). These pieces of evidence are all consistent with the idea that LpHD1 is involved in the photoperiodic control of the flowering phenotype. We have therefore used this to search for allelic variants associated with HD. The importance of population structure is illustrated in the association analysis described here in which populations from throughout Europe were selected to maximize variation in HD (and to some degree also WSC) and analyzed for association of these traits with allelic variation in two candidate genes, LpHD1 and LpcAI, respectively.
MATERIALS AND METHODS
Plant material:
In total, 96 genotypes from each of nine populations of L. perenne were used in this work, of which seven are natural or seminatural and two were varieties. The populations all originate from Europe. They were primarily selected to provide the maximum possible variation in HD and details of their origin are listed in Table 1. Second, wherever possible, populations within the same flowering time category, but from more than one distinct geographic origin, were represented. This was done to minimize the risk of spurious correlations with latitude. Third, populations with variation in HD are also likely to vary in WSC content, so have the potential to provide useful material for association analysis of forage quality traits. Seeds were planted in 6-in. diameter pots in potting compost in 2003, and plants left in a polytunnel to vernalize. HD was recorded in 2004 while still maintained as single genotypes in individual pots in the polytunnel. After flowering, above ground plant material was harvested, dried, and prepared for Near Infrared Reflectance Spectroscopy (NIRS) analysis of WSC, nitrogen content (N), and dry matter digestibility (DMD) as described (Lister and Dhanoa 1998). Tillers were planted as spaced plants in a field near the Institute of Grassland and Environmental Research in a fully randomized design in two replicates. The following year HD, WSC, N, and DMD were recorded or measured as in 2004. The HD data for 2005 represent the mean of two replicates.
TABLE 1.
List of L. perenne populations used in this work
| Accession | Accession status | Latitude | Longitude | MASL | Flowering time |
|---|---|---|---|---|---|
| Ba9955 | Semi-natural | 52°56′N | 3°03′W | 100 | Very late |
| Ba10113 | Semi-natural | 61°10′N | 6°40′E | 75 | Intermediate |
| Ba10158 | Semi-natural | 51°41′N | 9°27′W | 70 | Very early |
| Ba10278 | Semi-natural | 47°20′N | 9°25′E | 830 | Very early |
| Ba10284 | Semi-natural | 47°27′N | 8°51′E | 720 | Very early |
| Ba10732 | Semi-natural | 53°34′N | 1°34′W | Intermediate | |
| Ba10870 | Variety | Very late | |||
| Ba11304 | Variety | Very early | |||
| Ba12945 | Semi-natural | 52°24′N | 6°49′W | 90 | Late |
MASL, meters above sea level.
DNA, sequencing, and SNP analysis:
Extraction of DNA was performed as described previously (Skøt et al. 2005). Sequencing was carried out using an ABI 3100 genetic analyzer according to the manufacturer's instructions (Applied Biosystems, Warrington, UK). Primers for amplification of PCR fragments for sequencing within the LpHD1 and LpcAI are listed in Table 2. The primers were designed from sequences deposited in the EMBL/GenBank data libraries under accession numbers AM489608 and AM489692, respectively. A total of 5604 bp of the LpHD1 locus was resequenced, corresponding to base pair numbers 10720–16323 in AM489608. This included a putative peroxidise precursor gene located upstream of LpHD1, as well as the sequence between the two genes and exon 1 of LpHD1. Despite numerous primer designs and PCR reaction modifications, we were unable to obtain reliable PCR amplification for SNP discovery in the 3′ region of the LpHD1 gene, including the second exon. In the LpcAI locus, three segments were resequenced: base pair numbers 61–1263, 1927–2730, and 4635–5465, covering a total of 2796 bp (starting at base number 701 in AM489692). The sequenced segments cover ∼800 bases upstream of the codon start site and exon 1, then exons 2 and 3, and finally exons 5 and 6. This resequencing strategy was used to enhance the chances of detecting SNPs in the upstream regulatory sequence and coding sequence. The PCR reactions were performed in a total volume of 50 μl containing 30 ng DNA, 1× Roche Taq buffer with Mg2+, 200 mm of each dNTP, 200 μm of each primer, and 0.05 units of Taq polymerase (Roche Diagnostics, West Sussex, UK). The PCR reactions were carried out using an ABI 9700 thermocycler (Applied Biosystems). The conditions were as follows: a 2-min 94° denaturation step followed by 40 cycles of 94° for 30 sec, annealing temperature for 30 sec, and 72° for 1 min, followed by a final 7-min extension step at 72°. The annealing temperature varied between 55° and 60° depending on the melting temperature of the specific primer pair.
TABLE 2.
Primer sequences used for PCR amplification of segments of the two genes LpHD1 and LpcAI
| Gene | Pair | Primer name | Primer sequence (5′–3′) |
|---|---|---|---|
| LpHD1 | 1 | HD1F | CAGAATGAAACAGGTGCTGA |
| HD1R | AGGAATAGGCCAGGTTCATT | ||
| 2 | HD2F | TGTTTGCTAGGTCAAGACTTGC | |
| HD2R | TGAAGCCACCAAACACTG | ||
| 3 | HD3F | AGCAAGCAGAAAGTATCTGTAG | |
| HD3R | TTCCTCGGGTATTTTGATC | ||
| 4 | HD4F | CTGACGGGGATAAGATATTTTC | |
| HD4R | GTTTTTTTGCCATTCATTGG | ||
| 5 | HD5F | ACTATCTAGTGACATGGCATGG | |
| HD5R | TAAAGAAGCAGTCGGAATGG | ||
| 6 | HD6F | CAAGCCACAAGGCCTCCTT | |
| HD6R | GGAGTGGCTATGACGCAGTTCT | ||
| LpcAI | 1 | AI1F | GCTTTTCTGTTAGCCCAATG |
| AI1R | AAGGTCATCAATCCTCTGGC | ||
| 2 | AI2F | GCATCCAGTTTACCCCTCAG | |
| AI2R | CGTACAATTTCATACTCCCCC | ||
| 3 | AI3F | TTCCATCCTCTCTTCCCTTC | |
| AI3R | CATGGCTTGACTTTTGACAAAC | ||
| 4 | AI4F | GGCAACAACCCAACAATCAC | |
| AI4R | GCATTCCTCCATCAAAAGCAC |
The discovery of SNPs in the LpHD1 gene was performed by sequencing a subset of two genotypes from each of the nine populations. The eight SNP loci selected for the full data set were analyzed for polymorphism using the TaqMan assay. Primers and fluorescent probes were designed using the Primer Express version 2 program (Applied Biosystems) and are listed in Table 3. The allelic discrimination assay was performed using the ABI 7500 Real Time PCR system (Applied Biosystems), with the default settings on the PCR program. The reaction mix consisted of 1× Taqman universal buffer, 0.9 μm of each primer, 0.1 μm of each probe, and 10 ng of genomic DNA. For SNP discovery in the LpcAI gene, 19 genotypes were used, 7 of which were from the 9 populations described here. The remaining 12 were genotypes from different populations previously described (Skøt et al. 2005). LD and neutrality tests were performed on these two subsets using the program DnaSP (http://www.ub.es/dnasp) (Rozas et al. 2003). Due to the heterozygous nature of the sequence data, haplotype pairs were inferred using the PHASE version 2.0 program (Stephens et al. 2001; Stephens and Donnelly 2003), so that two sequences were entered for each individual. Fisher's exact test was used to calculate the significance of pair-wise LD, and Tajima's D test was used to estimate neutrality of the SNP polymorphisms. Fifteen of the 92 SNPs in the LpcAI gene were selected for subsequent genotyping in 450 of the 864 genotypes (50 from each population). Three factors determined which SNPs were selected: the availability of a sufficient length of monomorphic sequence to allow primer design, inclusion of amino acid changing polymorphisms wherever possible, and representation of all the inferred haplotypes. The genotyping analysis was carried out by K-Biosciences (Hoddesdon, UK).
TABLE 3.
Primers and fluorescently labeled probes that were used in the TaqMan assay of the eight SNP polymorphisms assessed in the LpHD1 locus
| SNP | Forward and reverse primers (5′–3′) | TaqMan probe |
|---|---|---|
| 320 | ACCACATCTCCTGGTCAGAGTTG | AGGCC[C/T]TCGCCCT |
| ACCTTCGATAACGCTTATTTGCA | ||
| 513 | ATGTCATCTCGAAGTCAC | CCCG[G/A]TGCGCCA |
| ATTAAGGATCCCACACGA | ||
| 1475 | ATCTCCGTCCGGACCACAT | TTCCACA[T/A]TCGGGC |
| GAACCTGAGCCTGGAGCACTA | ||
| 2118 | GCCTCATATGTTCCTTTGTTAGATCA | TGAACA[C]GTGAAATT |
| GATGGATGCATCTGTGTTAACACTT | CCATGAACA[T]GTGAAAT | |
| 2389 | ATCTGGAAAGGAAAGAGACATGAGG | AGACAGCTA[C]AGTACAA |
| TGCACCATATCCTCCTTGGC | CAGACAGCTA[T]AGTACAAG | |
| 4443 | GACACTCTACTATTAGTACCCTGCACTGA | ACTGCCAA[C]ATAGC |
| CTGCATATGTGAGTGTGGAGGAA | ACTGCCAA[A]ATAG | |
| 4717 | CAGGCTGTGTGATGGATGTTG | TGCCAAGC[G/A]TGGTGT |
| ATCACAAGACGCGCAGAGGTAT | ||
| 5443 | CAACAACAGCGTGAGTTCATCTATT | CATGCTATAATTT[G]GTTAAT |
| GGTCTTTCAAATGAAGTAGCATTAACTC | TCATGCTATAATTT[T]GTTAAT |
AFLP analysis was performed as described (Skøt et al. 2005), except that an ABI 3130xl Genetic analyzer (Applied Biosystems) was used to separate the fluorescently labeled fragments, and Genemapper version 3.7 (Applied Biosystems) rather than Genotyper version 3.7 was used to analyze the data.
Data analysis:
The AFLP molecular marker data were analyzed for basic population genetics parameters including genetic diversity and population differentiation using the method of Lynch and Milligan (1994) as implemented in the AFLP-SURV 1.0 program (Vekemans 2002). Markers from individual primer pairs were analyzed separately to avoid the creation of too large data files. Since AFLP markers are dominant, we had to assume Hardy–Weinberg equilibrium. Analysis of variance (ANOVA) and linear mixed model analysis was performed using Genstat Release 8.11 (http://www.vsni.co.uk). In the latter, SNP genotypes were fitted as fixed terms, and population structure was incorporated by fitting inferred clusters as a random term. The population structure was estimated using the program STRUCTURE version 2.1 (Pritchard et al. 2000). Since the input data for that analysis consisted of dominant AFLP markers a no-admixture model was employed, in which allele frequencies were considered independent among populations. Only markers with a band frequency ≥0.05 were used. The length of the burn-in period and the number of MCMC replications after the burn-in was 50,000 for each. The given number of populations (K) was varied between 2 and 10. This choice was based on the fact that the genotypes consisted of seven geographically distinct populations within Europe, and the remaining two populations were varieties developed at the Institute of Grassland and Environmental Research (Table 1).
Haplotypes in the two genes were inferred using a Bayesian approach implemented in the program PHASE version 2.0 (Stephens et al. 2001; Stephens and Donnelly 2003). The default settings were used since the haplotype frequencies and the goodness-of-fit measure were consistent between runs. The inferred diplotypes obtained from running this program was used as input in the program TREESCAN version 0.9 (http://darwin.uvigo.es) (Templeton et al. 2005). This software was used to perform a tree-scanning analysis of the phenotypic data against haplotype trees constructed from the haplotypes inferred in PHASE 2.0. The list of haplotypes in the best reconstruction from the PHASE version 2.0 output file was used for the construction of the haplotype trees, employing the phylogenetic program PHYLIP version 3.6 (Felsenstein 1993) using maximum parsimony. The default setting was used in which the first haplotype was used as the outgroup root. In the execution of the TREESCAN program the probability threshold was set at 0.05 for the corrected permutational P-value after enforcement of monotonicity. The number of permutations was 1000, and the minimum class size was set to 5.
RESULTS
Phenotype analysis:
The phenotypic data for all four traits are summarized in Table 4. The two-way ANOVA showed that there were not only significant differences between populations and years, but also population × year interaction for all four response variables (Table 5). The difference in phenotype between years, particularly for the three quality traits, can be attributed partly to the contrasting plant growth conditions (pots vs. field). Second, a larger proportion of the harvested plant material from the pots probably consisted of leaf bases compared to leaf blades, than that from the field. The leaf bases contain significantly more WSC than the blades (Gallagher et al. 2004, 2007). The HD phenotype values agreed in general with previous classifications of the accessions as shown in Table 1, but also here there was significant population by year interaction (Table 5). Particularly the very early flowering populations Ba10278, Ba10284, and Ba11304 were variable between years.
TABLE 4.
Phenotypic data for the nine L. perenne populations used in this work
| Year
|
||||||||
|---|---|---|---|---|---|---|---|---|
| 2004
|
2005
|
|||||||
| Population | HD | WSC | N | DMD | HD | WSC | N | DMD |
| Ba9955 | 110.6 | 21.1 | 2.5 | 73.1 | 104.3 | 10.5 | 3.5 | 64.9 |
| Ba10113 | 81.3 | 15.7 | 2.7 | 74.9 | 69.9 | 8.6 | 3.7 | 59.9 |
| Ba10158 | 68.9 | 20.7 | 2.3 | 73.4 | 63.8 | 10.2 | 3.5 | 62.6 |
| Ba10278 | 81.2 | 19.9 | 2.4 | 72.3 | 50.3 | 10.1 | 3.7 | 66.2 |
| Ba10284 | 70.2 | 22.0 | 2.7 | 76.2 | 47.8 | 10.9 | 3.4 | 63.8 |
| Ba10732 | 91.9 | 17.7 | 2.7 | 75.3 | 80.9 | 10.1 | 3.2 | 62.6 |
| Ba10870 | 110.2 | 21.0 | 2.5 | 73.9 | 104.0 | 11.5 | 3.4 | 65.2 |
| Ba11304 | 72.1 | 25.6 | 2.6 | 74.6 | 41.0 | 10.0 | 3.6 | 64.6 |
| Ba12945 | 107.4 | 22.8 | 2.6 | 76.8 | 101.2 | 10.6 | 3.4 | 64.7 |
| LSD (0.05) | 1.05 | 2.15 | 0.23 | 1.24 | 1.03 | 1.31 | 0.16 | 1.80 |
LSD, least significant difference (P < 0.05); HD, days after March 1st; WSC, N, and DMD, % of dry matter. For the HD analysis N = 96 genotypes per population; for WSC, N, and DMD analyses N = 50.
TABLE 5.
Two-way ANOVA of phenotypic data collected from pot and field experiments
| Mean square
|
Variance component estimates (%)
|
||
|---|---|---|---|
| Source of variation | d.f. | HD | HD |
| Population | 8 | 83,604.6*** | 437.39 (73) |
| Year | 1 | 90,824.6*** | 105.59 (18) |
| Population × year | 8 | 5,524.6*** | 56.68 (9) |
| Residual | 1700 | 115.5 | |
| Total | 1717 |
| Mean square
|
Variance component estimates (%)
|
||||||
|---|---|---|---|---|---|---|---|
| Source of variation | d.f. | WSC | N | DMD | WSC | N | DMD |
| Population | 8 | 266.8*** | 0.96*** | 87.3*** | 2.5 (4) | 0.01 (2) | 0.73 (1) |
| Year | 1 | 24,186.9*** | 200.23*** | 25,291.9*** | 54.3 (92) | 0.45 (94) | 56.74 (93) |
| Population × year | 8 | 155.7*** | 1.15*** | 185.7*** | 2.7 (4) | 0.02 (4) | 3.44 (6) |
| Residual | 873 | 20.52 | 0.25 | 15.4 | |||
| Total | 890 | ||||||
Nine populations were used, and data were collected for 2 years. For the HD data, 96 genotypes per population were analyzed (864 in total), for the quality data 50 genotypes per population (450 in total) were analyzed. ***P < 0.001.
LD in candidate genes:
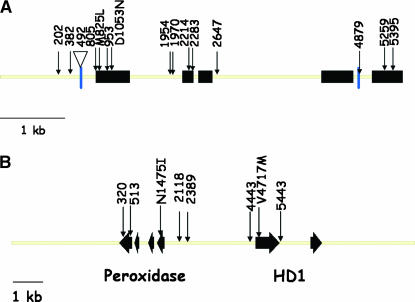
The degree of polymorphism in the LpcAI and LpHD1 genes differed greatly, as 92 were found in the former and only 12 in the latter (Table 6). This difference is further underlined by the fact that 2796 bp were sequenced for SNP discovery in the LpcAI locus and 5604 bp in LpHD1. Maps of the two loci are shown in Figure 1 including the SNP polymorphisms analyzed for association with phenotypes. The strategy for selecting the 15 SNPs in the LpcAI gene (Figure 1A) was described in materials and methods. Figure 1B shows that the LpHD1 locus also included a putative peroxidise precursor-like gene. We had no a priori reason to believe it is involved in the control of flowering time. We included SNPs from this gene, as it might inform us on the extent of LD in the region. The overall and within-population allele frequencies at the loci used in the association analysis are shown in Table 7. All SNPs were polymorphic overall in both loci, but some were monomorphic within some populations, particularly in the LpHD1 locus. Nucleotide diversity (π) in LpcAI was determined for each of the three segments that were resequenced. In the first segment (61–1263 bp) π = 0.01138; in the second (1927–2730 bp) π = 0.00822, and in the third (4635–5465 bp) π = 0.00605. The Tajima's D values were 0.1974, 0.3593, and 1.0318, respectively. All three were nonsignificant (P > 0.10), indicating that there was no evidence to suggest a significant deviation from neutrality. However, in one localized window of the sequence (939–1038 bp) the Tajima's D value was 2.25, which was significant (P < 0.05), suggesting an excess of intermediate allele frequencies. In the LpHD1 gene a continuous segment of 5603 bp was resequenced. The nucleotide diversity was π = 0.00500, and Tajima's D = 0.8795 (P > 0.10), also indicating a trend toward excess of intermediate allele frequencies, but the effect was not statistically significant.
TABLE 6.
Summary of SNP and indel polymorphisms in the two loci LpcAI (cytosolic alkaline invertase) and LpHD1 (HD1)
| Polymorphism type | LpcAI | LpHD1 |
|---|---|---|
| Noncoding (indels) | 62 (9) | 8 (0) |
| Coding | 21 | 4 |
| Synonymous | 13 | 2 |
| Replacement | 8 | 2 |
| Total | 92 | 12 |
Figure 1.—
Structure of the LpcAI and LpHD1 loci investigated in this work. The black rectangular boxes indicate exons, and arrows represent the SNPs investigated here. (A) Alkaline invertase gene. The upside down triangle indicates a 67-bp indel in the 5′ UTR region. M825L and D1053N indicate amino acid changing polymorphisms. (B) The HD1 gene. The arrowed rectangles indicate the direction of transcription of the peroxidise-like gene and the HD1 gene. N1475I and V4717M indicate amino acid changing polymorphisms.
TABLE 7.
Overall and within-population allele frequencies of the SNP polymorphisms investigated in this work
| SNP | Overall | Ba9955 | Ba10113 | Ba10158 | Ba10278 | Ba10284 | Ba10732 | Ba10870 | Ba11304 | Ba12945 |
|---|---|---|---|---|---|---|---|---|---|---|
| LpHD1 | ||||||||||
| 320(C/T) | 0.88 | 0.95 | 0.82 | 0.88 | 0.86 | 0.79 | 0.88 | 0.92 | 0.93 | 0.88 |
| 513(C/T) | 0.95 | 0.86 | 0.97 | 0.95 | 0.96 | 0.79 | 1.00 | 1.00 | 1.00 | 1.00 |
| 1475(T/A) | 0.79 | 0.70 | 0.96 | 0.80 | 0.81 | 0.60 | 0.75 | 0.67 | 0.86 | 0.92 |
| 2118(T/C) | 0.62 | 0.57 | 0.23 | 0.92 | 0.78 | 0.92 | 0.36 | 0.68 | 0.70 | 0.37 |
| 2389(C/T) | 0.75 | 0.78 | 0.50 | 0.49 | 0.65 | 0.98 | 1.00 | 0.79 | 0.62 | 0.90 |
| 4443(C/A) | 0.81 | 0.58 | 0.90 | 0.99 | 0.97 | 1.00 | 0.72 | 0.77 | 0.98 | 0.43 |
| 4717(G/A) | 0.97 | 1.00 | 0.77 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 5443(G/T) | 0.79 | 0.70 | 0.95 | 0.80 | 0.82 | 0.60 | 0.77 | 0.66 | 0.86 | 0.92 |
| LpcAI | ||||||||||
| 202(G/A) | 0.61 | 0.30 | 0.64 | 0.68 | 0.75 | 0.87 | 0.53 | 0.39 | 0.85 | 0.37 |
| 382(C/T) | 0.50 | 0.15 | 0.58 | 0.58 | 0.66 | 0.56 | 0.53 | 0.34 | 0.78 | 0.34 |
| 492(I/D) | 0.48 | 0.14 | 0.58 | 0.55 | 0.57 | 0.55 | 0.53 | 0.33 | 0.76 | 0.32 |
| 805(T/C) | 0.87 | 0.97 | 0.80 | 0.89 | 0.65 | 0.78 | 1.00 | 0.98 | 0.79 | 0.97 |
| 825(A/T) | 0.60 | 0.29 | 0.62 | 0.67 | 0.74 | 0.86 | 0.61 | 0.45 | 0.84 | 0.36 |
| 953(T/C) | 0.51 | 0.14 | 0.57 | 0.64 | 0.71 | 0.56 | 0.53 | 0.43 | 0.77 | 0.28 |
| 1053(A/G) | 0.57 | 0.15 | 0.62 | 0.66 | 0.74 | 0.81 | 0.60 | 0.42 | 0.81 | 0.34 |
| 1954(C/G) | 0.89 | 0.86 | 0.94 | 0.93 | 0.97 | 0.69 | 0.90 | 0.96 | 0.94 | 0.92 |
| 1970(T/G) | 0.53 | 0.16 | 0.61 | 0.63 | 0.59 | 0.73 | 0.59 | 0.33 | 0.76 | 0.28 |
| 2214(C/T) | 0.91 | 0.87 | 0.95 | 0.93 | 0.97 | 0.68 | 0.90 | 0.94 | 0.94 | 0.97 |
| 2283(G/A) | 0.52 | 0.15 | 0.62 | 0.61 | 0.62 | 0.73 | 0.57 | 0.34 | 0.78 | 0.29 |
| 2647(T/C) | 0.62 | 0.29 | 0.64 | 0.71 | 0.76 | 0.89 | 0.63 | 0.49 | 0.83 | 0.35 |
| 4879(G/A) | 0.92 | 0.79 | 0.94 | 0.96 | 0.98 | 1.00 | 0.95 | 0.84 | 0.95 | 0.86 |
| 5259(G/C) | 0.52 | 0.86 | 0.42 | 0.45 | 0.57 | 0.44 | 0.47 | 0.68 | 0.25 | 0.71 |
| 5395(A/G) | 0.52 | 0.86 | 0.41 | 0.45 | 0.43 | 0.44 | 0.47 | 0.67 | 0.24 | 0.71 |
The frequencies refer to the first allelic variant at each locus. I/D, insertion/deletion.
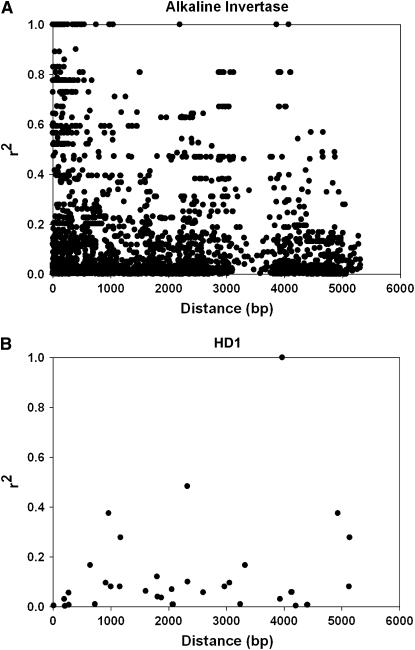
The pattern of LD in the two genes is shown in Figure 2. In the LpcAI gene LD decayed to below 0.2 within 1–2 kb, although there was still significant LD between some loci at larger distances (Figure 2A). Of the 3655 pair-wise comparisons 996 were significant (P < 0.05), and of those, 177 were still significant after Bonferroni correction for multiple testing. The observed P-values were plotted against the expected P-values, expressed as −log(i/(L + 1)), where i is the ith smallest P-value, and L is the number of pair-wise comparisons. The values deviated significantly from the expected 1 to 1 ratio (L. Skøt, unpublished data), suggesting that population structure or other systematic forces were influencing the result (Balding 2006). Nevertheless, Figure 2A shows that there are many significant pair-wise LD values at distances >4000 bp. The small number of SNPs in the LpHD1 locus made it difficult to draw firm conclusions about decay of LD with distance. Only 9 of the 12 polymorphisms were included in this analysis, as the remaining 3 were singletons. Of the 36 pair-wise comparisons, 8 were significant before Bonferroni correction, and 2 were significant after. Within-population LD patterns in the LpHD1 locus is summarized in Figure 3. They show that the proportion of locus-pairs in LD tended to be largest in Ba9955, Ba10732, Ba10870, and Ba12945, all intermediate to very late flowering populations (see Tables 1 and 4).
Figure 2.—
Pattern of LD in the LpcAI (Alkaline invertase) (A) and the LpHD1 (HD1) (B) loci, as detected in the test set of genotypes used for resequencing. The r2 value for pairwise LD is plotted against physical distance.
Figure 3.—
Within-population LD pattern in the LpHD1 locus. The top right diagonal represents the LD expressed as r2. Black squares, r2 ≥ 0.5; dark gray squares, 0.2 ≤ r2 ≤ 0.5; light gray squares, 0.05 ≤ r2 ≤ 0.2; white squares, r2 ≤ 0.05. The bottom left diagonal represents Fisher's exact test probabilities. Black squares, P ≤ 0.0001; gray squares, 0.0001 ≤ P ≤ 0.001; white squares, 0.001 ≤ P. Gray and black squares in the bottom left diagonal are all significant (P < 0.05) after Bonferroni correction for multiple testing.
Population structure:
The population structure was investigated using AFLP markers. A total of 506 markers with a band frequency ≥0.05 were produced from amplification with six selective primer pair combinations. They were analyzed with the AFLP-SURV version 1.0 software for basic population genetics parameters including gene diversity (expected heterozygosity) and F statistics (population differentiation). The results are summarized in Table 8 and show that within-population heterozygosity accounted for 85–91% of the total heterozygosity, which is consistent with many previous assessments in L. perenne. This confirms previous work that within-population genetic diversity is generally much larger than between populations in this species (Roldan-Ruiz et al. 2000; Cresswell et al. 2001; Skøt et al. 2005). The FST values indicate that ∼10–15% of the total genetic variation is due to population structure. The presence of population substructure is not surprising, given the diverse geographic origins and the deliberate selection of accessions with the widest possible range of variation in flowering time (see Table 1).
TABLE 8.
Basic population genetic data from the nine populations
| Primer pair | Loci | HT | HW | HB | FST |
|---|---|---|---|---|---|
| ACA_CAC | 89 | 0.2403 | 0.2118 (0.0051) | 0.0285 (0.0019) | 0.1184 (0.0594) |
| ACA_CTA | 113 | 0.2906 | 0.2596 (0.0081) | 0.0309 (0.0015) | 0.1064 (0.0575) |
| ACT_CAC | 65 | 0.2298 | 0.2057 (0.0057) | 0.0241 (0.0027) | 0.1050 (0.0975) |
| ACT_CTA | 104 | 0.2665 | 0.2410 (0.0053) | 0.0255 (0.0018) | 0.0958 (0.0648) |
| ACG_CTA | 56 | 0.2604 | 0.2202 (0.0072) | 0.0401 (0.0049) | 0.1543 (0.1131) |
| ACG_CTG | 79 | 0.2399 | 0.2181 (0.0058) | 0.0218 (0.0000) | 0.0908 (0.0140) |
A total of 864 genotypes derived from AFLP marker data. The genetic diversity and population subdivision measures follow the notation of Lynch and Milligan (1994). HW is the within-population genetic diversity, HB is the between-population diversity, and HT = HW + HB. FST = HB/HT is Wright's measure of population subdivision. Numbers in brackets are standard errors based on 1000 permutations.
The AFLP marker data were also used in a more detailed analysis of population structure with the STRUCTURE version 2.1 software program. Of the 506 markers, 73 were also polymorphic in an F2 mapping family described elsewhere (Turner et al. 2006). While AFLP marker distribution can be clustered (Bert et al. 1999), we found that the 73 mapped markers were distributed fairly randomly on each of the seven linkage groups of L. perenne (between 6 and 17 per linkage group). If extrapolated to the unmapped markers, it would be between 40 and 120 per linkage group. Data representing AFLP markers for each primer pair were analyzed individually, but they all gave similar results. When we included a prior assumption of nine populations, and K was varied from 2 to 10, the number of inferred clusters with the highest probability was eight or nine, depending on the primer pair, and they coincided well with the nine given. Moreover, for each genotype, there was little evidence of ancestry from more than one inferred cluster. An almost identical result was obtained without a priori assuming the presence of nine populations. When each of the nine populations were analyzed separately, with K varied from 2 to 6, the proportion of ancestry from the different clusters was approximately evenly distributed between the clusters, suggesting little or no population structure within the nine populations (see documentation for STRUCTURE software version 2). The within-population analysis was based on fewer polymorphic markers than 506 owing to absence of markers in some populations. Nevertheless, it was still between 325 and 418, depending upon the population, sufficient to detect within-population structure, if it was present. Taken together, these results led us to the conclusion that the 864 genotypes were clustered in nine groups, coinciding with the nine accessions used in this work.
Association analysis:
An initial association analysis was performed using one-way ANOVA without taking population structure into account. There was no significant association between any of the SNPs in the LpcAI gene and WSC, N, or DMD in 2004, but four loci associated with WSC and DMD in 2005 (SNPs 1970, 2283, 5259, and 5395). The latter trait also associated with two further loci in the 5′ untranslated region of the gene (SNPs 382 and 492) (P < 0.05). There was highly significant association between HD and three SNPs in the LpHD1 locus in both years (SNPs 2118, 2389, and 4443) (P < 0.0001). The three SNPs are located in the intergenic region between the putative peroxidise precursor gene and LpHD1. One of them (4443) is located 265 bp upstream of the translational start site of LpHD1.
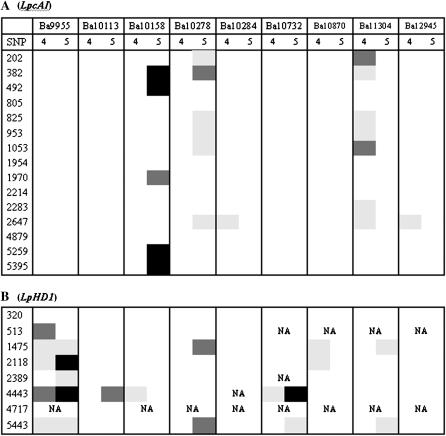
The presence of population structure makes it likely that some of these associations are spurious. Three strategies were used to correct for this. First, a one-way ANOVA was performed on data from individual populations separately, assuming no within-population substructure. In the LpcAI locus, seven SNPs associated with WSC in Ba11304 in 2004 (Figure 4A). Six of those SNPs associated with WSC in Ba10278. Five SNPs were also significant in Ba10158 in 2005, but only SNP_382 was significant in all three populations. For DMD, 11 SNPS were significant in Ba10732 in 2004. Five of those were also significant in Ba10284, and six were significant in Ba10158 in 2005 (L. Skøt, unpublished data). A more consistent pattern emerged from the association analysis of the LpHD1 gene (Figure 4B). First, there were a large number of associations with HD in the Ba9955 population (six in 2004 and five in 2005, with a high degree of overlap). Second, the SNPs 4443 and 5443 were both significantly associated with HD in six samples. Both SNPs are located in the HD1 gene or immediately upstream, rather than in the putative peroxidase precursor gene. Furthermore, the degree of significance was particularly high for the 4443 SNP (P = 0.0078 for Ba9955 in 2004, and P = 0.0001 in 2005; for Ba10732 in 2005 P = 8.71 × 10−8).
Figure 4.—
One-way ANOVA of association of WSC (A) with SNPs in the LpcAI gene and HD (B) with SNPs in individual populations in 2004 and 2005. Phenotypic values represent the response variate, and genotypes are the independent variables. The numbers 4 and 5 in A represent years. The shading represents different levels of significance: Light gray, P < 0.05; intermediate gray, P < 0.01; black: P < 0.001. NA, not analyzed owing to monomorphism at that locus.
The second strategy consisted of a linear mixed model analysis of the data, in which population was incorporated by including the nine inferred groups (i.e., populations) as random effects. Only one SNP (2647) in the LpcAI gene associated with WSC in 2004 (P = 0.05), but not in 2005. In the LpHD1 gene however, SNP 4443 was significantly associated with HD in both 2004 and 2005 (P = 0.05 and P < 0.001, respectively).
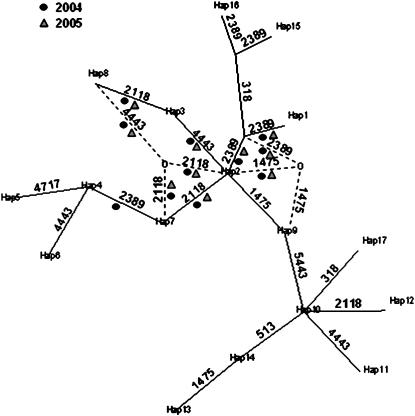
Recently, the use of haplotype trees has been advocated for the analysis of genotype/phenotype associations, as they have the potential to uncover associations with extended haplotypes, particularly if the level of LD is significant (Buntjer et al. 2005; Templeton et al. 2005). Since there was evidence of significant LD in both loci investigated here over the whole of each gene, we carried out the tree-scanning analysis as described by Templeton et al. (2005) as the third analysis. The heterozygous nature of L. perenne meant that phases had to be inferred. This was performed using the program PHASE version 2.0 as described above and resulted in 17 haplotypes over the eight loci of the LpHD1 gene (Table 9). They were used to produce a maximum parsimony tree in the program PHYLIP. Changing the haplotype used as an outgroup root did not alter the outcome of the association analysis significantly. The information from this plus the most likely diplotypes of the 864 genotypes and the phenotype data were all used in the TREESCAN program. The result of the first round of tree scanning for the whole data set is shown in Figure 5. The significant branch points are connecting the same haplotypes in both of the alternative haplotype trees. For reasons of simplicity we will therefore focus on the results of the tree-scanning analysis of the first tree. Figure 5 illustrates the consistency of the results between the 2 years. The only discrepancy was the transition between haplotype 4 and 7, which was only significant in 2004. The significant branch points all involve three polymorphisms, namely 2118, 2389, and 4443. The single exception is the branch between haplotype 2 and an intermediate haplotype, not present in the sample, joining haplotype 9 in the second haplotype tree (Figure 5). This is the 1475 SNP, which changes an asparagine to isoleucine in the first exon of the putative peroxidase precursor protein (Figure 1B). The 2118 and 4443 SNPs were one of only two pair-wise SNPs, which were in significant LD after Bonferroni correction for multiple testing. These three polymorphisms were also highly significantly associated with HD in the single SNP ANOVA test. However, this tree-scanning analysis was performed on all 864 genotypes without consideration of population structure. We therefore did the analysis again on each population separately. Table 10 shows that Ba9955, Ba10732, and Ba10870 had significant associations at branch points involving the 2118 and 4443 polymorphisms, but not 2389 in all three populations. In addition, Ba9955 was significant at the branch between haplotypes 10 and 14 (polymorphism 513) in 2004. The reduced number of significant branches could be caused by loss of power due to smaller number of genotypes in each analysis, and/or by false positives in the analysis of the full data set ignoring population structure.
TABLE 9.
Haplotypes of the LpHD1 gene
| Haplotype no. | Haplotype | Inferred no. in sample |
|---|---|---|
| 1 | CCTTTCGG | 342 |
| 2 | CCTTCCGG | 146 |
| 3 | CCTTCAGG | 1 |
| 4 | CCTCTCGG | 44 |
| 5 | CCTCTCAG | 44 |
| 6 | CCTCTAGG | 1 |
| 7 | CCTCCCGG | 259 |
| 8 | CCTCCAGG | 315 |
| 9 | CCATCCGG | 3 |
| 10 | CCATCCGT | 272 |
| 11 | CCATCAGT | 3 |
| 12 | CCACCCGT | 1 |
| 13 | CTTTCCGT | 1 |
| 14 | CTATCCGT | 87 |
| 15 | TCTTTCGG | 7 |
| 16 | TCTTCCGG | 199 |
| 17 | TCATCCGT | 3 |
Haplotypes of the LpHD1 gene and the number of times they are estimated to occur in the sample in the best reconstruction as implemented in the PHASE version 20 program and described in materials and methods.
Figure 5.—
Haplotype tree of the SNP polymorphisms from the LpHD1 gene. The numbers connecting the haplotype nodes represent the position of the SNP polymorphism. The haplotypes are shown in Table 9. The circular and triangular symbols indicate transitions that were statistically significant in the tree-scanning analysis in 2004 and 2005, respectively. The broken lines refer to the alternative branches of the second haplotype tree that could be resolved, because of the phylogenetic ambiguity. The 0's represent intermediate haplotypes not present in the sample. The tree was obtained from the program Treeview (Page 1996) on the basis of the treefile output from PHYLIP.
TABLE 10.
Tree-scanning analysis within populations
| Population | Year | Branch | F-statistic | Pvk | PSim | PMon |
|---|---|---|---|---|---|---|
| Ba9955 | 2004 | 10–14 | 7.545 | 0.066 | 0.0120 | 0.0330 |
| Ba9955 | 2005 | 10–14 | 3.792 | 0.028 | 0.0560 | 0.2570 |
| Ba9955 | 2004 | 2–3 | 5.139 | 0.082 | 0.0030 | 0.0880 |
| Ba9955 | 2005 | 2–3 | 9.616 | 0.152 | 0.0000 | 0.0100 |
| Ba9955 | 2004 | 3–8 | 5.139 | 0.082 | 0.0050 | 0.0880 |
| Ba9955 | 2005 | 3–8 | 9.616 | 0.152 | 0.0000 | 0.0100 |
| Ba10732 | 2004 | 2–3 | 3.943 | 0.058 | 0.0270 | 0.2390 |
| Ba10732 | 2005 | 2–3 | 17.657 | 0.258 | 0.0000 | 0.0000 |
| Ba10732 | 2004 | 3–8 | 3.943 | 0.058 | 0.0220 | 0.2390 |
| Ba10732 | 2005 | 3–8 | 17.657 | 0.258 | 0.0000 | 0.0000 |
| Ba10870 | 2004 | 2–7 | 6.505 | 0.055 | 0.0150 | 0.0350 |
| Ba10870 | 2005 | 2–7 | 6.205 | 0.052 | 0.0080 | 0.0370 |
Branches with significant association with HD after correction for multiple testing and enforcement of monotonicity (PMon) in at least 1 year are shown. PSim is the permutational probability before correction for multiple testing. Pvk is the proportion of the trait variation explained by the partition. There were no significant associations in the second round of tree scanning.
In the LpcAI gene the PHASE program identified 37 haplotypes in the best reconstruction of the sample. However, the phylogenetic ambiguity of the data meant that it could be resolved in 75 possible haplotype trees. A subset of 10 randomly chosen trees was used in the TREESCAN analysis, and in every case the same branch between haplotype 7 and 31 was significant for WSC in 2004 and N in 2005. This involves the SNP_202 polymorphism (Figure 1A). When the analysis was performed on individual populations no significant branches were found.
DISCUSSION
Polymorphism in LpcAI and LpHD1:
The large difference between LpcAI and LpHD1 in the number of polymorphisms (Table 6) and nucleotide diversity, particularly in the first segment of the LpcAI gene, may at first glance suggest that the LpHD1 gene is functionally more important and has been subject to stronger selective pressure than the LpcAI gene. However, the proportion of nonsynonymous SNPs is actually higher in the LpHD1 locus (16.7%) than in LpcAI (8.7%). Admittedly, this comparison is based on very different total numbers of SNPs in the two genes (Table 6). Nevertheless, the vast majority of SNPs in LpcAI are located in the 5′ upstream region. It is also interesting to note that the overall Tajima's D value in one window (939–1038) of the LpcAI gene was significantly positive, while it was nonsignificant throughout the LpHD1 locus. The positive Tajima's D values in both genes indicate a trend toward excess of intermediate frequency alleles. This effect can be caused by population bottlenecks, structure, or balancing selection (Biswas and Akey 2006). In view of the strong evidence for significant population structure in the plant material used here, this may be a contributing factor.
The presence of a high degree of polymorphism raises the possibility that the PCR-based resequencing of the LpcAI gene may have amplified different genes of a gene family. It is well established that temperate grasses, including L. perenne, have a number of closely related genes involved in fructan and sucrose metabolism, including invertases (Gallagher et al. 2004; Chalmers et al. 2005). We therefore developed a sequence-characterized RFLP marker on the basis of one of the SNPs, which was located in a SfoI restriction site. This mapped to the same location as the original invertase RFLP in the F2 mapping family described by Turner et al. (2006). In the case of the LpHD1 locus, the sequencing was based on a BAC clone isolated from a L. perenne library (Farrar et al. 2007) and identified on the basis of the S2539 marker primers, which span the unique putative peroxidise precursor gene adjacent to the HD1 gene (Armstead et al. 2005). The synteny with the rice HD1 locus on chromosome 6 was confirmed by further sequencing of the BAC clone beyond the peroxidise precursor gene (accession no: AM489608).
Association analysis:
Although there were significant associations identified within populations, none of the analyses identify consistent associations between the polymorphisms in the LpcAI gene and WSC, N, or DMD phenotypes. A possible explanation could of course be that there is no causal link between the allelic variants in LpcAI and these traits. Although the data from different years were analyzed separately to minimize the year effect, the significance of the population × year interaction is most likely a contributory factor.
In the LpHD1 locus, both the ANOVA and tree-scanning analyses of the total set of 864 genotypes identified SNPs 2118, 2389, and 4443 as highly significant. These results could be spurious due to population structure, but the linear mixed model, within-population ANOVA and tree-scanning analyses all identified the 4443 SNP as significantly associated with HD. The latter two methods identified both SNP_2118 and SNP_4443 as highly significant in the Ba9955 population, as well as SNP_4443 in Ba10732. The relative loss of power of the tree-scanning method compared to the within-population ANOVA of individual SNPs, due to the multiple testing issue (Templeton et al. 2005), probably explains why the ANOVA identified more significant associations (Table 10, Figure 4B). Nevertheless, the haplotype tree-based analysis has the potential to add a further dimension to the association analysis by identifying potentially interesting haplotype clusters, which single SNP analysis may miss. In this context it is interesting to note that the PHASE and the tree-scanning analyses show that, of the 320 haplotypes containing the A allele in the 4443 polymorphism, 315 were haplotype 8 (CCTCCAGG) (Table 9). In contrast, the 544 genotypes with the C allele were distributed over 13 haplotypes. It may suggest that this haplotype as a whole, rather than just the 4443-A allele, is associated with late flowering, illustrating the kind of result that the haplotype tree-based analysis has the potential to highlight. However, there are too few genotypes of the other 4443-A allele haplotypes to verify this. As pointed out by Templeton et al. (2005), the tree-scanning method distinguishes between the same allelic variant in different haplotypes and may thus be able to identify a significant specific haplotype with the allelic variant, while the single SNP analysis of the same allelic variant might be diluted by its presence in other nonsignificant haplotypes. This potential advantage of tree-scanning disappears if the allelic variant in question is functional, since the haplotype setting in that case is unimportant. There is as yet no evidence to suggest if the C-4443-A polymorphism is functional or simply in LD with a functional variant.
The within-population ANOVA and tree-scanning analyses show that individual populations differ in their ability to identify associations (Figure 4B, Table 10). This has potential implications for choosing which populations to use for association mapping, although there seems to be no clear pattern to guide the choice of populations for analysis. Some of the differences between populations can be attributed to small or zero minor allele frequencies (Table 7), but Figures 3 and 4B show that the populations in which most associations were found with HD (Ba9955, Ba10732, and Ba10870) were all intermediate or late flowering and also tended to have most locus pairs in significant LD. The Ba12945 population is an exception by not having any loci significantly associated with HD, but a high proportion of locus pairs are in LD, as well as being late flowering, so the significance of this is not clear.
Aranzana et al. (2005) discuss the issue of discarding genuine associations by accounting for population structure, due to association between the polymorphism and population stratification as well. Although all three analysis strategies, which took account of population structure, identified the 4443 polymorphism as significant in this work, Table 7 shows that allele frequency differences exist between populations, particularly in the LpHD1 gene. The 4717 SNP was only polymorphic in the Norwegian population Ba10113, and even in that, it was only homozygous for the G allele or heterozygous (A/G). This SNP was included because it was the only nonsynonymous polymorphism in the LpHD1 coding sequence, changing a valine to methionine (amino acid number 33 in the translated sequence) within the B-box 1 motif of the zinc finger domain in the translated sequence (Yano et al. 2000; Martin et al. 2004; Armstead et al. 2005). The functional importance of this polymorphism is not known, but its rare occurrence would suggest that it is either a very recent mutation or a functionally important one, or both. It would be interesting to obtain genotypes that were homozygous for the methionine allele and assess the impact on HD.
In view of the results of the association analysis, it seemed reasonable to assume that the 4443 polymorphism could be a potentially useful polymorphism for marker-assisted selection for HD. Uniformity of flowering date is essential for variety registration, and fixing the genes with major effect on flowering time directly through SNP allele selection will increase uniformity in this out-crossing species. Flowering time is a relatively time-consuming trait to measure accurately, as it involves recording days after a particular date until ear emergence of the whole breeding population at two-day intervals throughout the flowering period. The marker may be particularly useful in the turf-grass breeding program where one of the goals is to obtain elite varieties with an earlier flowering time, since this is likely to enhance seed yield as harvesting conditions will be more favorable earlier in the year. However, it is crucial to verify the usefulness of markers identified in association analyses as illustrated by Andersen et al. (2005) and Camus-Kulandaivelu et al. (2006), who undertook a second association analysis of the Dwarf8 polymorphisms and flowering time in order to validate the results of Thornsberry et al. (2001). We have therefore carried out crosses between genotypes from the Ba10732 population, which were homozygous for the “early” C allele, with either heterozygous or homozygous “late” A allele turf-grass varieties. Sixty genotypes of the turf-grass cultivar AberElf were screened for the 4443 SNP and were all found to be homozygous A/A. As well as being extremely late heading, AberElf also has low seed set. The LpHD1 gene underlies a highly significant QTL, presumably influenced by unrelated but linked gene(s), that increases seed setting two- and fourfold in two unrelated perennial ryegrass mapping families (L. Skøt, unpublished data); and the 4443 SNP is a potentially useful marker for jointly bringing forward flowering date and improving seed setting, a major component of seed yield in out-crossing crops in a backcross breeding program. The segregating progeny will be evaluated for HD and seed setting.
In conclusion, a potential candidate SNP (4443) was identified in the LpHD1 locus, which consistently associated with HD. Its usefulness as a marker is currently being assessed by crosses with turf-grass varieties. While haplotype tree-based association mapping has been described in the model species A. thaliana (Olsen et al. 2004; Aranzana et al. 2005), this is, to our knowledge, the first use of the tree-scanning analysis in a crop species. It also identified the 4443 polymorphism as in the single SNP analysis, but extended it to the 2118 SNP, despite the loss of power due to correction for a larger number of multiple tests compared to single SNP analyses (Templeton et al. 2005). As stated by Buntjer et al. (2005), haplotype tree-based association analysis may have further potential to infer unobserved haplotypes in the sample and predicting their phenotype. In combination with single SNP analyses it could assist in distinguishing between functional and nonfunctional allelic variants merely linked to a functional variant.
Acknowledgments
We thank Zewei Luo, Michael Kearsey, and Kuruvilla Abraham at the School of Biosciences, The University of Birmingham, United Kingdom for very useful discussions about this work. We are grateful to Sue Heywood for technical assistance, Kirsten Skøt for sequencing and AFLP analysis, Sue Lister for the NIRS analysis, and Mark Hirst for advice about the linear mixed model analysis. This work was funded by responsive mode grant no. 203/D18078 and a competitive strategic grant from the Biotechnology and Biological Sciences Research Council.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AM489608 and AM489692bb
References
- Andersen, J. R., T. Schrag, A. E. Melchinger, I. Zein and T. Lubberstedt, 2005. Validation of Dwarf8 polymorphisms associated with flowering time in elite European inbred lines of maize (Zea mays L.). Theor. Appl. Genet. 111: 206–217. [DOI] [PubMed] [Google Scholar]
- Aranzana, M. J., S. Kim, K. Zhao, E. Bakker, M. Horton et al. 2005. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstead, I. P., L. B. Turner, M. Farrell, L. Skøt, P. Gomez et al., 2004. Synteny between a major heading-date QTL in perennial ryegrass (Lolium perenne L.) and the Hd3 heading-date locus in rice. Theor. Appl. Genet. 108: 822–828. [DOI] [PubMed] [Google Scholar]
- Armstead, I. P., L. Skøt, L. B. Turner, K. Skøt, I. S. Donnison et al., 2005. Identification of perennial ryegrass (Lolium perenne (L.)) and meadow fescue (Festuca pratensis (Huds.)) candidate orthologous sequences to the rice Hd1(Se1) and barley HvCO1 CONSTANS-like genes through comparative mapping and microsynteny. New Phytol. 167: 239–247. [DOI] [PubMed] [Google Scholar]
- Balding, D. J., 2006. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 7: 781–791. [DOI] [PubMed] [Google Scholar]
- Bao, J. S., H. Corke and M. Sun, 2006. a Microsatellites, single nucleotide polymorphisms and a sequence tagged site in starch-synthesizing genes in relation to starch physicochemical properties in nonwaxy rice (Oryza sativa L.). Theor. Appl. Genet. 113: 1185–1196. [DOI] [PubMed] [Google Scholar]
- Bao, J. S., H. Corke and M. Sun, 2006. b Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor. Appl. Genet. 113: 1171–1183. [DOI] [PubMed] [Google Scholar]
- Bert, P. F., G. Charmet, P. Sourdille, M. D. Hayward and F. Balfourier, 1999. A high-density molecular map for ryegrass (lolium perenne) using AFLP markers. Theor. Appl. Genet. 99: 445–452. [DOI] [PubMed] [Google Scholar]
- Biswas, S., and J. M. Akey, 2006. Genomic insights into positive selection. Trends Genet. 22: 437–446. [DOI] [PubMed] [Google Scholar]
- Breseghello, F., and M. E. Sorrells, 2006. a Association analysis as a strategy for improvement of quantitative traits in plants. Crop Sci. 46: 1323–1330. [Google Scholar]
- Breseghello, F., and M. E. Sorrells, 2006. b Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172: 1165–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buntjer, J. B., A. P. Sorensen and J. D. Peleman, 2005. Haplotype diversity: the link between statistical and biological association. Trends Plant Sci. 10: 466–471. [DOI] [PubMed] [Google Scholar]
- Camus-Kulandaivelu, L., J. B. Veyrieras, D. Madur, V. Combes, M. Fourmann et al., 2006. Maize adaptation to temperate climate: relationship between population structure and polymorphism in the Dwarf8 gene. Genetics 172: 2449–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers, J., A. Lidgett, N. Cummings, Y. Cao, J. Forster et al., 2005. Molecular genetics of fructan metabolism in perennial ryegrass. Plant Biotechnol. J. 3: 459–474. [DOI] [PubMed] [Google Scholar]
- Cornish, M. A., M. D. Hayward and M. J. Lawrence, 1979. Self incompatibility in ryegrass. 1. Genetic control in diploid Lolium perenne L. Heredity 43: 95–106. [Google Scholar]
- Cresswell, A., N. R. Sackville Hamilton, A. K. Roy and B. M. F. Viegas, 2001. Use of amplified fragment length polymorphism markers to assess genetic diversity of Lolium species from Portugal. Mol. Ecol. 10: 229–241. [DOI] [PubMed] [Google Scholar]
- Farrar, K., T. Asp, T. Lubberstedt, M. Xu, A. Thomas et al., 2007. Construction of two Lolium perenne BAC libraries and identification of BACs containing candidate genes for disease resistance and forage quality. Mol. Breed. 19: 15–23. [Google Scholar]
- Felsenstein, J., 1993. PHYLIP (Phylogenetic Inference Package) version 3.5c. Department of Genetics, University of Washington, Seattle.
- Flint-Garcia, S. A., J. M. Thornsberry and E. S. Buckler, 2003. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54: 357–374. [DOI] [PubMed] [Google Scholar]
- Flint-Garcia, S. A., A. C. Thuillet, J. Yu, G. Pressoir, S. M. Romero et al., 2005. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant Journal 44: 1054–1064. [DOI] [PubMed] [Google Scholar]
- Gallagher, J., and C. Pollock, 1998. Isolation and characterization of a cDNA clone from Lolium temulentum L. encoding for a sucrose hydrolytic enzyme which shows alkaline/neutral invertase activity. J. Exp. Bot. 49: 789–795. [Google Scholar]
- Gallagher, J. A., A. J. Cairns and C. J. Pollock, 2004. Cloning and characterization of a putative fructosyltransferase and two putative invertase genes from the temperate grass Lolium temulentum L. J. Exp. Bot. 55: 557–569. [DOI] [PubMed] [Google Scholar]
- Gallagher, J. A., A. J. Cairns and L. B. Turner, 2007. Fructan in temperate forage grasses, agronomy, physiology and molecular biology, pp. 15–46 in Recent Advances in Fructooligosaccharides Research, edited by N. Benkeblia. Research Signpost, Kerala, India.
- Gaut, B. S., and A. D. Long, 2003. The lowdown on linkage disequilibrium. Plant Cell 15: 1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, P. K., S. Rustgi and P. L. Kulwal, 2005. Linkage disequilibrium and association studies in higher plants: Present status and future prospects. Plant Mol. Biol. 57: 461–485. [DOI] [PubMed] [Google Scholar]
- Humphreys, M. W., R. S. Yadav, A. J. Cairns, L. B. Turner, J. Humphreys et al., 2006. A changing climate for grassland research. New Phytol. 169: 9–26. [DOI] [PubMed] [Google Scholar]
- Ivandic, V., C. A. Hackett, E. Nevo, R. Keith, W. T. B. Thomas et al., 2002. Analysis of simple sequence repeats (SSRs) in wild barley from the Fertile Crescent: associations with ecology, geography and flowering time. Plant Mol. Biol. 48: 511–527. [DOI] [PubMed] [Google Scholar]
- Jones, E. S., N. L. Mahoney, M. D. Hayward, I. P. Armstead, J. G. Jones et al., 2002. An enhanced molecular marker based genetic map of perennial ryegrass (Lolium perenne) reveals comparative relationships with other Poaceae genomes. Genome 45: 282–295. [DOI] [PubMed] [Google Scholar]
- Kraakman, A., F. Martinez, B. Mussiraliev, F. van Eeuwijk and R. Niks, 2006. Linkage disequilibrium mapping of morphological, resistance, and other agronomically relevant traits in modern spring barley cultivars. Mol. Breed. 17: 41–58. [Google Scholar]
- Lister, S. J., and M. S. Dhanoa, 1998. Comparison of calibration models for the prediction of forage quality traits using near infrared spectroscopy. J. Agric. Sci. 131: 241–242. [Google Scholar]
- Lynch, M., and B. G. Milligan, 1994. Analysis of population genetic structure with RAPD markers. Mol. Ecol. 3: 91–99. [DOI] [PubMed] [Google Scholar]
- Martin, J., M. Storgaard, C. H. Andersen and K. K. Nielsen, 2004. Photoperiodic regulation of flowering in perennial ryegrass involving a CONSTANS-like homolog. Plant Mol. Biol. 56: 159–169. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., and S. Tavare, 2002. Linkage disequilibrium: what history has to tell us. Trends Genet. 18: 83–90. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., J. O. Borevitz, J. Bergelson, C. C. Berry, J. Chory et al., 2002. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30: 190–193. [DOI] [PubMed] [Google Scholar]
- Olsen, K. M., S. S. Halldorsdottir, J. R. Stinchcombe, C. Weinig, J. Schmitt et al., 2004. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167: 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R. D. M., 1996. An application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12: 357–358. [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill, J., F. Robson, K. Lee, R. Simon and G. Coupland, 1995. The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Rafalski, A., and M. Morgante, 2004. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 20: 103–111. [DOI] [PubMed] [Google Scholar]
- Roldan-Ruiz, I., J. Dendauw, E. V. Bockstaele, A. Depicker and M. D. Loose, 2000. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breed. 6: 125–134. [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Skøt, L., M. O. Humphreys, I. Armstead, S. Heywood, K. P. Skøt et al., 2005. An association mapping approach to identify flowering time genes in natural populations of Lolium perenne (L.). Mol. Breed. 15: 233–245. [Google Scholar]
- Stephens, M., and P. Donnelly, 2003. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73: 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, M., N. J. Smith and P. Donnelly, 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalma, S. J., E. S. Buckler, M. E. Snook and M. D. McMullen, 2005. Association analysis of candidate genes for maysin and chlorogenic acid accumulation in maize silks. Theor. Appl. Genet. 110: 1324–1333. [DOI] [PubMed] [Google Scholar]
- Templeton, A. R., T. Maxwell, D. Posada, J. H. Stengard, E. Boerwinkle et al., 2005. Tree scanning: A method for using haplotype trees in phenotype/genotype association studies. Genetics 169: 441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielsen et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289. [DOI] [PubMed] [Google Scholar]
- Turner, L. B., A. J. Cairns, I. P. Armstead, J. Ashton, K. Skøt et al., 2006. Dissecting the regulation of fructan metabolism in perennial ryegrass (Lolium perenne) with quantitative trait locus mapping. New Phytol. 169: 45–58. [DOI] [PubMed] [Google Scholar]
- Vekemans, X., 2002. AFLP-SURV version 1.0. Laboratoire de Génétique et Ecologie Végétale, Université Libre de Bruxelles, Brussels, Belgium.
- Wilkins, P. W., and M. O. Humphreys, 2003. Progress in breeding perennial forage grasses for temperate agriculture. J. Agric. Sci. 140: 129–150. [Google Scholar]
- Wilson, L. M., S. R. Whitt, A. M. Ibanez, T. R. Rocheford, M. M. Goodman et al., 2004. Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16: 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., Y. Katayose, M. Ashikari, U. Yamanouchi, L. Monna et al., 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., and E. S. Buckler, 2006. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 17: 155–160. [DOI] [PubMed] [Google Scholar]
- Yu, J., G. Pressoir, W. H. Briggs, I. Vroh Bi, M. Yamasaki et al., 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]