Abstract
The study of DNA double-strand break (DSB) repair has been greatly facilitated by the use of rare-cutting endonucleases, which induce a break precisely at their cut sites that can be strategically placed in the genome. We previously established such a system in Drosophila and showed that the yeast I-SceI enzyme cuts efficiently in Drosophila cells and those breaks are effectively repaired by conserved mechanisms. In this study, we determined the genetic requirements for the repair of this I-SceI-induced DSB in the germline. We show that Drosophila Rad51 and Rad54 are both required for homologous repair by gene conversion, but are dispensable for single-strand annealing repair. We provided evidence suggesting that Rad51 is more stringently required than Rad54 for intersister gene conversion. We uncovered a significant role of DNA ligase IV in nonhomologous end joining. We conducted a screen for candidate mutations affecting DSB repair and discovered novel mutations in genes that include mutagen sensitive 206, single-strand annealing reducer, and others. In addition, we demonstrated an intricate balance among different repair pathways in which the cell differentially utilizes repair mechanisms in response to both changes in the genomic environment surrounding the break and deficiencies in one or the other repair pathways.
A eukaryotic cell employs a variety of conserved mechanisms to repair double-strand breaks (DSBs), which threaten the integrity of its genome. These mechanisms can be grossly grouped into two pathways: homologous recombinational (HR) repair and nonhomologous end joining (NHEJ). Gene conversion (GC) is a common outcome of HR both in mitotic and in meiotic cells (reviewed in Paques and Haber 1999). In GC, the DSB is repaired by DNA synthesis templated from a homologous segment. GC is generally conservative, resulting in no net loss of DNA sequences. If the template for GC is located on the sister chromatid, such repair precisely restores the original sequence at the break. Many factors play important roles in regulating GC, notably the Rad52 epistasis group in budding yeast and their homologs in other organisms (Symington 2002). These include Rad50, Rad51, Rad52, Rad54, Rad59, Mre11, and others. Single-strand annealing (SSA) repair is commonly used to repair DSBs that occur between direct repeats (Paques and Haber 1999). SSA is nonconservative, resulting in the loss of one of the repeats as well as the segment between the repeats. The budding yeast Rad52 and Rad59 proteins are essential for SSA (Ivanov et al. 1996; Sugawara et al. 2000; Davis and Symington 2001), but Drosophila homologs for neither protein can be identified by sequence homology searches. The identification of their functional homologs in flies would have important implications since a similar situation exists for both Caenorhabditis elegans and Arabidopsis.
In NHEJ, the two ends of the DSB are ligated with little or no homology requirement between them. NHEJ is intrinsically mutagenic in that it can lead to sequence alteration at the site of DSB. On the other hand, precise end joining can be a predominant pathway if the ends have complementary single-stranded overhangs (Boulton and Jackson 1996). Several conserved proteins have been shown to regulate NHEJ, which include the Ku70–Ku80 heterodimer and DNA ligase IV (reviewed in Daley et al. 2005). However, recent studies in Drosophila shed doubts on the importance of ligase IV in regulating end joining (Bi et al. 2004; McVey et al. 2004a; Romeijn et al. 2005).
Our understanding of repair mechanisms has been greatly enhanced by studies using site-specific endonucleases, especially rare cutters. The advantages of being able to induce site-specific DSBs on demand are manyfold. One can control the timing and severity of DSB generation by manipulating endonuclease production. One can control the number and genomic location of the DSB by strategically placing the enzyme cut site. Last, one can engineer a specific genomic environment surrounding the DSB site so that a particular mode(s) of repair can be studied in detail. These advantages have been best exemplified by the use of the HO endonuclease in the yeast Saccharomyces cerevisiae (reviewed in Paques and Haber 1999), which leads to our detailed understanding of important molecular events during DSB repair in vivo, such as 5′–3′ break resection (White and Haber 1990), sequential loading of repair proteins (Sugawara et al. 2003; Wolner et al. 2003), and de novo telomere formation (Diede and Gottschling 1999). Modeled after the success of the HO system, the yeast rare-cutting I-SceI enzyme was successfully introduced to both plant and mammalian cells to induce site-specific DSBs (Puchta et al. 1993; Rouet et al. 1994) and subsequently to conduct functional studies of repair factors, especially those that may be specific to higher eukaryotes (e.g., Moynahan et al. 1999, 2001; Tauchi et al. 2002).
We and others have successfully introduced the I-SceI site-specific DSB system to Drosophila (Bellaiche et al. 1999; Rong and Golic 2000). We showed that I-SceI cuts very efficiently in the fly genome and the single DSB at its cut site can be effectively repaired by a variety of mechanisms, which include SSA, GC, and imprecise NHEJ (Rong and Golic 2003). Since a DSB can be repaired by either HR or NHEJ, this creates a potentially competitive situation. Competition between precise end joining and GC for the same DSBs has been demonstrated in yeast (Frank-Vaillant and Marcand 2002). In other yeast studies, competition between NHEJ and HR was not observed, casting doubts on the generality of interpathway competitions (Karathanasis and Wilson 2002; Zhang and Paull 2005). Within each pathway, either HR or NHEJ, a similar competitive situation could also exist. Recently, we showed that interhomolog GC competes effectively with SSA in the germline of male Drosophila (Rong and Golic 2003). More recently, differential usage of repair pathways during normal Drosophila development has also been demonstrated (Preston et al. 2006a,b). However, mutational studies have been scarce in metazoans in which one investigates whether defects in one repair mechanism can be compensated by a higher utilization of other mechanisms (e.g., Johnson-Schlitz and Engels 2006; Johnson-Schlitz et al. 2007). In this study, we combine the use of several versatile I-SceI-based repair assays and the use of known repair-defective mutants to demonstrate an intricate balance among repair pathways.
Defects in DNA damage repair often lead to cellular sensitivity to DNA damaging agents (Game and Mortimer 1974). Mutagen-sensitive (mus) mutations in Drosophila were first reported >30 years ago (Smith 1973; Boyd and Setlow 1976; Graf and Wurgler 1978). Subsequent genetic screens, especially a recent one conducted by Laurencon et al. (2004), have led to a large collection of Drosophila mutants sensitive to DNA damaging agents. Over the years, some of these mutations were shown to affect both well-characterized DSB repair functions [e.g., mus209 = PCNA (Henderson et al. 1994); mus309 = Bloom RecQ (Kusano et al. 2001)] as well as ones that were novel [e.g., mus312 (Yildiz et al. 2002)]. Therefore, molecular and functional characterization of these mutations will continue to yield important insights into DNA repair mechanisms especially in areas that relate to the development of multicellular organisms. We screened a collection of existing mus mutations with the I-SceI-based repair assays and succeeded in identifying several mutations that have various defects in DSB repair. To our knowledge, this is the first of such screens conducted in a metazoan.
MATERIALS AND METHODS
Drosophila stocks:
Description of stocks not provided here can be found in FlyBase (http://flybase.net; Drysdale et al. 2005). The heat-shock protein 70 (hsp70) promoter-driven I-SceI transgene (70I-SceI) has been described previously (Rong and Golic 2000). The lines used in this study were [70I-SceI]2B on chromosome 2 and [70I-SceI]1A on chromosome 3. The wIw reporter construct has been described (Rong and Golic 2003). The lines used were [wIw]4A on chromosome 2 and [wIw]2 on chromosome 3. The lines [wIw]8z and [wIw]yellow were derived from [wIw]2 by imperfect NHEJ (Rong and Golic 2003) and used in combination with [wIw]2 in the homozygous assays. An X chromosome hsp70-driven FLP line (70FLP3F) has been described previously (Golic et al. 1997). All the mutant lines except the following were obtained from the Bloomington Stock Center (Indiana): mei-4129D from Tin Tin Su at the University of Colorado (Laurencon et al. 2003); mus209B1 and mus2092735 from Darryl Henderson at SUNY of Stony Brook (Henderson et al. 1994); mio2 from Mary Lilly at NIH (Iida and Lilly 2004); okrWS, okrRU, and okrAA from Trudi Schüpbach at Princeton University (Ghabrial et al. 1998); sir22A-7-11 from Kent Golic at the University of Utah (Xie and Golic 2004); and mus309D2 and mus309D3 from Jeff Sekelsky at the University of North Carolina (Adams et al. 2003). For deletion mapping of ssar, deficiencies from Bloomington's “Deficiency Kit” were used that cover the region from 85 to 91.
Our repair assays require male fertility, and we were unable to get enough fertile test males for the following mutagen-sensitive (mus) mutations: mus106, mus108, mus111, mus115, mus302, mus310, mus315, mus318, and mus320. Some of the chromosome 3 mus mutants recovered from a recent screen were also excluded due to the loss of mutagen sensitivity (Laurencon et al. 2004; R. Hawley and K. Burtis, personal communication to FlyBase). In addition, our repair assays are based on eye pigmentation. This made it difficult to test mutations on a cinnabar (cn) and brown (bw) doubly mutant chromosome 2 since cn bw double homozygotes have white eyes regardless of the state of the w gene. These include mus204, mus205, and the entire chromosome 2 collection from the Laurencon screen.
Generation of a DNA ligase IV (lig4) mutant:
Line EP(X)0385 has a w+-marked P element inserted upstream of the X chromosome gene CG12176, which encodes the Drosophila lig4 homolog. EP0385 males with the Δ2–3 P transposase gene on a Stubble(Sb)-marked chromosome 3 were singly mated to C(1)DX, y f/Y females. White-eyed and Sb+ males were recovered, from which 13 lines were established. PCR tests were performed to detect genomic alteration covering the lig4 region. The sequences of the primers used are available upon request. In the lig411 mutation, nt13501164–nt13501714 were deleted, which included the first 148 codons of lig4 (for nucleotide numbering see FlyBase).
Drosophila genetics for I-SceI assays:
Test males for various repair assays were produced as shown in the crossing schemes (appendix). The parents were transferred every 3 days and the progeny were immediately heat-shocked for 1 hr at either 38° or 32°. The test males were testcrossed as shown in the appendix. To score the “recut” phenotype, white+ (w+) progeny were directly examined for eye mosaicism if they have inherited the Scutoid(Sco)-marked [70I-SceI]2B transgene, which has leaky somatic expression. Alternatively, w+ progeny were crossed individually to flies with [70I-SceI]2B and scored for mosaicism in the next generation. Interhomolog GC events from the [wIw]8z homozygous assay were scored by allelic PCR as described (Rong and Golic 2003). Briefly, DNA from single w+ recut− flies was PCR amplified with w7926u and 8z-minus to score interhomolog GC and with w7926u and w14178d as a control.
Testing the effect of mus307D1 on the heat-shock response:
In wIw, the mini-w is flanked by direct repeats of FLP recombination targets, FRTs (for a more detailed description of wIw, see Rong and Golic 2003). FLP can excise a portion of w+ from the chromosome leading to its inactivation. The 70FLP transgene that we used to induce FLP production was identically constructed as 70I-SceI except for the enzyme coding regions. Therefore, a mutation that reduces heat-shock-induced transcription ought to similarly decrease 70FLP expression, which in turn would lead to a reduced rate of w+ excision. We measured FLP-induced w+ loss in the germline of both wild-type (WT) and mus307D1 males that had been heat-shocked at 38° during early development and obtained essentially identical frequencies for both genotypes.
Preliminary mapping of ssar1:
We constructed flies that were heterozygous for both mus307D1 and individual deficiencies that uncover overlapping segments of the region between 85 and 91. These flies were tested with the hemizygous assay to measure SSA repair. In no combination, did we observe a drop of SSA rate beyond the one for mus307D1 heterozygotes (data not shown), suggesting that the mutation that reduces SSA is not located in the tested interval. The map position of mus307D1 was 3-59 (Boyd et al. 1981), placing it close to the Sb (3-60) mutation. We attempted to separate our SSA-reducing mutation from mus307D1 by meiotic recombination. We placed mu307D1 over a cu- and Sb-marked chromosome and recovered recombinants that had a crossover between the two markers in the region between 86 and 89. Eight lines were established from these recombinants, representing four lines for each of the reciprocal classes. These lines were tested over the original mus307D1 chromosome for their ability to inhibit SSA repair. All of the Sb-marked lines but none of the cu-marked lines were able to reduce SSA to ∼0.40, the rate recovered from original mus307D1 homozygotes (Table 1 and data not shown). This suggests that the SSA-reducing mutation is likely located to the left of cu, consistent with our earlier mapping results with overlapping deficiencies. Boyd et al. (1981) recovered a second mus mutation to the left of cu on the mus307D1 chromosome. It remains to be determined whether our SSA-reducing mutation is allelic to this mus mutation.
TABLE 1.
Hemizygous assays on known mutants
| 38°a
|
32°a
|
|||||
|---|---|---|---|---|---|---|
| Genotype | Nb | SSA freq.c ± SEM | Pd | Nb | SSA freq.c ± SEM | Pd |
| Chromosome X | ||||||
| +e | 33 (1858, 2032) | 0.893 ± 0.023 | 19 (771, 928) | 0.241 ± 0.044 | ||
| lig411 | 36 (2062, 2409) | 0.946 ± 0.012 | 0.0340 | 26 (1587, 1791) | 0.284 ± 0.035 | 0.4522 |
| Chromosome 2 | ||||||
| +/+e | 29 (1456, 1376) | 0.867 ± 0.015 | 27 (936, 895) | 0.290 ± 0.041 | ||
| okrWS/Dff | 14 (736, 692) | 0.770 ± 0.052 | 0.0530 | 22 (969, 911) | 0.494 ± 0.044 | 0.0016 |
| okrRU/Dff | 27 (1372, 1348) | 0.900 ± 0.025 | 0.2648 | 19 (1193, 1143) | 0.537 ± 0.028 | <0.0001 |
| okrAA/Dff | 35 (2218, 2174) | 0.884 ± 0.017 | 0.4614 | 29 (1611, 1529) | 0.602 ± 0.030 | <0.0001 |
| Chromosome 3 | ||||||
| +/+e | 46 (2766, 2474) | 0.847 ± 0.014 | 27 (2314, 1977) | 0.318 ± 0.042 | ||
| spnA1/Dfg | 29 (2148, 2032) | 0.948 ± 0.010 | <0.0001 | 40 (3167, 2929) | 0.548 ± 0.027 | <0.0001 |
| spnA1 | 10 (628, 641) | 0.935 ± 0.030 | 0.0096 | Not done | ||
The heat-shock temperatures for I-SceI induction.
N: sample size, the number of male parents tested followed by the number of progeny with and without the I-SceI cut chromosome scored in parentheses.
SSA frequency, the proportion of white-eyed progeny. SEM: standard error of the mean.
The two-tailed P-value calculated using the permutation test for the null hypothesis that the median SSA frequencies are the same between a particular mutant and the WT control. The underlined numbers indicate that the two samples were statistically different.
w1118.
Df=Df(2L)JS17.
Df=Df(3R)X3F.
Characterization of NHEJ deletions in lig4 mutant:
DNA from independent w+ recut− lines was PCR amplified using primers w7514u (5′-caactgaaggcggacattga) and w14178d (5′-tgtgtgtttggccgaagtat), which generates a 600- bp product. For negative samples, a control PCR was carried out with w7728d (5′-aaacacccatctgccgagca) and w11678u (5′-tcatcgcagatcagaagcgg), which generates a 1-kb product. All negative samples were positive for the control PCR. All samples that were negative for PCR with w7514u and w14178d were amplified with w7514u and one of the following primers: w13623d (5′-cgtagttgctctttcgctgt), w12254d (5′-acaacggtgagtggttccag), or PE5′ (5′-gatagccgaagcttaccgaagt). PCR products, if any, were sequenced to localize the NHEJ junction.
Statistics:
Under our experimental conditions, DSB repair induced by I-SceI cutting occurs in the premeiotic male germline (Rong and Golic 2003). A single DSB repair event could be amplified, leading to multiple progeny with that event. Therefore, individual progeny from a single test male cannot be considered as having independent events. We determined, for every test male, the percentage of progeny that harbor a particular class of repair event (i.e., SSA, GC, or both). We used the permutation test developed by William Engels at the University of Wisconsin (Preston et al. 2006a) to compare the means of these ratios from WT vs. mutant males.
RESULTS
We set out to identify new factors that are important for DSB repair in Drosophila by screening a collection of existing mus mutations with several repair assays that are based on the I-SceI site-specific DSB system. In these assays, a single DSB has been induced in different genomic environments. Using these assays, we measured the effect of different mutant backgrounds on the repair of that single DSB in the Drosophila germline. As a proof of principle, we first tested a few known DSB repair mutations.
The repair assays:
All repair assays were based on the wIw P-element construct described previously (Rong and Golic 2003) and diagramed in Figure 1. An I-SceI cut site was placed between two copies of the w gene. The w copy to the right of the cut site was functional, whereas the copy to the left of the cut site was not, containing only the 3′ portion of w. In other words, two direct w repeats, each ∼3-kb in size, flanked the future DSB. Since all the genetic experiments were performed in the w1118 null background except otherwise noted, the integrity of this mini-w gene dictated eye pigmentation.
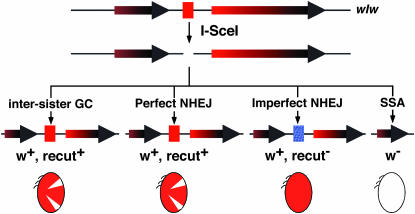
Figure 1.—
The hemizygous assay. The wIw insertion has two w genes: the copy to the left of the I-SceI cut site (red box) is nonfunctional (shorter arrow), and the copy to the right is a functional mini-w (longer arrow). The shading helps illustrate the part of w that is repeated. Four possible repair mechanisms are given below the I-SceI-generated DSB (middle), with the names of the mechanism on top and the phenotypic classifications at the bottom of the diagrams that depict the molecular structures of the different repair products. The blue box represents a mutated I-SceI cut site due to imperfect NHEJ. The ovals represent eyes with different degrees of pigmentation. A mosaic eye has both white and red areas.
We termed the first assay “the hemizygous assay” on the basis of the chromosomal configuration of the wIw insertion. Males with a single copy of wIw and a heat-inducible 70I-SceI transgene were generated by crossing and heat-shocked. These males displayed eye color mosaicism due to somatic w+ loss induced by I-SceI cutting. They were mated individually to w females. By scoring their progeny, we estimated the contributions from different repair pathways in the male premeiotic germline.
As described previously (Rong and Golic 2003), we recovered three classes of phenotypically distinct progeny, which are attributable to different types of repair of the I-SceI-generated DSB (Figure 1). Molecular analyses confirmed that the white-eyed progeny were the result of SSA repair, which led to the loss of one copy of the w repeats as well as all the intervening sequences. The rest of the progeny had pigmented eyes (w+), and they could be further divided into two classes on the basis of whether they had inherited an intact I-SceI cut site (recut+) or a mutated one (recut−). The recut+ progeny displayed eye color mosaicism in the presence of I-SceI as described earlier, whereas the recut− offspring showed solid pigmented eyes even in the presence of I-SceI. These w+ recut− progeny were the result of imperfect NHEJ repair. On the other hand, the w+ recut+ offspring could arise from perfect end joining since I-SceI generates a DSB with complementary 5′ overhangs or GC using an intact sister chromatid as the template, both restoring the cut site. Alternatively, they could be the result of I-SceI failing to cut.
We primarily used the hemizygous assay to screen different mutations. For certain mutations with a suspected effect on GC, we used two additional assays to estimate repair contributions from GC between the homologous chromosomes. We termed these two assays “the homozygous assay,” since the wIw insertion was in a homozygous state. Only one homolog carries an I-SceI cleavable wIw (Figure 2), and the other carries a [wIw]2 derivative with a mutated I-SceI cut site. We could correctly identify the I-SceI-cut chromosome in the progeny because (1) one of the wIw chromosomes was marked with the dominant Sb mutation, (2) Drosophila males do not have meiotic recombination, and (3) mitotic DSB repair under our experimental conditions seldom leads to crossing over (Rong and Golic 2003).
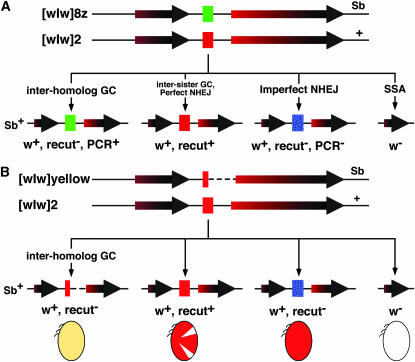
Figure 2.—
The homozygous assays. (A) The assay with [wIw]8z as a template for interhomolog GC. The red box represents a normal I-SceI cut site. The green box represents the mutated cut site in [wIw]8z. The four possible outcomes of this assay are given at the bottom. The phenotypic classification of the progeny was done the same way as it was done for the hemizygous assay, except that the products of interhomolog GC were distinguished from those of imperfect NHEJ by an allelic-specific PCR reaction. (B) The assay with [wIw]yellow as the GC template. The [wIw]yellow chromosome has half of an I-SceI cut site remaining (the narrower red box), and a 333-bp deletion (dotted line), which includes part of the mini-w gene (see main text). The interhomolog GC products can be scored directly by the presence of yellow-colored eyes (yellow oval).
The [wIw]8z template has a small mutation at the I-SceI cut site. We recovered three classes of progeny similar to the ones from the hemizygous assay: w−, w+ recut+, and w+ recut− (Figure 2A). The w+ recut− progeny could be further categorized into two subclasses: those that had inherited a mutated I-SceI cut site identical to the one in [wIw]8z due to interhomolog GC and those that had inherited a randomly mutated cut site due to imperfect NHEJ. By an allele-specific PCR method based on the known [wIw]8z sequences (Rong and Golic 2003), we identified the first subclass as w+ recut− and PCR+ and the second subclass as w+ recut− PCR−.
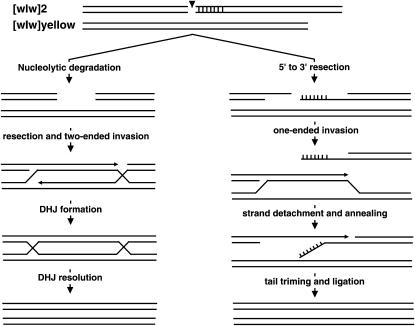
In the above [wIw]8z homozygous assay, the detection of interhomolog GC events relied on sampling by PCR. We developed a second homozygous assay in which interhomolog GC events could be visually identified (Figure 2B). During the course of studying imperfect NHEJ at the [wIw]2 insertion, we recovered a derivative that we named [wIw]yellow, which had a 333-bp deletion to the right of the I-SceI cut site including the right half of the cut site. Flies with the original [wIw]2 have orange eyes due to the hypomorphic nature of the mini-w gene. Flies with [wIw]yellow have a light yellow eye color. Presumably, the small deletion eliminated upstream regulatory elements of mini-w, further weakening its expression. In the new assay, the right end of the I-SceI-generated DSB does not have immediate homology to the [wIw]yellow homolog. We imagine two ways that interhomolog GC could still occur (Figure 3). First, nucleolytic degradation of double-stranded DNA to the right end would expose homology to allow GC to proceed by the traditional DSB repair model (Szostak et al. 1983). Second, interhomolog GC could occur by the synthesis- dependent strand-annealing (SDSA) mechanism (Nassif et al. 1994). In SDSA, the left end of the I-SceI-induced DSB invades [wIw]yellow to initiate repair synthesis. After synthesis has gone past the right side of the 333-bp deletion, the invading strand could detach from the [wIw]yellow template and anneal with the complementary single-stranded region from the right end of the DSB. This annealing would be followed by single-strand tail trimming and ligation. For both [wIw]8z and [wIw]yellow homozygous assays, SSA repair requires the same amount of end processing, yet more extensive end processing is needed for GC in the [wIw]2/[wIw]yellow setting. We therefore predicted a decrease in the GC/SSA ratio in the [wIw]yellow assay when compared to the [wIw]8z assay.
Figure 3.—
Two possible mechanisms for interhomolog GC with [wIw]yellow. Horizontal lines represents DNA single strands. The region marked by vertical lines represents the 333-bp region that is not present on the [wIw]yellow chromosome. To the left is the traditional “Double Holliday Junction (DHJ)” model for GC. First, the DSB is enlarged to a double-strand gap removing heterology between the homologs (area marked by vertical lines). Both ends invade the template and initiate DNA synthesis with the direction indicated by the arrow. DHJ formation and resolution complete GC repair. To the right is the SDSA model for GC. Following single-strand resection, only the left end of the DSB invades the template and initiates DNA synthesis. Once synthesis has passed the heterologous region marked by vertical lines, the invading strand detaches from the template and anneals with the right end of the DSB. Other mechanisms might also be possible.
Drosophila Rad51 is essential for GC but dispensable for SSA:
The spindleA (spnA) gene encodes the Drosophila homolog of bacterial RecA and eukaryotic Rad51 proteins (Staeva-Vieira et al. 2003), which is essential for GC repair. Interestingly, in cases where GC and SSA compete for the same set of DSBs, yeast rad51 mutations lead to an increase of SSA repair at the expense of GC (Ivanov et al. 1996; Osman et al. 2000). A similarly competitive situation exists for our hemizygous assay (Figure 1). In the S/G2 phase of the cell cycle, the I-SceI-generated DSB can be repaired by either SSA or GC from an intact sister chromatid. Therefore, we predicted that spnA mutations would lead to an elevated SSA frequency due to their inhibiting effects on intersister GC. As shown in Table 1, a WT male, when heat-shocked at 38°, produced a germline SSA frequency of 0.847. For males either homozygous or hemizygous for the spnA1 mutation, the SSA frequency was significantly elevated, approaching 0.95. We established six independent white-eyed lines from the spnA1/Df(3R)X3F experiment. Southern blot analyses conducted as previously described confirmed that they arose from SSA repair (data not shown; Rong and Golic 2003). Taken collectively, our results suggest that Rad51 deficiency causes an increased usage of SSA repair at the expense of intersister GC.
Although the results from the hemizygous assay suggest that Rad51 is essential for GC, the evidence remains indirect, since intersister GC cannot be directly measured in this assay. To directly measure the frequency of GC, we used the [wIw]8z homozygous assay that permits efficient interhomolog GC (Figure 2A) (Rong and Golic 2003). In this assay, the I-SceI-induced DSB can be repaired to produce the same three classes of progeny as the hemizygous assay. An additional fourth class is expected to arise as a result of interhomolog GC. From WT males that had been heat-shocked at 38°, 43.4% of the progeny were the result of SSA repair and an equal portion was derived from interhomolog GC (Table 2A). In sharp contrast, spnA mutant combinations led to elevated SSA frequencies (≥0.919), which are virtually identical to the ones obtained from the hemizygous assay (Table 1). None of the w+ recut− progeny was positive for allele-specific PCR for detecting interhomolog GC. These results suggest that both interhomolog and intersister GC are abolished in the absence of Rad51. For the spnA1/Df(3R)X3F1 combination (Table 2A), 95.5% of the progeny were w−, the result of SSA. The rest (4.5%) of the progeny were w+. Of those, 8 of 38 were also recut−, which translates to an imperfect NHEJ frequency of 0.009. Taken together, these results suggest that cutting under this condition is very efficient, approaching 100%, and that imperfect NHEJ is very inefficient in the male germline when SSA repair is feasible.
TABLE 2.
Homozygous assay
| Genotype | Na | SSA freq.b ± SEM | Pc | GC freq.d | P | N for recute | SSA + GC | N for PCRf | P |
|---|---|---|---|---|---|---|---|---|---|
| A. With [wIw]8z | |||||||||
| Chromosome 2 at 38°g | |||||||||
| +/+ | 30 (1819, 1981) | 0.384 ± 0.029 | 0.415 | Not done | 141 (94) | ||||
| okrWS/Dfh | 26 (2112, 2050) | 0.728 ± 0.045 | <0.0001 | Not done | |||||
| okrRU/Df | 24 (2064, 2110) | 0.822 ± 0.026 | <0.0001 | Not done | |||||
| okrAA/Df | 26 (1946, 1950) | 0.869 ± 0.018 | <0.0001 | Not done | |||||
| Chromosome 3 at 38° | |||||||||
| +/+ | 27 (1445, 1700) | 0.434 ± 0.047 | 0.426 | 411 (366) | 100 (83) | ||||
| spnA1/+ | 28 (1407, 1358) | 0.463 ± 0.043 | 0.6147 | 0.399 | 372 (297) | 100 (95) | |||
| spnA1 | 30 (1965, 1925) | 0.919 ± 0.018 | <0.0001 | Not done | |||||
| spnA1/Dfi | 30 (1647, 1770) | 0.955 ± 0.010 | <0.0001 | <0.001j | 38 (8) | 8 (0) | |||
| B. With [wIw]yellow | |||||||||
| Chromosome X at 38°g | |||||||||
| + | 30 (4739, 4618) | 0.505 ± 0.035 | 0.295 ± 0.024 | 0.800 ± 0.029 | |||||
| lig411 | 29 (4064, 4199) | 0.663 ± 0.047 | 0.0094 | 0.229 ± 0.034 | 0.1177 | 0.892 ± 0.028 | 0.0251 | ||
| Chromosome 2 at 38° | |||||||||
| +/+ | 40 (4424, 4135) | 0.509 ± 0.017 | 0.306 ± 0.017 | 0.815 ± 0.016 | |||||
| mus206A1 | 41 (3809, 3703) | 0.585 ± 0.023 | 0.0093 | 0.295 ± 0.022 | 0.6953 | 0.878 ± 0.013 | 0.0028 | ||
| okrAA/Dfh | 30 (3368, 3956) | 0.868 ± 0.017 | <0.0001 | 0.017 ± 0.007 | <0.0001 | 0.885 ± 0.013 | 0.0016 | ||
| okrRU/Df | 40 (5096, 5181) | 0.829 ± 0.013 | <0.0001 | 0.009 ± 0.004 | <0.0001 | 0.838 ± 0.012 | 0.2453 | ||
| Chromosome 2 at 32° | |||||||||
| +/+ | 40 (3383, 2998) | 0.149 ± 0.017 | 0.306 ± 0.017 | 0.261 ± 0.019 | |||||
| okrAA/Df | 32 (2995, 3098) | 0.543 ± 0.017 | <0.0001 | 0.019 ± 0.006 | <0.0001 | 0.562 ± 0.019 | <0.0001 | ||
| okrRU/Df | 30 (2608, 2470) | 0.431 ± 0.026 | <0.0001 | 0.020 ± 0.008 | <0.0001 | 0.451 ± 0.027 | <0.0001 | ||
N: sample size, the number of male parents tested followed by the number of progeny with and without the I-SceI cut chromosome scored in parentheses.
SSA frequency, the proportion of white-eyed progeny. SEM: standard error of the mean.
The two-tailed P-value calculated using the permutation test for the null hypothesis that the median SSA frequencies are the same between a particular mutant and the WT control. The underlined numbers indicate that the two samples were statistically different.
GC frequencies for the [wIw]8z assay are calculated as follows: 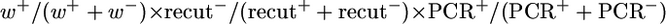 , for testing both the lig4 and spnA mutations with the homozygous assay using the [wIw]8z template. GC frequencies for second chromosome mutations are calculated as follows:
, for testing both the lig4 and spnA mutations with the homozygous assay using the [wIw]8z template. GC frequencies for second chromosome mutations are calculated as follows: 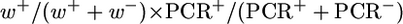 , since w+ progeny were taken for allelic PCR directly without scoring for the recut phenotypes. Since these GC frequencies are derived from complex calculations, no P-values were given for these samples. GC events for the [wIw]yellow assay were scored directly as progeny with light yellow eyes (Figure 2B). GC frequencies are calculated as the proportion of these yellow-eyed flies in total progeny.
, since w+ progeny were taken for allelic PCR directly without scoring for the recut phenotypes. Since these GC frequencies are derived from complex calculations, no P-values were given for these samples. GC events for the [wIw]yellow assay were scored directly as progeny with light yellow eyes (Figure 2B). GC frequencies are calculated as the proportion of these yellow-eyed flies in total progeny.
The number of flies tested for eye mosaicism in the presence of I-SceI to assay the integrity of the I-SceI cut site. The numbers in parentheses are the number of flies producing nonmosaic progeny.
The number of flies tested with allelic PCR to detect interhomolog GC with the [wIw]8z template. The numbers in parentheses are the number of flies producing a positive PCR product.
The heat-shock temperatures for I-SceI induction.
Df=Df(2L)JS17.
Df=Df(3R)X3F.
Since none of the eight w+ recut− events was PCR+, the highest possible GC rate was calculated as 0.001 (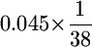 ).
).
Drosophila Rad54's role in GC and SSA repair:
The okra (okr) gene encodes the fly homolog of Rad54 (Ghabrial et al. 1998), which intimately interacts with Rad51 to promote its function in GC. In both budding and fission yeast, GC is also abolished by rad54 mutations (reviewed in Paques and Haber 1999). We predicted that okr mutations, similar to spnA, would lead to an elevated SSA frequency due to their inhibiting effects on intersister GC. Surprisingly, we observed SSA frequencies similar to the WT level for all three okr mutations tested in a hemizygous configuration (Table 1). We considered two possible explanations for this result. First, the okr alleles that we tested may be weak so that they retain substantial ability to repair DSBs by GC. We considered this model unlikely. Mutations in okr also cause female sterility partly due to patterning defects in the egg (Ghabrial et al. 1998). The okrAA and okrRU mutations are caused by premature stop codons and have been characterized as null or strong loss-of-function alleles in terms of their effects on oogenesis. Second, we hypothesized that Drosophila Rad54 might have a less significant role in intersister GC than Rad51. In the hemizygous assay, the majority of the DSBs were eventually repaired via SSA. A smaller enhancing effect of okr on SSA may not acquire statistical significance.
We then used the homozygous assays to measure the effect of okr mutations on GC in a different way. In the [wIw]8z homozygous assay (Figure 2A), okr mutant males had SSA frequencies (0.728–0.869) similar to the combined frequency of SSA and GC from WT males (0.799). We infer from this result that interhomolog GC is inhibited by the okr mutations leading to the increase of SSA. However, these SSA frequencies from okr males do not differ from the ones observed for 38°-treated WT (0.867) or okr males (0.770–0.900) in the hemizygous assay, which suggests that intersister GC in the homozygous assay was not greatly affected by okr, similar to the situation in the hemizygous assay under the same heat-shock condition.
We also used the [wIw]yellow homozygous assay (Figure 2B) to directly measure the effect of the okr mutations on GC. When comparing the [wIw]yellow and the [wIw]8z assays, we discovered that WT males had a reduced interhomolog GC frequency [from 0.415 to 0.306 and a corresponding increase of SSA from 0.384 to 0.509 (P = 0.0001 for comparing SSA frequencies)]. This confirms our earlier hypothesis that the more extensive end processing needed for GC would favor SSA in this new homozygous assay. We examined 20 independent yellow-eyed progeny by PCR and sequencing and confirmed that they were the result of copying the small deletion onto the original [wIw]2 chromosome (data not shown). For okr mutants, we obtained SSA rates that are similar to ones from the [wIw]8z homozygous assay (Table 2B). Interestingly, we also detected a small but significant frequency of interhomolog GC under this condition (0.017 for okrAA and 0.009 for okrRU). By PCR and sequencing, we verified the presence of the 333-bp deletion in 10 independent events. We believed that these were genuine GC repair products, but not the rare events caused by mitotic crossing over between the Sb marker and the [wIw]yellow insertion since we recovered 103 potential interhomolog GC events without recovering a single reciprocal product. Therefore, okr mutant germlines can support a very low level of GC. This may be attributed to maternal Rad54 contribution or Rad54 independent repair.
In summary, Drosophila Rad54 is essential for interhomolog GC, but may have a less important role in intersister GC than Drosophila Rad51.
Intersister GC can be better estimated with less I-SceI cutting:
Intersister GC restores the cut site rendering the chromosome susceptible to a second round of cutting, and repeated I-SceI cutting would “select” repair products that have a mutated I-SceI cut site such as those from SSA repair. In addition, when the nuclease is abundant, both sisters can be cut, which prevents intersister GC. Therefore, intersister GC events are likely underrepresented in progeny after a 38° induction. To better estimate the contribution from intersister GC, we repeated the hemizygous assay with a less severe heat shock of 1 hr at 32°. With this condition, WT SSA frequency drops to ∼0.30. As we expected, the SSA frequency was elevated from 0.319 to 0.548 in spnA1 hemizygotes, an increase of >80% from the WT level. In addition, all three okr mutations led to an SSA frequency increase of as high as 100% of the WT level (Table 1). The similar effects of spnA and okr mutations on SSA repair under this condition suggest that Drosophila Rad54 is indeed important for intersister GC.
We also repeated the [wIw]yellow homozygous assay with a 32° heat induction. WT males gave rise to reduced SSA (0.149) and interhomolog GC (0.112) frequencies (Table 2). Interestingly under the lower I-SceI inducing condition, we recovered close to equal proportions of SSA and interhomolog GC whereas the more severe 38° heat shock led to almost twice as much SSA as interhomolog-GC repair products (0.509 vs. 0.306). This suggests that some of the 38° SSA repair events were the results of repeated cutting of a chromosome with a restored I-SceI cut site.
We tested the okr mutations with the [wIw]yellow homozygous assay at 32°. Consistent with previous 38° results, we observed a decrease of interhomolog GC frequency to ∼0.02 and an increase of SSA repair up to 0.543 when compared to WT males (Table 2). In okr mutants, “visible” repair events (SSA plus interhomolog GC) account for up to 56.3% of the progeny vs. 26.1% in WT, suggesting that the “invisible” intersister GC accounts for at least 30% of the total repair events.
In summary, by lowering the severity of I-SceI induction, we have prevented some of the intersister GC events from being converted to SSA or interhomolog GC, thus providing a better way to estimate the contribution for this important mode of DSB repair.
DNA ligase IV facilitates end-joining repair:
Having demonstrated that mutants defective in HR could be readily identified by our assays, we tested the feasibility of using the same assays to uncover mutants defective in NHEJ. For that purpose, we tested a Drosophila ligase4 (lig4) mutation.
In our hemizygous assay with a 38° heat shock, the lig411 deletion mutation led to an increase in SSA frequency from 0.893 to 0.946 (Table 1). Ten independent lines were set up from white-eyed progeny. Southern blot analyses confirmed that they were the result of SSA repair (data not shown; Rong and Golic 2003). The increase in SSA repair was also evident in lig411 mutant soma. The loss of white by SSA leads to the appearance of white patches in the eye on an otherwise pigmented background. Eyes from lig411 mutants with hemizygous wIw and 70I-SceI were mostly or completely white, yet similar lig4+ eyes had larger pigmented areas and were almost never completely white (data not shown).
To provide further support for our hypothesis that some DSBs in lig4-deficient cells were channeled to SSA and perhaps GC, we measured SSA and GC in the lig411 germline using the [wIw]yellow homozygous assay with a 38° induction. We observed an overall increase of visible repair products from 80.0% in WT to 89.2% in lig411. Interestingly, the increase of visible repair events in the lig411 mutant can be entirely accounted for by the increase in SSA repair, again suggesting that SSA is favored over interhomolog GC under this condition. Our results are consistent with those from a recent report (Johnson-Schlitz et al. 2007).
The most obvious reason for the increase in SSA repair in a lig4-deficient background would be that imperfect NHEJ is inhibited, resulting in a greater number of the I-SceI- generated DSBs being repaired by SSA. We investigated whether imperfect NHEJ was significantly impaired by the lig4 mutation. In our assays, imperfect NHEJ events can be identified as w+ recut− (Figure 1). Since we were assaying repair events in mitotic germline cells, a single imperfect NHEJ event could be replicated, leading to multiple progeny with nonindependent events. In addition, there were very few progeny in the w+ recut− class (<5% of the relevant progeny), which suggests that all imperfect NHEJ events from a single father were likely the result of a single-germline repair event. We thus considered only the number of WT or lig411males that gave rise to at least 1 imperfect NHEJ progeny. In the hemizygous assay, 14 of 17 WT males produced imperfect events, while 8 of 29 lig411 males did so. The difference is highly significant (P = 0.0004 from a one-tailed Fisher's exact test). In a later experiment using the homozygous assay, we recovered 9 independent imperfect NHEJ events from 30 lig411 males and 19 events from 30 WT males. The difference is again significant (P = 0.0095). By combining these two sets of results, which are not statistically different, we conclude that there are over twice as many WT males (33/47) as lig4 males (17/59) producing imperfect NHEJ events. Therefore, imperfect NHEJ is significantly impaired but not eliminated by the lig4 deficiency, which is consistent with earlier results (McVey et al. 2004a; Romeijn et al. 2005; Johnson-Schlitz et al. 2007).
We were interested in how an NHEJ-defective mutant would behave under moderate I-SceI induction, and we repeated the hemizygous assay with a milder 32° heat shock. We observed no increase of SSA frequency, suggesting that NHEJ was not a major repair pathway, possibly because intersister GC repair is highly efficient under this condition.
End degradation is more extensive in a lig4-mutant background:
We imagine that inefficient end joining due to the loss of lig4 function may render DSBs a longer half-life, making them more susceptible to nucleolytic digestion. We obtained supporting evidence for this hypothesis from our molecular analyses of imperfect NHEJ events. From independent w+ recut− lines obtained from the hemizygous and homozygous assays, we amplified DNA fragments with a pair of primers ∼300 bp to either side of the I-SceI cut site. Interestingly, 5 of the 20 events from lig411 failed to yield a PCR product. Further analyses revealed that they all harbor a large deletion to the left of the I-SceI cut site. One deletion ends at the P element. The other four extend beyond the P element and into neighboring genomic regions (see materials and methods). Therefore, they are at least 4 kb in size. On the contrary, 31 of the 32 events recovered from WT males yielded a PCR product with the same pair of primers. The one exception had a ∼500-bp deletion to the left of the I-SceI cut site. Therefore, large deletions were preferentially recovered in a lig4 mutant background (P = 0.0060 for deletions >4 kb and P = 0.0263 for deletions >500 bp, both from one-tailed Fisher's exact tests). Our results are consistent with previous observations that larger deletions during NHEJ are preferentially recovered from yeast, worm, and fly lig4 mutants (Wilson et al. 1997; Morton et al. 2006; Johnson-Schlitz et al. 2007). Although this would be consistent with our idea that lig4 mutations render breaks more susceptible to degradation, more analyses are needed to establish the exact role of lig4 in preventing end degradation.
Novel mutations that affect DSB repair efficiency:
Having established that a combination of our repair assays can be used to uncover mutations affecting both HR and NHEJ repair, we set out to screen the existing mus mutants (Table 3). We expected that a mutation affecting SSA would have a reduced proportion of white-eyed progeny when tested with the hemizygous assay at both 38°- and 32° heat shock; a mutation affecting GC would have an elevated proportion of white-eyed progeny at 38°, but more so at 32°; and a mutation affecting NHEJ would have an increased proportion of white-eyed progeny only at 38°.
TABLE 3.
Mutant screen with the hemizygous assay
| 38°a
|
32°a
|
|||||
|---|---|---|---|---|---|---|
| Genotype | Nb | SSA freq.c ± SEM | Pd | Nb | SSA freq.c ± SEM | Pd |
| Chromosome X | ||||||
| +e | 33 (1858, 2032) | 0.893 ± 0.023 | 19 (771, 928) | 0.241 ± 0.044 | ||
| mus101D1 | 28 (1712, 1725) | 0.786 ± 0.056 | 0.0694 | 30 (1582, 1690) | 0.150 ± 0.035 | 0.1033 |
| mus101D2 | 24 (1726, 2015) | 0.811 ± 0.045 | 0.0896 | 18 (788, 889) | 0.318 ± 0.066 | 0.3364 |
| mus102D1 | 23 (1592, 1824) | 0.909 ± 0.019 | 0.6580 | 25 (1033, 1129) | 0.325 ± 0.059 | 0.2931 |
| mus105A1 | 25 (1305, 1373) | 0.919 ± 0.015 | 0.4433 | Not done | ||
| mus109D1 | 25 (1797, 1955) | 0.945 ± 0.014 | 0.0791 | 22 (997, 1114) | 0.272 ± 0.039 | 0.6077 |
| mus109D2 | 30 (1153, 1158) | 0.891 ± 0.024 | 0.9521 | 30 (773, 850) | 0.246 ± 0.033 | 0.9360 |
| mus112RT2 | 30 (1024, 1063) | 0.892 ± 0.017 | 0.9812 | 24 (497, 535) | 0.201 ± 0.037 | 0.4793 |
| mus114RT1 | 26 (848, 917) | 0.891 ± 0.023 | 0.9647 | 20 (729, 670) | 0.182 ± 0.036 | 0.2943 |
| mei-9A2 | 21 (1415, 1649) | 0.890 ± 0.047 | 0.9518 | 24 (1699, 1864) | 0.301 ± 0.052 | 0.4099 |
| mei-412 | 24 (798, 844) | 0.769 ± 0.033 | 0.0020 | Not done | ||
| mei-4129D | 28 (1377, 1386) | 0.871 ± 0.016 | 0.5035 | 31 (1683, 1883) | 0.271 ± 0.050 | 0.6917 |
| Chromosome 2 | ||||||
| +/+e | 29 (1456, 1376) | 0.867 ± 0.015 | 27 (936, 895) | 0.290 ± 0.041 | ||
| mus201D1 | 35 (1961, 1931) | 0.808 ± 0.030 | 0.1024 | 30 (1848, 1764) | 0.335 ± 0.048 | 0.4720 |
| mus206A1 | 30 (1033, 981) | 0.935 ± 0.016 | 0.0027 | 22 (993, 921) | 0.436 ± 0.042 | 0.0172 |
| mus207A1 | 23 (858, 864) | 0.928 ± 0.018 | 0.0114 | 23 (1040, 976) | 0.278 ± 0.033 | 0.8296 |
| mus209B1/2735 | 27 (1362, 1338) | 0.830 ± 0.024 | 0.1746 | 25 (1561, 1585) | 0.261 ± 0.035 | 0.6086 |
| rad2011 | 23 (1019, 935) | 0.865 ± 0.027 | 0.9544 | 23 (948, 797) | 0.300 ± 0.025 | 0.8431 |
| sir22A | 29 (1646, 1704) | 0.840 ± 0.022 | 0.3060 | 26 (1402, 1411) | 0.230 ± 0.026 | 0.2352 |
| mio2 | 26 (1295, 1323) | 0.870 ± 0.021 | 0.8976 | 30 (1486, 1470) | 0.177 ± 0.025 | 0.0193 |
| Chromosome 3 | ||||||
| +/+e | 46 (2766, 2474) | 0.847 ± 0.014 | 27 (2314, 1977) | 0.318 ± 0.042 | ||
| mus301D1/D4 | 18 (1080, 1027) | 0.840 ± 0.029 | 0.8188 | 17 (728, 634) | 0.330 ± 0.060 | 0.8647 |
| mus304D1/D3 | 20 (973, 994) | 0.804 ± 0.023 | 0.1092 | 14 (494, 418) | 0.308 ± 0.051 | 0.8903 |
| mus304D3 | 13 (815, 715) | 0.876 ± 0.030 | 0.3432 | Not done | ||
| mus305D1/D2 | 26 (1551, 1507) | 0.878 ± 0.032 | 0.3221 | 28 (1550, 1511) | 0.265 ± 0.042 | 0.3849 |
| mus306D1 | 27 (1943, 1728) | 0.776 ± 0.046 | 0.0135 | 24 (1784, 1637) | 0.364 ± 0.028 | 0.4079 |
| mus308D2 | 22 (1870, 1693) | 0.811 ± 0.051 | 0.3921 | 21 (1417, 1283) | 0.367 ± 0.047 | 0.4424 |
| mus309D2/D3 | 23 (1350, 1245) | 0.792 ± 0.041 | 0.1221 | 10 (650, 569) | 0.232 ± 0.047 | 0.2654 |
| mus311D1/D2 | 22 (1033, 982) | 0.902 ± 0.018 | 0.1220 | Not done | ||
| mus312D1/D2 | 22 (986, 845) | 0.793 ± 0.037 | 0.1039 | 27 (1224, 1025) | 0.319 ± 0.042 | 0.9791 |
| mus324ZIII4325/ZIII5997 | 30 (4236, 4168) | 0.884 ± 0.025 | 0.1735 | 25 (2653, 1843) | 0.210 ± 0.025 | 0.0062 |
| mus327ZIII5906 | 29 (3623, 3479) | 0.869 ± 0.031 | 0.4647 | 24 (2772, 2615) | 0.238 ± 0.034 | 0.1575 |
| mu21 | 23 (3069, 3073) | 0.889 ± 0.012 | 0.0530 | 31 (3803, 3731) | 0.462 ± 0.021 | 0.0040 |
| mu21/Dff | 26 (3678, 3467) | 0.891 ± 0.017 | 0.0515 | 30 (3852, 3806) | 0.425 ± 0.024 | 0.0328 |
| spnB1 | 13 (714, 661) | 0.884 ± 0.038 | 0.2623 | Not done | ||
| spnD1 | 14 (1048, 950) | 0.790 ± 0.031 | 0.0706 | Not done | ||
| spnE1/Dfg | 17 (684, 614) | 0.846 ± 0.036 | 0.9905 | Not done | ||
| SSAR1 mus307D1 | 26 (2257, 2226) | 0.428 ± 0.040 | <0.0001 | 30 (2210, 1809) | 0.139 ± 0.030 | 0.0012 |
| SSAR1 mus307D1/+ + | 42 (3041, 2667) | 0.698 ± 0.026 | <0.0001 | Not done | ||
| SSAR1 +/SSAR1 mus307D1 | 22 (1605, 1388) | 0.407 ± 0.043 | <0.0001 | Not done | ||
ssar inhibits SSA repair:
We discovered that in the presence of the homozygous mus307D1 chromosome, the frequency of SSA repair induced by a 38° heat shock was reduced from 0.847 to 0.423 (Table 3). When induced by 32°, SSA repair in the same mutant background was reduced by a similar degree from 0.318 to 0.139. Interesting, this SSA-inhibiting mutation acted in a semidominant way so that heterozygous mutant males had a 38°-induced SSA frequency intermediately reduced to 0.698. It is possible that the SSA-inhibiting effect of mus307D1 was due to the indirect effect of a mutation that attenuated the heat-shock response. We ruled out this hypothesis by comparing the rates of site-specific recombination catalyzed by the FLP recombinase expressed from a hsp70-FLP transgene in WT vs. mus307D1 backgrounds (100% for WT, n = 13 males; 99% for mus307D1, n = 14 males; see materials and methods).
We were interested in whether a drop of SSA repair was accompanied by an increase in another repair mechanism in mus307D1 males. We measured imperfect NHEJ by scoring the recut− phenotype in 331 randomly selected w+ progeny from mus307 males (see materials and methods). Fifty-six of them were w+ recut−, which translates to an overall imperfect NHEJ frequency of 0.10, which is similar to the normal one of 0.06. Therefore, mus307D1 did not markedly increase imperfect NHEJ at the expense of SSA repair.
We mapped this SSA-inhibiting mutation outside of the region between the curled and stripe mutations (see materials and methods), which is where mus307D1 was previously mapped (Boyd et al. 1981; Drysdale et al. 2005). Therefore, this mutation can be separated from mus307D1 and likely represents a new repair mutation. We name the locus single-strand annealing reducer (ssar) and the allele ssar1.
mus206A1 promotes SSA:
We identified the mus206A1 mutation as having a similar effect on SSA as the spnA or okr mutations. With a 38° heat shock, mus206 males produced a higher SSA frequency than WT males (0.935 vs. 0.867). The milder 32° heat shock also led to a greater SSA increase in mus206 males (from 0.290 to 0.436). We performed the homozygous assay with the [wIw]yellow template to directly measure the effect of mus206A1 on GC. Different from what was observed with spnA and okr, mus206 males produced more SSA repair products (58.5%) than WT males (50.9%), but with no reduction in interhomolog GC products (Table 2B). The lack of an effect of mus206A1 on interhomolog GC is consistent with the fact that mus206A1 homozygous females are fertile, whereas strong mutations affecting GC inevitably lead to female sterility (e.g., Ghabrial et al. 1998; Abdu et al. 2003; Staeva-Vieira et al. 2003).
mei-9 encodes the Drosophila Rad1 homolog (Sekelsky et al. 1995). In budding yeast, the Rad1–Rad10 endonuclease complex serves to cleave the 3′ protruding tails during SSA (Sugawara et al. 1997). Mutations in rad1 or rad10 reduce SSA efficiency in yeast. However, we did not observe any inhibition of SSA by the mei-9A2 null mutation. Therefore, the Drosophila Rad1 is not required for the trimming of 3′ tails in our SSA assay.
mei-41 encodes the Drosophila homologs of mammalian ATR (Hari et al. 1995). We observed an allelic-specific inhibition of SSA (Table 1), which suggests that the effect was specific to that allele or to X-linked second-site mutations.
missing oocytes (mio), mutator2 (mu2), and mus324 behaved similarly in reducing SSA efficiency only with the milder 32° heat shock. mio encodes a protein that was implicated to function in meiotic DSB repair (Iida and Lilly 2004). mu2 encodes a putative Drosophila homolog of mammalian MDC1 DNA damage checkpoint protein (Kasravi et al. 1999; J. Mason, personal communication). The effect of these mutations may be complex and will be the subject of future studies.
mus207A1 and mus306D1 behaved similarly to lig411; i.e., we observed a significant increase of SSA repair with the 38° induction but not with the 32° induction, suggesting that both mutants might be defective in end joining. To investigate whether mus207 behaves similarly to lig4 under a different condition, we conducted the [wIw]yellow homozygous assay, but did not observe a significant effect.
DISCUSSION
In this study, we further develop our I-SceI-based germline assays in two important areas: (1) we showed that the important contribution of intersister GC can be better estimated with reduced I-SceI cutting and (2) we developed a versatile assay using the [wIw]yellow chromosome in which interhomolog GC events can be phenotypically recognized. Using a series of repair assays combined with different degrees of I-SceI induction, we determine some of the genetic requirements for DSB repair in the Drosophila germline.
We demonstrated that Drosophila Rad51 is essential for both intersister and interhomolog GC repair, but dispensable for SSA. Our results are consistent with those from earlier studies (McVey et al. 2004b; Johnson-Schlitz et al. 2007). Contrary to results from McVey et al. (2004b), we did not observe any effect of spnA heterozygosity on either intersister or interhomolog GC (Table 2A). This could be due to the fact that different assays were employed in the two studies: McVey et al. studied the repair of a large gap induced by P transposase while we create a simple break with I-SceI. The abundance of spnA may be more important for the repair of a large gap.
We also showed that Drosophila rad54 (okr) mutations behaved very similarly to spnA in our repair assays suggesting that Rad54 is also essential for GC repair. We were first perplexed by the apparent lack of an effect of okr on what we inferred as intersister GC events with a 38° I-SceI induction, whereas okr mutations clearly inhibited both intersister and interhomolog GC at 32°. We suspected that the acute heat treatment might have been the cause. As we were preparing this article, Johnson-Schlitz et al. (2007) made several interesting observations on okr using I-SceI-based repair assays that are very similar to ours. A ubiquitin promoter-driven I-SceI source was used by Johnson-Schlitz et al., which provides strong and ubiquitous I-SceI expression that might be similar to our 38° heat induction. In their “cross 1,” similar to our hemizygous assay, spnA, but not okr, led to an increased usage of SSA. The results from the two studies suggest that the lack of an effect of okr on intersister GC is specific to the situation in which intersister GC is selected against due to excessive I-SceI cutting. We suggest that Drosophila Rad54 is less important than Rad51 in intersister GC. On the other hand, we showed that both Rad51 and Rad54 were essential for interhomolog GC. Perhaps Rad54 promotes pairing between the broken DNA and its repair template. This function may be more stringently required for interhomolog GC than for intersister GC since sister chromatids are held together by cohesins.
The important function of DNA ligase IV in genome maintenance was not as clearly defined in Drosophila as for HR repair factors. It was concluded that lig4 mutants were generally not sensitive to DNA damaging agents (Gorski et al. 2003; McVey et al. 2004a) and that they were not grossly deficient for imperfect joining of broken ends (McVey et al. 2004a; Romeijn et al. 2005). In addition, our lig4 mutation did not suppress end fusion between unprotected telomeres (Bi et al. 2004). Our current results are more consistent with those from a recent publication (Johnson-Schlitz et al. 2007). We provided several pieces of evidence suggesting that lig4 is important for end joining in Drosophila: (1) the number of test males producing progeny with imperfect end-joining repair was significantly reduced under the lig4 mutant background in both the hemizygous and homozygous assays, (2) imperfect NHEJ with large deletions were preferentially recovered in a lig4 mutant background, and (3) loss of lig4 function led to elevated frequencies of homology-based repair (SSA). This compensatory relationship between NHEJ and HR was also evident in an earlier study in which Romeijn et al. (2005) suggested that a lig4 mutation caused an increased utilization of somatic interhomolog GC for the repair of a P transposase-induced DSB. However, the authors did not estimate the frequencies of NHEJ in WT or the lig4-deficient germline. In another study, McVey et al. (2004a) estimated the frequency of end joining in the germline but did not observe any effect from a lig4 mutation. In that study, DSBs were induced by P transposase at a low frequency, making it difficult to uncover a significant but not essential role of lig4 in end joining. In addition, P transposase creates a large gap of a few kilobases in that study whereas I-SceI creates a simple break in the current study. Moreover, P transposase generates long (17-bp) and noncomplementary overhangs, while I-SceI generates short (4-bp) and complementary overhangs. These differences in break configuration may have altered the stringency for the requirement of lig4 function.
A recurring observation that we made during the course of this study was the competition between different repair mechanisms. We demonstrated an intricate balance among different repair mechanisms by manipulating the genetic control of these mechanisms, or the immediate genomic environment surrounding the DSB, or both simultaneously. We showed earlier that interhomolog GC effectively competes with SSA given that homology is provided immediately adjacent to the DSB on the homolog (Rong and Golic 2003). By inhibiting GC with a spnA or okr mutation or by limiting the immediate homology to only one side of the DSB with the [wIw]yellow GC template, we can tilt the balance back in favor of SSA.
GC using the sister chromatid is generally believed to be the most efficient way of DSB repair (Engels et al. 1990; Kadyk and Hartwell 1992; Johnson and Jasin 2000). However, the contribution from intersister GC is difficult to measure due to the lack of genetic consequence for these events. We were able to get around this problem by taking advantage of the competitive nature of DSB repair in the Drosophila germline so that these invisible events can be converted to visible ones by the use of mutations. With a 38° heat induction, ∼85% of the progeny from a WT male experienced SSA, while ∼95% of those from spnA males did so, suggesting that intersister GC accounts for at least 10% of the progeny from a WT male. This is an underestimation since intersister GC renders the chromosome susceptible to a second round of cutting. Support for the above reasoning came from experiments in which the same repair assay was carried out but with a milder 32° heat shock to produce less I-SceI. Under this new condition, SSA frequency was elevated from ∼0.30 in WT to 0.55 in spnA and up to 0.60 in okr, suggesting that intersister GC accounts for at least 25–30% of the progeny in a WT male. From these data we suggest that lesser I-SceI induction is a better way to estimate the contributions from intersister GC repair. In addition, considering that SSA but not intersister GC is permitted throughout most of the cell cycle, the frequency ratio (close to 1:1) between SSA and intersister GC measured with a 32° heat shock suggests that there might be at least as many S/G2 cells as G1 cells in the male germline during the first 3 days of development. This extrapolation may be experimentally tested using cell-cycle-specific markers.
We screened a collection of existing mutations with the I-SceI-based repair assays and succeeded in identifying several mutations that had various defects in DSB repair. We discovered ssar as a locus necessary for normal SSA repair. There are a limited number of mutations identified in budding yeast that are defective for SSA (Ivanov and Haber 1995; Ivanov et al. 1996; Sugawara et al. 1997, 2000; Davis and Symington 2001). Rad52 and Rad59 have the most important function of promoting strand annealing, while the Rad1–Rad10 endonuclease and the Msh2–Msh3 mismatch repair proteins play important roles under certain conditions. However, what is known about yeast SSA-defective mutants was little help for us in identifying ssar. Homologs of Drosophila Rad52, Rad59, and Msh3 cannot be identified by sequence homology (Sekelsky et al. 2000). Our results suggest that Drosophila mei-9/Rad1 is dispensable for SSA repair in our assay. Drosophila homologs for Msh2 and Rad10 are both located on a different chromosome. Therefore, ssar likely encodes a novel repair protein important for SSA repair. Perhaps ssar encodes a Drosophila functional homolog of the fungal Rad52/Rad59 protein. This predicts that ssar1 would also inhibit GC. However, we have not been able to recover recombinant chromosomes with both ssar1 and [wIw]2 or its derivatives, most likely due to their close genetic linkage. The effect of ssar1 on GC repair awaits future investigation.
We identified mus206A1 as a new mutation that might enhance SSA. By using the hemizygous assay, we showed that mus206A1 causes an increase in SSA usage under both 38° and 32° heat-shock conditions. By using the homozygous assay, we showed that mus206A1 did not affect interhomolog GC, suggesting that its effect on SSA might be direct. It is also possible that mus206A1 weakens sister chromatid cohesion, specifically affecting intersister but not interhomolog GC. We have not excluded the possibility that the mutation affecting SSA is different from the one conferring mutagen sensitivity since second-site mutations could have accumulated over the years.
Johnson-Schlitz et al. (2007) tested many of the same mutations that we have tested in this study. The two studies led to consistent results for some genes (i.e., spnA, okr, and lig4), but not others. We did not detect an effect of mei-9, mus101, and mus301 mutations on SSA repair. We suspect that the different results were mainly caused by the difference in the repair assays employed. In our assays, the white duplication is ∼3000 bp, which is significantly longer than the 147-bp repeat used by Johnson-Schlitz et al. In addition, the intervening sequence flanked by the repeats is ∼1.5 kb in our assay, again significantly longer than the one used in the other study. Either one or both of these features may have rendered our assay insensitive to those mutations tested. Nevertheless, we uncovered the ssar1 mutation, which causes the largest drop of SSA efficiency among all mutations tested in the two studies. The two studies also came to different conclusions regarding Drosophila ATR's role in SSA repair. Although we observed a drop in SSA with the mei-412 allele, which is similar to what was observed with the mei-41D5 allele by Johnson-Schlitz et al., we observed no effect from the mei-4129D allele. In addition, LaRocque et al. (2007) observed a drop of SSA using mei-4129D in our hemizygous assay. However, the decrease in SSA efficiency seen in the LaRocque study seems less severe than the one observed by Johnson-Schlitz et al., even though mei-4129D was classified as a null (Laurencon et al. 2003) and that mei-41D5 is likely a weaker allele evidenced by the fertility of mei-41D5 females. A plausible explanation for these different results on the effect of mei-41 on SSA repair is that the mei-4129D chromosome may have an SSA-enhancing mutation masking the effect of mei-4129D. The exact role of Drosophila ATR in SSA repair may require additional investigation.
The most significant factor that may have limited the success of our screen is that our assays require male fertility. Therefore, we would have missed genes whose products are required not for the viability of single cells but for the development of complex organisms. For examples, mre11 and nbs null mutations are viable in yeast but lethal in flies (Bi et al. 2004, 2005). So are strong loss-of-function atm mutations in Drosophila, while a weak atm mutation is male sterile (Bi et al. 2004; Gong et al. 2005). In the future, somatic reporter constructs expressing fluorescent proteins could be used to assay lethal or sterile mutations (Preston et al. 2006a).
Acknowledgments
We thank Carl Wu at the National Cancer Institute for supporting D.S.W.; Kent Golic, in whose lab this project was initiated; Deepa Srikanta and Rongye Shi for technical assistance; and K. G. Golic, D. Henderson, M. Lilly, J. J. Sekelsky, T. T. Su, and T. Schüpbach for sharing fly stocks. We thank Michael Lichten and Dhruba Chattoraj at the National Cancer Institute (NCI) for comments on the manuscripts and help with statistical analyses. We are especially grateful to W. Engels for creating the permutation program and for making it freely available to us. Research in our lab is supported by the intramural research program of the NCI.
APPENDIX: CROSSING SCHEMES
Hemizygous assay:
-
Chromosome X mutants (mutX):
w mutX; [wIw]2 Sb/+ males ⊗⊗ C(1)M3, y/Y; [70I-SceI]1A/TM6, Ubx e females
⇓⇓
w mutX; [wIw]2 Sb/[70I-SceI]1A male ⊗⊗ w1118 females
⇓⇓
Score repair products in Sb progeny.
-
Chromosome 2 mutants (mut2):
w; mut2/Sco; [70I-SceI]1A Sb/+ males ⊗⊗ w; mut2/CyO; [wIw]2 /+ females
⇓⇓
w; mut2/mut2; [wIw]2 /[70I-SceI]1A Sb males ⊗⊗ w1118 females
⇓⇓
Score repair products in Sb+ progeny.
-
Chromosome 3 mutants (mut3):
w; [70I-SceI]2B Sco/+; mut3/Sb males ⊗⊗ w; [wIw]4A/+; mut3/TM6 females
⇓⇓
w; [wIw]4A/[70I-SceI]2B Sco; mut3/mut3 males ⊗⊗ w1118 females
⇓⇓
Score repair products in Sco+ progeny.
Homozygous assay:
-
lig411:
w; [70I-SceI]2B Sco/+; [wIw]8z Sb/+ males ⊗⊗ w lig411; [wIw]2/TM6 females
⇓⇓
w lig411; [70I-SceI]2B Sco/+; [wIw]8z Sb/[wIw]2 males ⊗⊗ w1118 females
⇓⇓
Score repair products in Sb+ progeny.
-
Chromosome 2 mutants (mut2: okr, mus206, mus207):
w; mut2/Sco; [wIw]8z [70I-SceI]1A Sb/+ males ⊗⊗ w; mut2/CyO; [wIw]2/+ females
⇓⇓
w; mut2/mut2; [wIw]2 /[wIw]8z [70I-SceI]1A Sb males ⊗⊗ w1118 females
⇓⇓
Score repair products in Sb+ progeny.
-
spnA1 and Df(3R)X3F (Df):
w; [70I-SceI]2B Sco/+; [wIw]8z e spnA1/+ males ⊗⊗ w; [wIw]2 Sb e Df/TM6 females
⇓⇓
w; [70I-SceI]2B Sco/+; [wIw]8z e spnA1/[wIw]2 Sb e Df males ⊗⊗ w1118 females
⇓⇓
Score repair products in Sb progeny.
Notes:
We scored all the progeny from a cross regardless of whether the progeny have inherited a [wIw] chromosome. In general, progeny with or without the [wIw] chromosome are of equal proportion, which suggests that most of the DSBs were repaired. The control males for each chromosome were all derived from w1118.
For the hemizygous assay, test males were generated by mass mating and heat- shocked early in development, but they were testcrossed to two to three w females individually. For X chromosome mutants, the [70I-SceI]1A transgene came from the mother so that there could be maternal contribution of the I-SceI enzyme. For mutations on a w+ X chromosome, only male progeny, which inherited the w1118 chromosome from the mother, were scored for repair events.
For the homozygous assay, test males were generated by mass mating, but tested individually. For testing WT, lig411, mus206A1, mus207A1, and okr with the new homozygous assay, the crossing scheme was identical except that the [wIw]yellow insertion was used in place of [wIw]8z. okr mutations were tested over Df(2L)JS17.
References
- Abdu, U., A. González-Reyes, A. Ghabrial and T. Schüpbach, 2003. The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis. Genetics 165: 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, M. D., M. McVey and J. J. Sekelsky, 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. [DOI] [PubMed] [Google Scholar]
- Bellaiche, Y., V. Mogila and N. Perrimon, 1999. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics 152: 1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, X., S. C. Wei and Y. S. Rong, 2004. Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr. Biol. 14: 1348–1353. [DOI] [PubMed] [Google Scholar]
- Bi, X., D. Srikanta, L. Fanti, S. Pimpinelli, R. Badugu et al., 2005. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc. Natl. Acad. Sci. USA 102: 15167–15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24: 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, J. B., and R. B. Setlow, 1976. Characterization of postreplication repair in mutagen-sensitive strains of Drosophila melanogaster. Genetics 84: 507–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, J. B., M. D. Golino, K. E. Shaw, C. J. Osgood and M. M. Green, 1981. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics 97: 607–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley, J. M., P. L. Palmbos, D. Wu and T. E. Wilson, 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451. [DOI] [PubMed] [Google Scholar]
- Davis, A. P., and L. S. Symington, 2001. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede, S. J., and D. E. Gottschling, 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99: 723–733. [DOI] [PubMed] [Google Scholar]
- Drysdale, R. A, M. A. Crosby and FlyBase Consortium, 2005. FlyBase: genes and gene models. Nucleic Acids Res. 33: D390–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels, W. R., D. M. Johnson-Schlitz, W. B. Eggleston and J. Sved, 1990. High-frequency P element loss in Drosophila is homolog dependent. Cell 62: 515–525. [DOI] [PubMed] [Google Scholar]
- Frank-Vaillant, M., and S. Marcand, 2002. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell 10: 1189–1199. [DOI] [PubMed] [Google Scholar]
- Game, J. C., and R. K. Mortimer, 1974. A genetic study of x-ray sensitive mutants in yeast. Mutat. Res. 24: 281–292. [DOI] [PubMed] [Google Scholar]
- Ghabrial, A., R. P. Ray and T. Schüpbach, 1998. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 12: 2711–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, M. M., Y. S. Rong, R. B. Petersen, S. L. Lindquist and K. G. Golic, 1997. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 25: 3665–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, M., X. Bi and Y. Rong, 2005. Targeted mutagenesis of Drosophila atm and mre11 genes. Dros. Inf. Serv. 88: 79–83. [Google Scholar]
- Gorski, M. M., J. C. Eeken, A. W. de Jong, I. Klink, M. Loos et al., 2003. The Drosophila melanogaster DNA Ligase IV gene plays a crucial role in the repair of radiation-induced DNA double-strand breaks and acts synergistically with Rad54. Genetics 165: 1929–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf, U., and F. E. Wurgler, 1978. Mutagen-sensitive mutants in Drosophila: relative MMS sensitivity and maternal effects. Mutat. Res. 52: 381–394. [Google Scholar]
- Hari, K. L., A. Santerre, J. J. Sekelsky, K. S. McKim, J. B. Boyd et al., 1995. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell 82: 815–821. [DOI] [PubMed] [Google Scholar]
- Henderson, D. S., S. S. Banga, T. A. Grigliatti and J. B. Boyd, 1994. Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA. EMBO J. 13: 1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida, T., and M. A. Lilly, 2004. Missing oocyte encodes a highly conserved nuclear protein required for the maintenance of the meiotic cycle and oocyte identity in Drosophila. Development 131: 1029–1039. [DOI] [PubMed] [Google Scholar]
- Ivanov, E. L., and J. E. Haber, 1995. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 2245–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, E. L., N. Sugawara, J. Fishman-Lobell and J. E. Haber, 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R. D., and M. Jasin, 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19: 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Schlitz, D., and W. R. Engels, 2006. Template disruptions and failure of double Holliday junction dissolution during double-strand break repair in Drosophila BLM mutants. Proc. Natl. Acad. Sci. USA 103: 16840–16845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Schlitz, D. M., C. Flores and W. R. Engels, 2007. Multiple-pathway analysis of double-strand break repair mutations in Drosophila. PLoS Genet. 3: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk, L. C., and L. H. Hartwell, 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanasis, E., and T. E. Wilson, 2002. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161: 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasravi, A., M. F. Walter, S. Brand, J. M. Mason and H. Biessmann, 1999. Molecular cloning and tissue-specific expression of the mutator2 gene (mu2) in Drosophila melanogaster. Genetics 152: 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano, K., D. M. Johnson-Schlitz and W. R. Engels, 2001. Sterility of Drosophila with mutations in the Bloom syndrome gene–complementation by Ku70. Science 291: 2600–2602. [DOI] [PubMed] [Google Scholar]
- LaRocque, J. R., B. Jaklevic, T. T. Su and J. Sekelsky, 2007. Drosophila ATR in double-strand break repair. Genetics 175: 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencon, A., A. Purdy, J. Sekelsky, R. S. Hawley and T. T. Su, 2003. Phenotypic analysis of separation-of-function alleles of MEI-41, Drosophila ATM/ATR. Genetics 164: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencon, A., C. M. Orme, H. Peters, C. L. Boulton, E. K. Vladar et al., 2004. A large-scale screen for mutagen-sensitive loci in Drosophila. Genetics 167: 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey, M., D. Radut and J. J. Sekelsky, 2004. a End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics 168: 2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey, M., M. Adams, E. Staeva-Vieira and J. J. Sekelsky, 2004. b Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167: 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, J., M. W. Davis, E. M. Jorgensen and D. Carroll, 2006. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc. Natl. Acad. Sci. USA 103: 16370–16375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan, M. E., and M. Jasin, 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7: 263–272. [DOI] [PubMed] [Google Scholar]
- Moynahan, M. E., J. W. Chiu, B. H. Koller and M. Jasin, 1999. Brca1 controls homology-directed DNA repair. Mol. Cell 4: 511–518. [DOI] [PubMed] [Google Scholar]
- Nassif, N., J. Penney, S. Pal, W. R. Engels and G. B. Gloor, 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, F., M. Adriance and S. McCready, 2000. The genetic control of spontaneous and UV-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 38: 113–125. [DOI] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, C. R., C. C. Flores and W. R. Engels, 2006. a Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics 172: 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, C. R., C. C. Flores and W. R. Engels, 2006. b Age-dependent usage of double-strand-break repair pathways. Curr. Biol. 16: 2009–2015. [DOI] [PubMed] [Google Scholar]
- Puchta, H., B. Dujon and B. Hohn, 1993. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 21: 5034–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeijn, R. J., M. M. Gorski, M. A. van Schie, J. N. Noordermeer, L. H. Mullenders et al., 2005. Lig4 and rad54 are required for repair of DNA double-strand breaks induced by P-element excision in Drosophila. Genetics 169: 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet, P., F. Smih and M. Jasin, 1994. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. USA 91: 6064–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky, J. J., K. S. McKim, G. M. Chin and R. S. Hawley, 1995. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky, J. J., M. H. Brodsky and K. C. Burtis, 2000. DNA repair in Drosophila: insights from the Drosophila genome sequence. J. Cell Biol. 150: F31–F36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, P. D., 1973. Mutagen sensitivity of Drosophila melanogaster. Mutat. Res. 20: 215–220. [DOI] [PubMed] [Google Scholar]
- Staeva-Vieira, E., S. Yoo and R. Lehmann, 2003. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22: 5863–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein and F. W. Stahl, 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Sugawara, N., F. Paques, M. Colaiacovo and J. E. Haber, 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94: 9214–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, N., G. Ira and J. E. Haber, 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20: 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, N., X. Wang and J. E. Haber, 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12: 209–219. [DOI] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi, H., J. Kobayashi, K. Morishima, D. C. van Gent, T. Shiraishi et al., 2002. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 420: 93–98. [DOI] [PubMed] [Google Scholar]
- White, C. I., and J. E. Haber, 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T. E., U. Grawunder and M. R. Lieber, 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388: 495–498. [DOI] [PubMed] [Google Scholar]
- Wolner, B., S. van Komen, P. Sung and C. L. Peterson, 2003. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell 12: 221–232. [DOI] [PubMed] [Google Scholar]
- Xie, H. B., and K. G. Golic, 2004. Gene deletions by ends-in targeting in Drosophila melanogaster. Genetics 168: 1477–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz, O., S. Majumder, B. Kramer and J. J. Sekelsky, 2002. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell 10: 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., and T. T. Paull, 2005. The Mre11/Rad50/Xrs2 complex and non-homologous end-joining of incompatible ends in S. cerevisiae. DNA Repair 4: 1281–1294. [DOI] [PubMed] [Google Scholar]