Abstract
It has recently been demonstrated that activity of the essential JIL-1 histone H3S10 kinase is a major regulator of chromatin structure and that it functions to maintain euchromatic domains while counteracting heterochromatization and gene silencing. In the absence of JIL-1 kinase activity, the major heterochromatin markers histone H3K9me2 and HP1 spread in tandem to ectopic locations on the chromosome arms. In this study, we show that the lethality as well as some of the chromosome morphology defects associated with the null JIL-1 phenotype to a large degree can be rescued by reducing the dose of the Su(var)3-9 gene. This effect was observed with three different alleles of Su(var)3-9, strongly suggesting it is specific to Su(var)3-9 and not to second site modifiers. This is in contrast to similar experiments performed with alleles of the Su(var)2-5 gene that codes for HP1 in Drosophila where no genetic interactions were detectable between JIL-1 and Su(var)2-5. Taken together, these findings indicate that while Su(var)3-9 histone methyltransferase activity is a major factor in the lethality and chromatin structure perturbations associated with loss of the JIL-1 histone H3S10 kinase, these effects are likely to be uncoupled from HP1.
IN Drosophila initiation of heterochromatin formation and repression of transcription has been linked to the RNAi machinery (Pal-Bhadra et al. 2004) and involves covalent modifications of histone tails and/or the exchange of histone variants (Swaminathan et al. 2005). In addition, heterochromatin formation requires several nonhistone chromatin proteins (Schotta et al. 2002; Greil et al. 2003; Delattre et al. 2004). Two of these, Su(var)2-5 (HP1) and Su(var)3-9 (a histone methyltransferase), are predominantly found at pericentric heterochromatin (James et al. 1989; Schotta et al. 2002) and are important components for silencing of reporter genes by heterochromatic spreading (for review, see Weiler and Wakimoto 1995; Girton and Johansen 2007). Su(var)3-9 has been shown to catalyze most of the dimethylation of the histone H3K9 residue, which in turn can promote HP1 binding (Schotta et al. 2002). In addition, Su(var)3-9 and HP1 can directly interact, suggesting a model where interdependent interactions between Su(var)3-9, HP1, and histone H3K9 dimethylation lead to heterochromatin assembly and gene silencing (Lachner et al. 2001; Schotta et al. 2002; Elgin and Grewal 2003). However, this model does not address the mechanism for how heterochromatin formation is restricted to certain parts of the genome and for how heterochromatic spreading is regulated (Greil et al. 2003).
It has recently been demonstrated that activity of the JIL-1 histone H3S10 kinase (Jin et al. 1999; Wang et al. 2001) is a major regulator of chromatin structure (Deng et al. 2005) and that it functions to maintain euchromatic domains while counteracting heterochromatization and gene silencing (Ebert et al. 2004; Lerach et al. 2006; Zhang et al. 2006; Bao et al. 2007). In the absence of JIL-1 kinase activity, the major heterochromatin markers H3K9me2 and HP1 spread in tandem to ectopic locations on the chromosome arms with the most pronounced increase on the X chromosomes (Zhang et al. 2006). However, overall levels of the H3K9me2 mark and HP1 were unchanged, suggesting that the spreading was accompanied by a redistribution that reduces the levels in pericentromeric heterochromatin. Genetic interaction assays demonstrated that JIL-1 functions in vivo in a pathway with Su(var)3-9 and that JIL-1 activity and localization are not affected by the absence of Su(var)3-9 activity, indicating that JIL-1 is upstream to Su(var)3-9 in this pathway (Zhang et al. 2006). Furthermore, the results of Zhang et al. (2006) suggested the possibility that the lethality of JIL-1 null mutants may be due to repression of essential genes at these ectopic sites as a consequence of the spreading of Su(var)3-9 activity and HP1 recruitment. In this study, we have tested this hypothesis and examined the relative contributions of Su(var)3-9 and HP1. We show that while Su(var)3-9 histone methyltransferase activity is a major factor in the lethality and chromatin structure perturbations associated with loss of the JIL-1 histone H3S10 kinase, these effects are likely to be uncoupled from HP1.
MATERIALS AND METHODS
Drosophila melanogaster stocks:
Fly stocks were maintained according to standard protocols (Roberts 1998). Canton-S was used for wild-type preparations. The JIL-1z2 allele is described in Wang et al. (2001) and in Zhang et al. (2003). The Su(var)3-906, Su(var)2-504, and Su(var)2-505 alleles are described in Schotta et al. (2002) and in Eissenberg et al. (1992) and were generously provided by. L. Wallrath. The Su(var)3-91 and Su(var)3-92 stocks were obtained from the Umeå Stock Center. Recombinant JIL-1z2 Su(var)3-91, JIL-1z2 Su(var)3-92, and JIL-1z2 Su(var)3-906 chromosomes were identified by generating recombinants as described in Ji et al. (2005) except that the dominant Su(var)3-9 phenotype was selected for in a wm4 background and the presence of JIL-1z2 was confirmed by PCR as in Zhang et al. (2003). Balancer chromosomes and markers are described in Lindlsley and Zimm (1992).
Immunohistochemistry:
Polytene chromosome squash preparations were performed as in Kelley et al. (1999) using the 5-min fixation protocol and labeled with antibody as described in Jin et al. (1999). Primary antibodies used include affinity purified Hope rabbit antiserum raised against JIL-1 residues 886–1013 (Jin et al. 1999), H3K9me2 rabbit antiserum (Upstate Biotechnology), and anti-HP1 mAb C1A9 (Developmental Studies Hybridoma Bank, University of Iowa). DNA was visualized by staining with Hoechst 33258 or with propidium iodide (Molecular Probes, Eugene, OR) in PBS. The appropriate species- and isotype-specific Texas Red-, TRITC-, and FITC-conjugated secondary antibodies (Cappel/ICN, Southern Biotech) were used (1:200 dilution) to visualize primary antibody labeling. The final preparations were mounted in 90% glycerol containing 0.5% n-propyl gallate. The preparations were examined using epifluorescence optics on a Zeiss Axioskop microscope and images were captured and digitized using a high resolution Spot charge-coupled device camera. Images were imported into Photoshop where they were pseudocolored, image processed, and merged. In some images, nonlinear adjustments were made for optimal visualization of Hoechst labeling of chromosomes.
RESULTS
Viability and chromosome morphology in JIL-1 and Su(var)3-9 double mutants:
In a previous study, Zhang et al. (2006) used genetic assays to explore interactions between mutant alleles of Su(var)3-9 and JIL-1 by generating double mutant individuals. Since both Su(var)3-9 and JIL-1 are located on the third chromosome, a Su(var)3-91, JIL-1z60 chromosome was generated by recombination. However, in these experiments, only the +/Su(var)3-91 allelic combination was examined (Zhang et al. 2006) and the JIL-1z60 allele, although a strong hypomorph, still had low levels of histone H3S10 kinase activity (Zhang et al. 2003). We have therefore extended these studies by recombining the true null JIL-1z2 allele (Wang et al. 2001; Zhang et al. 2003) with three different loss-of-function Su(var)3-9 alleles. The Su(var)3-91 allele consists of a frameshift at the N terminus of the protein upstream of the chromo- and SET domains while the Su(var)3-92 allele has two missense mutations, and both alleles result in a null phenotype (Reuter et al. 1986; Tschiersch et al. 1994; Ebert et al. 2004). The null Su(var)3-906 allele is due to a DNA insertion and immunoblot analysis has shown that histone H3K9 dimethylation is greatly reduced in homozygous animals (Schotta et al. 2002). Homozygous null Su(var)3-9 mutants are viable and fertile (Tschiersch et al. 1994).
To determine whether reduction of Su(var)3-9 levels will rescue the lethality normally associated with a null JIL-1z2/JIL-1z2 mutant background, we crossed JIL-1z2 Su(var)3-9*/TM6 Sb Tb males [where Su(var)3-9* denotes the Su(var)3-91, Su(var)3-92, or Su(var)3-906 allele] with JIL-1z2/TM6 Sb Tb virgin females generating JIL-1z2 Su(var)3-9*/JIL-1z2 progeny identified as non-Sb (Table 1). In control experiments in which Su(var)3-9 activity was not altered, we crossed JIL-1z2/TM6 Sb Tb males with JIL-1z2/TM6 Sb Tb virgin females generating JIL-1z2/JIL-1z2 progeny. In the control crosses, of a total of 685 eclosed flies, we observed no flies of the JIL-1z2/JIL-1z2 genotype, indicating complete lethality (Table 1). However, in crosses that generate the double mutant combination [JIL-1z2/JIL-1z2 Su(var)3-9*] with one copy of either of the Su(var)3-9 mutant alleles, the number of surviving flies with the JIL-1z2/JIL-1z2 genotype increased dramatically. In these crosses, one-third of the eclosed flies would be expected to be of the JIL-1z2/JIL-1z2 Su(var)3-9* genotype, assuming full rescue, indicating that the reduction of Su(var)3-9 activity in these animals resulted in a 50.7–87.4% viability rate compared to a rate of 0% for JIL-1z2/JIL-1z2 flies without the reduction in Su(var)3-9 activity (Table 1). However, it should be noted that both rescued male and female flies are sterile. We also performed crosses to generate JIL-1z2 Su(var)3-91/JIL-1z2 Su(var)3-92 progeny. As indicated in Table 1, the further reduction in the dose of Su(var)3-9 did not lead to an additional increase in viability of homozygous JIL-1 null flies. Taken together, these results suggest that the lethality in null JIL-1 mutant backgrounds to a substantial degree is mediated by Su(var)3-9 activity. Furthermore, since this effect was observed with three different alleles of Su(var)3-9 it is likely to be specific to Su(var)3-9 and not to second site modifiers.
TABLE 1.
Genetic interaction between JIL-1 and Su(var)3-9 alleles
| Cross | Genotypes (no. of adult flies) | % expected ratioa | |
|---|---|---|---|
| JIL-1z2/TM6 × JIL-1z2/TM6 | JIL-1z2/TM6 (685) | JIL-1z2/JIL-1z2 (0) | 0.0 |
| JIL-1z2/TM6 × JIL-1z2 Su(var)3-91/TM6 | JIL-1z2/TM6 or JIL-1z2 Su(var)3-91/TM6 (350) | JIL-1z2/JIL-1z2 Su(var)3-91 (144) | 87.4 |
| JIL-1z2/TM6 × JIL-1z2 Su(var)3-92/TM6 | JIL-1z2/TM6 or JIL-1z2 Su(var)3-92/TM6 (280) | JIL-1z2/JIL-1z2 Su(var)3-92 (57) | 50.7 |
| JIL-1z2/TM6 × JIL-1z2 Su(var)3-906/TM6 | JIL-1z2/TM6 or JIL-1z2 Su(var)3-906/TM6 (209) | JIL-1z2/JIL-1z2 Su(var)3-906 (53) | 60.7 |
| JIL-1z2 Su(var)3-91/TM6 × JIL-1z2 Su(var)3-92/TM6 | JIL-1z2 Su(var)3-91/TM6 or JIL-1z2 Su(var)3-92/TM6 (892) | JIL-1z2 Su(var)3-91/JIL-1z2 Su(var)3-92 (204) | 55.8 |
In these crosses, the TM6 chromosome was identified by the Stubble marker. Consequently, the experimental genotypes could be distinguished from balanced heterozygotic flies by absence of the Stubble marker. The expected Mendelian ratio of non-Stubble to Stubble flies was 1:2 since TM6/TM6 is embryonic lethal. The percentage of expected genotypic ratios was calculated as observed non-Stubble flies × 300/total observed flies.
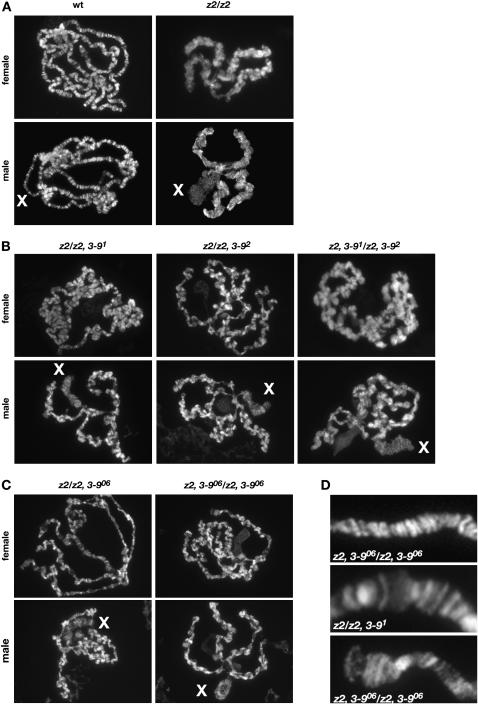
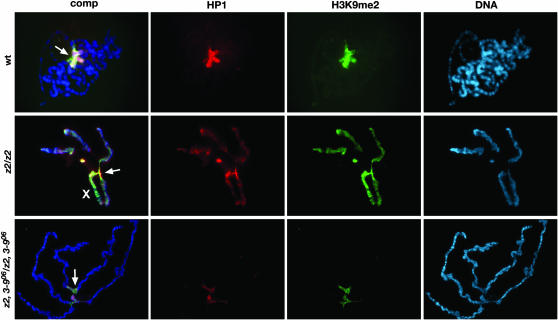
We further investigated whether a reduction in the dose of Su(var)3-9 would also affect the severely perturbed polytene chromosome morphology observed in null JIL-1z2 homozygous larvae (Wang et al. 2001; Deng et al. 2005). For this analysis, we prepared squashes of polytene chromosomes labeled with Hoechst or with propidium iodide from JIL-1z2 homozygous null and wild-type third instar larvae and compared them with squashes from double mutant homozygous JIL-1z2 larvae with various combinations of the Su(var)3-9 alleles described above. As illustrated in Figure 1A, loss of JIL-1 histone H3S10 kinase activity leads to misalignment of the interband chromatin fibrils, which is further associated with coiling of the chromosomes and an increase of ectopic contacts between nonhomologous regions. This results in a shortening and folding of the chromosomes with a nonorderly intermixing of euchromatin and the compacted chromatin characteristic of banded regions (Deng et al. 2005). The extreme of this phenotype is exhibited by the male X polytene chromosome where no remnants of coherent banded regions can be observed (Figure 1A). However, in homozygous JIL-1z2 double mutant combinations with a reduced dosage of Su(var)3-9, there was a marked improvement of polytene chromosome morphology of both male and female autosomes (Figure 1, B and C). The chromosome arms were to a large extent unfolded with reduced ectopic contacts and a clearly discernible banding pattern that in some cases attained near wild-type morphology (Figure 1D). In contrast, the morphology of the male X chromosome was largely unaffected by the reduction in Su(var)3-9, suggesting that ectopic Su(var)3-9 activity does not contribute to the “puffed” X chromosome phenotype in JIL-1 null mutant backgrounds. It is well documented that the male X chromosome is unique because of the activity of the MSL complex and the MOF histone acetyltransferase that leads to hyperacetylation of histone H4 (Bone et al. 1994; Hilfiker et al. 1997). Therefore, the effects on chromosome morphology by Su(var)3-9 may only be observable on autosomes and the female X chromosome. It has previously been shown that both morphology and JIL-1 localization in Su(var)3-9 null polytene chromosomes are indistinguishable from that observed in wild-type chromosomes (Zhang et al. 2006). Furthermore, HP1 binding is severely reduced at the chromocenter and does not spread to the chromosome arms in either a homozygous null Su(var)3-9 background (Schotta et al. 2002) or a homozygous null Su(var)3-9, JIL-1 background (Figure 2). Taken together, these data indicate that the lethality as well as some of the chromosome morphology defects observed in JIL-1 null mutant backgrounds may be mediated by ectopic Su(var)3-9 activity.
Figure 1.—
Morphology of polytene chromosomes in JIL-1 and Su(var)3-9 double mutant backgrounds. Polytene chromosome preparations from third instar larvae were labeled with Hoechst or with propidium iodide to visualize the chromatin. (A) Polytene chromosome preparations from wild-type (wt) male and female larvae and from male and female homozygous JIL-1z2 larvae (z2/z2). Note the misalignment and intermixing of interband and banded regions and the extensive coiling and folding of the chromosome arms in JIL-1z2/JIL-1z2 mutant chromosomes. The male X chromosome (X) is particularly affected and no remnants of banded regions are discernable. (B) Polytene chromosomes from male and female JIL-1z2 Su(var)3-91/JIL-1z2 (z2/z2, 3-91), JIL-1z2 Su(var)3-92/JIL-1z2 (z2/z2, 3-92), and JIL-1z2 Su(var)3-91/JIL-1z2 Su(var)3-92 (z2, 3-91/z2, 3-92) larvae. (C) Polytene chromosomes from male and female JIL-1z2 Su(var)3-906/JIL-1z2 (z2/z2, 3-906) and JIL-1z2 Su(var)3-906/JIL-1z2 Su(var)3-906 (z2, 3-906/z2, 3-906) larvae. Note the marked improvement of polytene chromosome morphology of both male and female autosomes in B and C. The chromosome arms were to a large extent unfolded with reduced ectopic contacts and a clearly discernible banding pattern that in some cases attained near wild-type morphology. In contrast, there was no discernible improvement in the morphology of the male X chromosome. (D) High magnification examples of regions of polytene chromosomes from JIL-1z2 Su(var)3-91/JIL-1z2 (z2/z2, 3-91) and JIL-1z2 Su(var)3-906/JIL-1z2 Su(var)3-906 (z2, 3-906/z2, 3-906) larvae with near wild-type morphology of band and interband regions.
Figure 2.—
Localization of HP1 and histone H3K9me2 in polytene chromosomes from JIL-1 and Su(var)3-9 mutant third instar larvae. The polytene squashes were triple labeled with antibodies to HP1 (red) and H3K9me2 (green) and with Hoechst (DNA, blue). The X chromosome is indicated by an X and the chromocenter by an arrow. Preparations from wild-type (wt), JIL-1z2 homozygous (z2/z2), and JIL-1z2 and Su(var)3-906 double homozygous (z2, 3-906/z2, 3-906) larvae are shown. In wild-type preparations, HP1 and H3K9me2 labeling was mainly localized to and abundant at the chromocenter; however, in the absence of the JIL-1 kinase, the HP1 and H3K9me2 labeling spread to the autosomes and particularly to the X chromosome (see also Zhang et al. 2006). In z2, 3-906/z2, 3-906 double mutant larvae, the HP1 and H3K9me2 labeling were greatly reduced and confined to the chromocenter.
Genetic interactions between JIL-1 and Su(var)2-5 (HP1):
The second component of the ectopic heterochromatic spreading observed in loss-of-function JIL-1 mutant animals is the dimethylated histone H3K9-binding protein, HP1 (Zhang et al. 2006), which is intrinsic to pericentric heterochromatin formation (reviewed in Elgin and Grewal 2003). In contrast to homozygous null Su(var)3-9 animals that are viable, transheterozygous null Su(var)2-5 animals die as third instar larvae (Eissenberg et al. 1992; Eissenberg and Hartnett 1993). Furthermore, tethering of HP1 to euchromatic sites has shown that HP1 is sufficient to nucleate the formation of silent chromatin and that it can cause the formation of ectopic chromatin associations (Seum et al. 2001; Li et al. 2003; Danzer and Wallrath 2004) similar to those observed in JIL-1 mutants. These findings suggest that the lethality as well as the disorganization of chromosomes in JIL-1 null mutant backgrounds may be affected by ectopic HP1 binding and that the partial rescue of animals with reduced Su(var)3-9 activity is a consequence of impaired HP1 recruitment resulting from the reduction in histone H3K9 dimethylation. To explore this possibility, we performed genetic interaction assays between JIL-1 and Su(var)2-5.
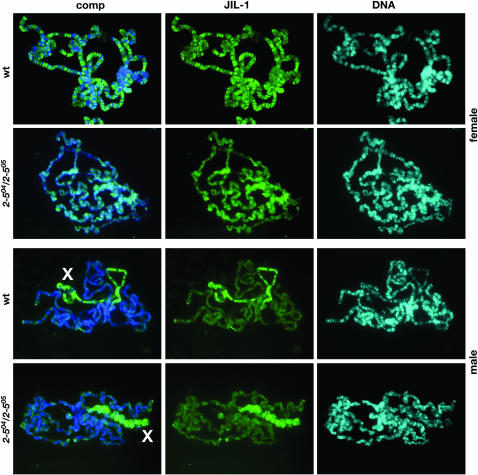
We first tested whether JIL-1 localization was affected in a functionally null mutant Su(var)2-5 background by generating Su(var)2-504/Su(var)2-505 heteroallelic third instar larvae (Eissenberg et al. 1992). The Su(var)2-504 allele is due to a nonsense mutation leading to a truncated HP1 protein that degrades, whereas the Su(var)2-505 allele is associated with a frameshift resulting in a nonsense peptide containing only the first 10 amino acids of HP1 (Eissenberg et al. 1992). Figure 3 shows polytene squashes from wild-type and Su(var)2-504/Su(var)2-505 heteroallelic larvae double labeled with Hoechst and JIL-1 antibody. Polytene chromosomes from loss-of-function HP1 mutant larvae partly lose the distinct pattern of band–interband regions of wild-type chromosomes, a phenotype that is especially pronounced for the male X chromosome (Spierer et al. 2005) (Figure 3). However, JIL-1 localizes to both male and female chromosomes and is upregulated on the male X chromosome, as in wild-type preparations (Figure 3), suggesting that JIL-1 distribution is not altered by loss of HP1 function.
Figure 3.—
JIL-1 localization in HP1 mutant larvae. Polytene chromosome preparations from male and female wild-type (wt) and Su(var)2-504/Su(var)2-505 (2-504/2-505) third instar larvae were double labeled with JIL-1 antibody and Hoechst to visualize the chromatin. The male X chromosome is indicated with an X. Although polytene chromosomes from loss-of-function HP1 mutant larvae partly lose the distinct pattern of band–interband regions of wild-type chromosomes, a phenotype that is especially pronounced for the male X chromosome, JIL-1 localizes to both male and female chromosomes and is upregulated on the male X chromosome as in wild-type preparations.
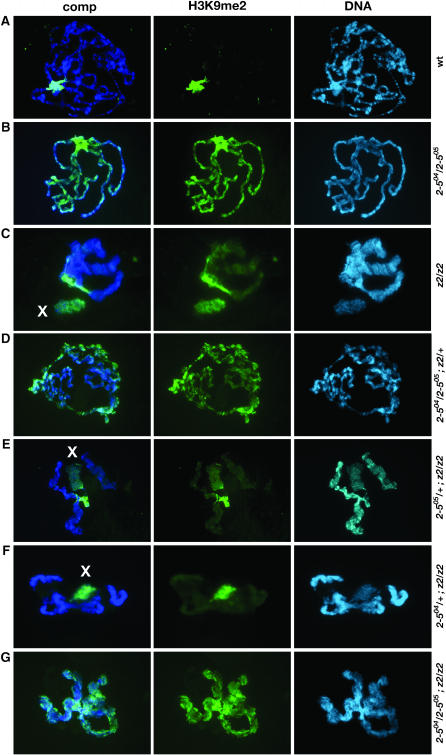
We further examined whether a reduction in HP1 levels could improve chromosome morphology of JIL-1 null animals and in reciprocal experiments whether a reduction in JIL-1 levels could improve chromosome morphology of HP1 null animals. Since the HP1 gene is located on the second chromosome while JIL-1 is on the third chromosome, such double mutant animals were generated by standard genetic crosses. Polytene squashes from third instar larval salivary glands from these double mutant combinations were double labeled with Hoechst and an antibody to histone H3K9me2 and compared to wild-type preparations (Figure 4). In HP1 null animals, histone H3K9 dimethylation is dramatically upregulated on all the chromosome arms (Schotta et al. 2002) (Figure 4B), whereas in JIL-1 null mutants, the upregulation is most pronounced on the X chromosome (Zhang et al. 2006) (Figure 4C). As illustrated in Figure 4D, a reduction in the dose of JIL-1 affected neither chromosome morphology nor the spreading of H3K9 dimethylation in JIL-1z2/+ ; Su(var)2-504/Su(var)2-505 mutant larvae. Likewise, chromosome morphology and H3K9me2 distribution in Su(var)2-504/+ ; JIL-1z2/JIL-1z2 (Figure 4F) and Su(var)2-505/+ ; JIL-1z2/JIL-1z2 (Figure 4E) double mutant larvae were indistinguishable from those of homozygous JIL-1z2 mutant larvae. In the double mutant null JIL-1 and Su(var)2-5 combination, the chromosome morphology resembled that of JIL-1 null polytene chromosomes, whereas the H3K9me2 distribution resembled that of Su(var)2-5 null chromosomes (Figure 4G). The similarity of the spreading of the H3K9me2 marker in HP1 loss-of-function mutants in both wild-type JIL-1 and JIL-1 null mutant backgrounds indicates that this ectopic redistribution is independent of JIL-1 kinase activity. Furthermore, in viability assays, there was neither rescue of JIL-1z2 homozygous lethality by reducing the dose of wild-type HP1 by either the Su(var)2-504 or the Su(var)2-505 allele nor rescue of Su(var)2-5 mutant lethality by reducing the dose of wild-type JIL-1 (Table 2). Unlike the situation for Su(var)3-9 in which loss of one wild-type allele results in significant rescue of the null JIL-1z2/JIL-1z2 lethality, loss of a Su(var)2-5 wild-type allele has no effect. Taken together, these results suggest that there are no genetic interactions detectable in these assays between JIL-1 and Su(var)2-5 and that HP1 in contrast to Su(var)3-9 does not contribute to the lethality or disruption of chromosome morphology observed in JIL-1 loss-of-function mutants.
Figure 4.—
Polytene chromosome morphology and distribution of the H3K9me2 marker in JIL-1 and Su(var)2-5 mutants. Polytene chromosome preparations from third instar larvae were double labeled with histone H3K9me2 antibody (green) and Hoechst (DNA in blue) to visualize the chromatin. The male X chromosome is indicated with an X. Preparations from wild-type (wt, A), Su(var)2-504/Su(var)2-505 (2-504/2-505; B), JIL-1z2/JIL-1z2 (z2/z2; C), Su(var)2-504/Su(var)2-505; JIL-1z2/+ (2-504/2-505; z2/+; D), Su(var)2-505/+; JIL-1z2/JIL-1z2 (2-505/+; z2/z2; E), Su(var)2-504/+; JIL-1z2/JIL-1z2 (2-504/+; z2/z2; F), and Su(var)2-504/Su(var)2-505; JIL-1z2/JIL-1z2 (2-504/2-505; z2/z2; G) larvae are shown.
TABLE 2.
Genetic interaction between JIL-1 and Su(var)2-5 alleles
| Cross | Genotypes (no. of adult flies) | % expected ratioa | |
|---|---|---|---|
| JIL-1z2/TM6 × JIL-1z2/TM6 | JIL-1z2/TM6 (685) | JIL-1z2/JIL-1z2 (0) | 0.0 |
| Su(var)2-504/CyO ; JIL-1z2/TM6 × Su(var)2-504/CyO ; JIL-1z2/TM6 | Su(var)2-504/CyO ; JIL-1z2/TM6 (231) | Su(var)2-504/CyO ; JIL-1z2/JIL-1z2 (0) | 0.0 |
| Su(var)2-505/CyO ; JIL-1z2/TM6 × Su(var)2-505/CyO ; JIL-1z2/TM6 | Su(var)2-505/CyO ; JIL-1z2/TM6 (328) | Su(var)2-505/CyO ; JIL-1z2/JIL-1z2 (0) | 0.0 |
| Su(var)2-504/CyO ; JIL-1z2/TM6 × Su(var)2-505/CyO ; JIL-1z2/TM6 | Su(var)2-504/CyO ; JIL-1z2/TM6 or Su(var)2-505/CyO ; JIL-1z2/TM6 (636) | Su(var)2-504/Su(var)2-505 ; JIL-1z2/JIL-1z2 or Su(var)2-504/CyO ; JIL-1z2/JIL-1z2 or Su(var)2-505/CyO ; JIL-1z2/JIL-1z2 (0) | |
In these crosses, the TM6 chromosome was identified by the Stubble marker. Consequently, the experimental genotypes could be distinguished from balanced heterozygotic flies by absence of the Stubble marker. The expected Mendelian ratio of non-Stubble to Stubble flies was 1:2 since TM6/TM6 and CyO/CyO are embryonic lethal and Su(var)2-504/Su(var)2-504, Su(var)2-505/Su(var)2-505, and Su(var)2-504/Su(var)2-505 die before or at the third instar larval stage. The percentage of expected genotypic ratios was calculated as observed non-Stubble flies × 300/total observed flies.
DISCUSSION
Interdependent interactions between the HP1 and Su(var)3-9 proteins as well as histone H3K9 dimethylation are thought to be major factors in heterochromatin formation and gene silencing in Drosophila (Lachner et al. 2001; Schotta et al. 2002; Elgin and Grewal 2003). Recently, it has been demonstrated that the essential JIL-1 histone H3S10 kinase antagonizes heterochromatization and functions to maintain the chromatin structure of euchromatic regions (Wang et al. 2001; Ebert et al. 2004; Zhang et al. 2006). In the absence of JIL-1 kinase activity, the heterochromatin markers H3K9me2 and HP1 spread in tandem to ectopic locations on the chromosomes (Zhang et al. 2006). Furthermore, loss of JIL-1 histone H3S10 kinase activity results in an increase of ectopic contacts between nonhomologous regions leading to a shortening and folding of the chromosomes with a nonorderly intermixing of euchromatic and compacted chromatin regions (Deng et al. 2005). In this study, we show that the lethality as well as some of the chromosome morphology defects associated with the null JIL-1 phenotype to a large degree can be rescued by reducing the dose of the Su(var)3-9 gene. This effect was observed with three different alleles of Su(var)3-9, strongly suggesting it is likely to be specific to Su(var)3-9 and not to second site modifiers. This is in contrast to similar experiments performed with alleles of the Su(var)2-5 gene that codes for HP1 in Drosophila. In these assays, no genetic interactions were detectable between JIL-1 and Su(var)2-5, suggesting that the lethality and disruption of chromosome morphology observed when JIL-1 levels are decreased are not due to increased HP1 on the chromosomal arms but rather are associated with ectopic Su(var)3-9 activity. How Su(var)3-9 may mediate these effects is unknown and will require additional studies.
While Su(var)3-9 and HP1 reciprocal interactions are well documented at pericentric regions, they are not universal (Schotta et al. 2002; Greil et al. 2003; Danzer and Wallrath 2004). For example, HP1 binding on the fourth chromosome has been shown to be independent of Su(var)3-9 (Schotta et al. 2002). Furthermore, a large scale survey of Su(var)3-9 and HP1 binding at coding regions demonstrated exclusive enrichment of Su(var)3-9 at the majority of such nonpericentric regions that map to the chromosome arms (Greil et al. 2003). Strikingly, while both Su(var)3-9 and HP1 preferentially associate with genes of low expression levels, this preference is more prominent for Su(var)3-9 than for HP1, suggesting that where Su(var)3-9 is actively involved in silencing of its target genes, these Su(var)3-9 complexes may be more potent silencers if they lack HP1 (Greil et al. 2003). Conversely, Danzer and Wallrath (2004), using a tethering system to recruit HP1 to euchromatic sites, have shown that HP1-mediated silencing can operate in a Su(var)3-9-independent manner. These findings indicate that although Su(var)3-9 and HP1 cooperate in heterochromatin formation and gene silencing at pericentric chromosome sites, they may function independently at other regions such as the chromosome arms. This is underscored by the finding that whereas Su(var)3-9 is necessary for HP1 recruitment to pericentric chromatin, Su(var)3-9 spreads and is dramatically upregulated on the chromosome arms in the absence of HP1 (Schotta et al. 2002). Interestingly, the spreading in HP1 loss-of-function mutants is independent of JIL-1 kinase activity, indicating that at least two different molecular mechanisms regulate Su(var)3-9 localization, one dependent on HP1 and one dependent on the JIL-1 kinase.
The results of this study suggest that the lethality of JIL-1 null mutants may be due to the repression of essential genes as a result of ectopic Su(var)3-9 activity unrelated to HP1 recruitment. At interphase, JIL-1 phosphorylates the histone H3S10 residue in euchromatic regions of polytene chromosomes (Jin et al. 1999; Wang et al. 2001), suggesting as a plausible model that this phosphorylation during interphase prevents recruitment of Su(var)3-9 to these sites. At present, we do not know whether the observed spreading of Su(var)3-9 in JIL-1 hypomorphic backgrounds occurred preferentially to specific euchromatic sites (Zhang et al. 2006). In JIL-1 null animals, the morphology of polytene chromosomes is greatly perturbed and there is not only an intermixing of euchromatin and the compacted chromatin characteristic of banded regions but also a looping of nonhomologous chromatid regions that become fused and confluent (Deng et al. 2005). However, in support for this model for JIL-1's function in counteracting heterochromatic spreading and gene silencing, it was recently demonstrated that loss-of-function JIL-1 alleles act as enhancers of position-effect-variegation (PEV) at centric sites, whereas the gain-of-function JIL-1Su(var)3-1 allele acts as a suppressor of PEV (Bao et al. 2007). The JIL-1Su(var)3-1 allele is one of the strongest suppressors of PEV so far described (Ebert et al. 2004) and it generates truncated proteins with COOH-terminal deletions that mislocalize to ectopic chromosome sites (Ebert et al. 2004; Zhang et al. 2006). Thus, the dominant gain-of-function effect of the JIL-1Su(var)3-1 alleles may be attributable to JIL-1 kinase activity at ectopic locations, leading to misregulated localization of the phosphorylated histone H3S10 mark counteracting the spreading of Su(var)3-9. In future experiments, it will be of interest to determine the molecular mechanisms for how Su(var)3-9 activity can effect the lethality and changes in chromatin structure observed in the absence of JIL-1.
Acknowledgments
We thank members of the laboratory for discussion, advice, and critical reading of the manuscript. We also acknowledge V. Lephart for maintenance of fly stocks and Laurence Woodruff for technical assistance. We especially thank L. Wallrath for providing the Su(var)3-906, Su(var)2-504, and Su(var)2-505 alleles. This work was supported by NIH grant GM62916 (to K.M.J.).
References
- Bao, X., H. Deng, J. Johansen, J. Girton and K. M. Johansen, 2007. Loss-of-function alleles of the JIL-1 histone H3S10 kinase enhance position-effect-variegation at pericentric sites in Drosophila heterochromatin. Genetics 176: 1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone, J. R., J. Lavender, R. Richman, M. J. Palmer, B. M. Turner et al., 1994. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 8: 96–104. [DOI] [PubMed] [Google Scholar]
- Danzer, J. R., and L. L. Wallrath, 2004. Mechanisms of HP1-mediated gene silencing in Drosophila. Development 131: 3571–3580. [DOI] [PubMed] [Google Scholar]
- Delattre, M., A. Spierer, Y. Jaquet and P. Spierer, 2004. Increased expression of Drosophila Su(var)3-7 triggers Su(var)3-9-dependent heterochromatin formation. J. Cell Sci. 117: 6239–6247. [DOI] [PubMed] [Google Scholar]
- Deng, H., W. Zhang, X. Bao, J. N. Martin, J. Girton et al., 2005. The JIL-1 kinase regulates the structure of Drosophila polytene chromosomes. Chromosoma 114: 173–182. [DOI] [PubMed] [Google Scholar]
- Ebert, A., G. Schotta, S. Lein, S. Kubicek, V. Krauss et al., 2004. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 18: 2973–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg, J. C., and T. Hartnett, 1993. A heat shock-activated cDNA rescues the recessive lethality of mutations in the heterochromatin-associated protein HP1 of Drosophila melanogaster. Mol. Gen. Genet. 240: 333–338. [DOI] [PubMed] [Google Scholar]
- Eissenberg, J. C., G. D. Morris, G. Reuter and T. Hartnett, 1992. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin, S. C., and S. I. Grewal, 2003. Heterochromatin: silence is golden. Curr. Biol. 13: R895–898. [DOI] [PubMed] [Google Scholar]
- Girton, J., and K. M. Johansen, 2007. Chromatin structure and regulation of gene expression: the lessons of PEV. Adv. Genet. (in press). [DOI] [PubMed]
- Greil, F., I. Van Der Kraan, J. Delrow, J. F. Smothers, E. De Wit et al., 2003. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev. 17: 2825–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker, A., D. Hilfiker-Kleiner, A. Pannuti and J. C. Lucchesi, 1997. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 16: 2054–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, T. C., J. C. Eissenberg, C. Craig, V. Dietrich, A. Hobson et al., 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell. Biol. 50: 170–180. [PubMed] [Google Scholar]
- Ji, Y., U. Rath, J. Girton, K. M. Johansen and J. Johansen, 2005. D-Hillarin, a novel W180-domain protein, affects cytokinesis through interaction with the septin family member Pnut. J. Neurobiol. 64: 157–169. [DOI] [PubMed] [Google Scholar]
- Jin, Y., Y. Wang, D. L. Walker, H. Dong, C. Conley et al., 1999. JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol. Cell 4: 129–135. [DOI] [PubMed] [Google Scholar]
- Kelley, R. L., V. H. Meller, P. R. Gordadze, G. Roman, R. L. Davis et al., 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98: 513–522. [DOI] [PubMed] [Google Scholar]
- Lachner, M., D. O'Carroll, S. Rea, K. Mechtler and T. Jenuwein, 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120. [DOI] [PubMed] [Google Scholar]
- Lerach, S., W. Zhang, X. Bao, H. Deng, J. Girton et al., 2006. Loss-of-function alleles of the JIL-1 kinase are strong suppressors of position effect variegation of the wm4 allele in Drosophila. Genetics 173: 2403–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., J. R. Danzer, P. Alvarez, A. S. Belmont and L. L. Wallrath, 2003. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 130: 1817–1824. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, New York.
- Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra et al., 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672. [DOI] [PubMed] [Google Scholar]
- Reuter, G., R. Dorn, G. Wustmann, B. Friede and G. Rauh, 1986. Third chromosome suppressor of position-effect variegation loci in Drosophila melanogaster. Mol. Gen. Genet. 202: 481–487. [Google Scholar]
- Roberts, D. B., 1998. Drosophila: A Practical Approach. IRL Press, Oxford.
- Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann et al., 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21: 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum, C., M. Delattre, A. Spierer and P. Spierer, 2001. Ectopic HP1 promotes chromosome loops and variegated silencing in Drosophila. EMBO J. 20: 812–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spierer, A., C. Seum, M. Delattre and P. Spierer, 2005. Loss of the modifiers of variegation Su(var)3-7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. J. Cell Sci. 118: 5047–5057. [DOI] [PubMed] [Google Scholar]
- Swaminathan, J., E. M. Baxter and V. G. Corces, 2005. The role of histone H2av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 19: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch, B., A. Hofmann, V. Krauss, R. Dorn, G. Korge et al., 1994. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13: 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., W. Zhang, Y. Jin, J. Johansen and K. M. Johansen, 2001. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 105: 433–443. [DOI] [PubMed] [Google Scholar]
- Weiler, K. S., and B. T. Wakimoto, 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577–605. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Y. Jin, Y. Ji, J. Girton, J. Johansen et al., 2003. Genetic and phenotypic analysis of alleles of the Drosophila chromosomal JIL-1 kinase reveals a functional requirement at multiple developmental stages. Genetics 165: 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., H. Deng, X. Bao, S. Lerach, J. Girton et al., 2006. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development 133: 229–235. [DOI] [PubMed] [Google Scholar]