Abstract
In the Abd-B 3′ cis-regulatory region, which is subdivided into a series of iab domains, boundary elements have previously been detected, including the Fab-7 element providing for the autonomous functioning of the iab-6 and iab-7 cis-regulatory domains. Here, it has been shown that a single copy of the 860-bp Fab-7 insulator effectively blocks the yellow and white enhancers. The eye and testis enhancers can stimulate the white promoter across the pair of Fab-7, which is indicative of a functional interaction between the insulators. Unexpectedly, Fab-7 has proved to lose the enhancer-blocking activity when placed near the white promoter. It seems likely that Fab-7 strengthens the relatively weak white promoter, which leads to the efficient enhancer–promoter interaction and insulator bypass.
AS enhancers exert long-distance effects, a question arises as to how an enhancer can specifically activate its target gene without affecting adjacent genes (Dorsett 1999; Bondarenko et al. 2003; West and Fraser 2005; Fraser 2006; Gaszner and Felsenfeld 2006). In this relation, of interest is the class of DNA sequence elements, named insulators, that contribute to the organization of independent gene function domains by restricting the functions of enhancers and silencers (Sun and Elgin 1999; Kuhn and Geyer 2003; Capelson and Corces 2004; Brasset and Vaury 2005; West and Fraser 2005; Gaszner and Felsenfeld 2006; Valenzuela and Kamakaka 2006). Insulators are characterized by two properties. First, they operate in a position-dependent manner, preventing the functioning of enhancers and silencers when inserted between these regulatory elements and a promoter but not when located upstream or downstream from them (Gyurkovics et al. 1990; Holdridge and Dorsett 1991; Geyer and Corces 1992; Kellum and Schedl 1992; Sigrist and Pirrotta 1997; Mallin et al. 1998; Comet et al. 2006). However, insulators do not inactivate enhancers, silencers, or promoters, which indicates that they interfere with signaling between these classes of control elements (Geyer and Corces 1992; Cai and Levine 1995; Scott and Geyer 1995). Second, insulators protect gene expression from the positive and negative effects of chromatin surrounding a gene (Ishii et al. 2002; Mutskov et al. 2002) and confer the capacity for position-independent transcription to transgenes reliably integrated into the genome (Bonifer et al. 1990; Kellum and Schedl 1991; Roseman et al. 1993, 1995; Burgess-Beusse et al. 2002). Most of the insulators that have been identified in multicellular organisms appear to be constitutive and remain active irrespective of developmental stage or tissue type.
Several constitutively active insulators/boundaries that have been found in the Drosophila Antennapedia (ANT-C) and bithorax (BX-C) complexes are crucial for the developmental functions of homeotic genes in each complex (Gyurkovics et al. 1990; Hagstrom et al. 1996; Zhou et al. 1996, 1999; Mihaly et al. 1998; Zhou and Levine 1999; Barges et al. 2000; Belozerov et al. 2003; Gruzdeva et al. 2005). The three homeotic genes of the bithorax complex—Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B)—are responsible for specifying the identity of parasegments 5–14 (PS5−PS14), which form the posterior half of the thorax and all abdominal segments of an adult fly (Lewis 1978; Sanchez-Herrero et al. 1985; Mihaly et al. 1998; Maeda and Karch 2006). The PS-specific expression patterns of Ubx, abd-A, and Abd-B are determined by a complex cis-regulatory region that spans a 300-kb DNA segment (Sipos and Gyurkovics 2005; Maeda and Karch 2006). For example, Abd-B expression in PS10, PS11, PS12, and PS13 is controlled by the iab-5, iab-6, iab-7, and iab-8 cis-regulatory domains, respectively (Lewis 1978; Karch et al. 1985; Duncan 1987; Celniker et al. 1990; Boulet et al. 1991; Sanchez-Herrero 1991). The current model suggests that boundaries flank each iab region and organize the Abd-B regulatory DNA into a series of separate chromatin loop domains (Gyurkovics et al. 1990; Galloni et al. 1993; Mihaly et al. 1998; Sipos and Gyurkovics 2005; Maeda and Karch 2006).
Among Abd-B boundaries, the best characterized is the Fab-7 element located between the iab-6 and iab-7 cis-regulatory domains. Mutations that inactivate Fab-7 lead to the fusion of the iab-6 and iab-7 domains, and this disrupts the specification of PS11 (Gyurkovics et al. 1990; Galloni et al. 1993; Karch et al. 1994; Mihaly et al. 1997). As with other known insulators, the insulating activity of the Fab-7 element is neither restricted to specific enhancer–promoter combinations nor stage or tissue specific (Galloni et al. 1993; Hagstrom et al. 1996; Zhou et al. 1996; Schweinsberg and Schedl 2004). The minimal Fab-7 boundary defined in different enhancer blocking assays is 1.2 kb long (Hagstrom et al. 1996; Zhou et al. 1996).
In this study, we have found that the Fab-7 insulator and its 0.86-kb subfragment block the yellow and white enhancers with similar efficiency. The 0.86-kb insulator has the same activity when placed at different sites between the yellow enhancers and promoter. Unexpectedly, the insulator has proved to lose the enhancer-blocking activity when inserted near the white promoter. As previously shown for several other insulators (Cai and Shen 2001; Muravyova et al. 2001; Conte et al. 2002; Kuhn et al. 2003; Gruzdeva et al. 2005; Kyrchanova et al. 2007), the interaction between the Fab-7 insulators leads to mutual neutralization of their enhancer-blocking activity.
MATERIALS AND METHODS
Transgenic constructs:
The 3-kb SalI–BamHI fragment containing the yellow regulatory region (yr) with the body and wing enhancers (−2873 to −1266 bp relative to the transcription start; Geyer and Corces 1987) was subcloned into the pGEM7 plasmid digested with BamHI and XhoI. The white eye enhancer (Ee, fragment −1465 to −1084 bp relative to the white transcription start; Qian et al. 1992) was then inserted at position −1868 from the yellow transcription start site (yr-Ee). The 5-kb BamHI–BglII fragment containing the yellow coding region (yc) was subcloned into CaSpeR2 (yc-C2). The I-SceI+126x2-Eye-yr plasmid containing the yellow and white enhancers between the I-SceI sites was described by Rodin and Georgiev (2005).
The 1.252-kb Fab-7 fragment (F71.2) was cloned by PCR amplification of the genomic DNA between primers 5′-ACTGCAGTGAAGACACGAACC-3′ and 5′-CGTGAGCGACCGAAACTC-3′; the 0.858-kb Fab-7 fragment was cloned by PCR amplification between primers 5′-GATTTCAAGCTGTGTGGCGGGG-3′ and 5′-CGTGAGCGACCGAAACTC-3′. Thereafter, these Fab-7 fragments were sequenced to confirm their identity and subcloned between lox [lox(F7) and lox(F71.2) plasmids] and frt [frt(F7) plasmid] sites. Sequences from e(y)3 cDNA were used as spacers. In particular, the 2.7-kb BamHI–NotI and the 1.4-kb PvuII–BglII fragments were cut from the coding region of the e(y)3 gene and cloned, respectively, between lox [lox(2.7)] and frt [frt(1.4)] sites.
To construct Eye(F71.2)YW and Eye(F7)YW, the lox(F71.2) and lox(F7) fragments were inserted into the yr-Ee plasmid digested with Eco47III at −893 from the yellow transcription start site [yr-Ee-lox(F71.2) and yr-Ee-lox(F7)]. Next, the yr-Ee-lox(F71.2) and yr-Ee-lox(F7) fragments were cloned into the yc-C2 plasmid digested with XbaI and BamHI.
To construct (Eyw)(F7)Y(F7)W and (Eyw)(F7R)Y(F7)W, the lox(F7) fragment was inserted into C2-yc between the yellow and white genes [C2-lox(F7)-yc]. Here and below, the orientation of the Fab-7 fragment relative to the direction of the yellow and white genes was verified by PCR. The frt(F7) fragment was inserted in the direct or reverse orientation into the I-SceI+126x2-Eye-yr plasmid digested with Eco47III [I-SceI+126x2-Eye-yr-frt(F7)]. The resulting fragment was subcloned into C2-yc-lox(F7) digested with XbaI and BamHI.
To construct (Eyw)Y(F7)W, the lox(F7) fragment was inserted in the direct orientation between the yellow and white genes [C2-yc-lox(F7)]. The I-SceI+126x2-Eye-yr fragment was subcloned into C2-yc-lox(F7) digested with XbaI and BamHI.
To construct (Eye)(2.7)(F7)YW, the frt(F7) fragment was inserted into the lox(2.7) plasmid digested with BamHI [lox(2.7)-frt(F7)]. The lox(2.7)-frt(F7) fragment was inserted into the I-SceI+126x2-Eye-yr plasmid digested with Eco47III [I-SceI+126x2-Eye-yr- lox(2.7)-frt(F7)]. The resulting fragment was subcloned into C2-yc cleaved with XbaI and BamHI.
To construct (Eyw)Y(F7R)(1.4)W, the frt(1.4) fragment was inserted into the lox(F7) plasmid digested with BamHI [lox(F7)-frt(1.4)]. The lox(F7)-frt(1.4) fragment was subcloned into the C2-yc digested with BglI [C2-yc-lox(F7)-frt(1.4)]. The I-SceI+126x2-Eye-yr fragment was subcloned into C2-yc-lox(F7)-frt(1.4) digested with XbaI and BamHI.
To construct EywF7Y(F7R)(1.4)W, the I-SceI+126x2-Eye-yr fragment was subcloned into C2-yc-lox(F7)-frt(1.4) digested with XbaI and BamHI.
To construct (Eye)F7−172YW, the 500-bp fragment was obtained by PCR amplification of the yr plasmid between primers 5′-CGCAAAGTTGGCCGATCTATGG-3′ and 5′-CAGGAAACAGCTATGAC-3′. After sequencing, the 500-bp fragment was cloned into F7 digested with PstI and SpeI (F7-500). The 770-bp fragment was obtained by PCR amplification of the yr plasmid between primers 3′-ATCCAGTTGATTTTCAGGGACCA-5′ and 5′-TGTCTTCCATGATTGATTTTCACGC-3′. After sequencing, the 770-bp fragment was cloned into F7-500 plasmid digested with HindIII and XhoI (F7-500-770). The F7-500-770 fragment was cloned into the pSK-I-SceI+126x2-Eye plasmid digested with HincII [(Eye)-F7−172]. Finally, the resulting DNA fragment was cloned into C2-yc digested with XbaI and BamHI.
To construct (Eye)(F7−343)YW the lox(F7) fragment was inserted in direct orientation into the yr pGEM7 plasmid digested with KpnI [yr-lox(F7−343)]. I-SceI+126x2-Eye-yr fragment was subcloned into yr-lox(F7−343) plasmid digested with XbaI-Eco47III [I-SceI+126x2-Eye-yr-lox(F7−343)]. The resulting fragment was subcloned into C2-yc cleaved with XbaI and BamHI.
Generation and analysis of transgenic lines:
All flies were maintained on the standard yeast medium at 25°. The mutant alleles and chromosomes used in this study and the balancer chromosomes are described elsewhere (Lindsley and Zimm 1992). The construct, together with P25.7wc, a P element having defective inverted repeats used as a transposase source (Karess and Rubin 1984), was injected into y ac w1118 preblastoderm embryos as described (Rubin and Spradling 1982; Spradling and Rubin 1982). The resulting flies were crossed with y ac w1118 flies, and the transgenic progeny were identified by the color of their eyes and cuticle structures. The transformed lines were tested for transposon integrity and copy number by Southern blot hybridization.
The lines with excisions were obtained by crossing the flies bearing the transposons with flies of Flp (w1118; S2 CyO, hsFLP, ISA/Sco; +) or Cre (y1, wi; CyO, P[w+,cre]/Sco; +) recombinase-expressing lines. A high level of FLP recombinase was produced by exposing late embryos and second or third instar larvae to heat shock at 37° for 2 hr. A high level of I-SceI endonuclease was achieved by heat-shock treatment for 2 hr on day 3 after hatching, as described (Rodin and Georgiev 2005). The excisions were confirmed by PCR analysis. The details of the crosses used for genetic analysis and for excision of functional elements are available upon request.
The yellow phenotype was determined from the level of pigmentation of the abdominal cuticle and wings in 3- to 5-day-old males developing at 25°. As a reference group, we used flies in which the y allele had been characterized previously. The level of pigmentation (i.e., of y expression) was estimated on an arbitrary five-grade scale: wild-type expression was assigned score 5, and the absence of expression, score 1. The white phenotype was determined from eye pigmentation and testis pigmentation in adult flies. Wild-type white expression in eyes determined bright-red eye color (R); in the absence of white expression, the eyes were white (W). Intermediate levels of white expression (in increasing order) were reflected in the eye color, ranging from pale yellow (pY) through yellow (Y), dark yellow (dY), orange (Or), and dark orange (dOr), to brown (Br) or brown-red (BrR). Males from different transgenic lines were allowed to age 10 days before dissection and visual inspection of the testes.
RESULTS
The 1.2-kb Fab-7 insulator and its 0.86-kb subfragment block the yellow and white enhancers with similar efficiency:
To test the enhancer-blocking activity of the Fab-7 insulators and functional interaction between them, we employed the test system with the yellow and white genes that have been extensively used in insulator studies (Geyer and Corces 1992; Kellum and Schedl 1992; Roseman et al. 1993; Muravyova et al. 2001; Kuhn et al. 2003; Schweinsberg et al. 2004; Schweinsberg and Schedl 2004; Gruzdeva et al. 2005; Savitskaya et al. 2006). The yellow gene (designated Y) is required for dark pigmentation of larval and adult cuticle and its derivatives. Two upstream enhancers (designated Ey) are responsible for yellow activation in the body cuticle and wing blades, while the enhancer responsible for yellow activation in bristles resides in the intron of the yellow gene (Geyer and Corces 1987; Martin et al. 1989). The white gene (designated W) is required for eye and testis pigmentation and is regulated by the eye- and testis-specific enhancers (designated Ew; Qian et al. 1992). These enhancers were inserted between the yellow wing and body enhancers (Eyw).
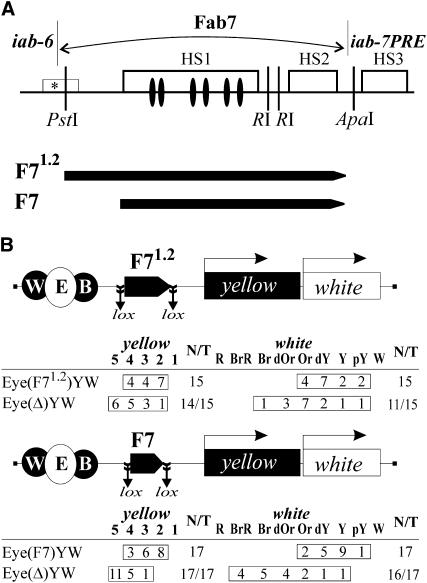
Previously, the minimal Fab-7 insulator was mapped in the 1.2-kb fragment between PstI and ApaI (Figure 1A). In our assay, we used the same 1.2-kb fragment (F71.2) and its 0.86-kb subfragment (F7) in which the 5′ sequences of F71.2 were deleted. The F7 subfragment contains all GAF binding sites related to the enhancer-blocking activity of the insulator (Schweinsberg et al. 2004).
Figure 1.—
Comparison of the enhancer-blocking activities of 1.2-kb (F71.2) and 0.86-kb (F7) fragments from the Fab-7 insulator. (A) Diagram shows Fab-7 region that includes one minor (marked with an *) and three major nuclease hypersensitive sites (HS1, HS2, and HS3) whose extent is indicated by open boxes (Karch et al. 1994) and the 1.2-kb and 0.86-kb Fab-7 fragments tested in the enhancer-blocking assay. GAF binding sites are shown as solid ovals. (B) Reductive schemes of transgenic constructs and analysis of F71.2 and F7 in an enhancer blocking assay. In schemes of constructs (not drawn to scale), Fab-7 fragments are shown as pentagons with apexes indicating their orientation. The yellow wing (W) and body (B) enhancers are shown as solid ovals, and the eye enhancer (E) inserted between them is shown as an open oval. The yellow (Y) and white (W) genes are shown as boxes with arrows indicating the direction of their transcription. Downward arrows indicate the target sites of Flp recombinase (frt) or Cre recombinase (lox); the same sites in construct names are denoted by parentheses. The “yellow” column shows the numbers of transgenic lines with the yellow pigmentation level in the abdominal cuticle (reflecting the activity of the body enhancer); in most of the lines, pigmentation levels in wing blades (reflecting the activity of the wing enhancer) closely correlated with these scores. The “white” column shows the numbers of transgenic lines with the white pigmentation level in eyes (reflecting the activity of the eye enhancer). The level of pigmentation (i.e., of y expression) was estimated on an arbitrary five-grade scale, with wild-type expression and the absence of expression assigned scores 5 and 1, respectively. The “white” column shows the numbers of transgenic lines with different levels of eye pigmentation. Wild-type white expression determined the bright red eye color (R); in the absence of white expression, the eyes were white (W). Intermediate levels of pigmentation with the eye color ranging from pale yellow (pY), through yellow (Y), dark yellow (dY), orange (Or), dark orange (dOr), and brown (Br), to brownish red (BrR) reflect the increasing levels of white expression. N is the number of lines in which flies acquired a new y phenotype upon deletion (Δ) of the specified DNA fragment; T is the total number of lines examined for each particular construct.
The F7 [Eyw(F7)YW] and F71.2 [Eyw(F71.2)YW] flanked by lox sites were inserted between the enhancers and promoters at −893 relative to the yellow transcription start site (Figure 1B). We obtained 15 transgenic lines carrying a single copy of Eyw(F71.2)YW and 17 Eyw(F7)YW transgenic lines. Both F71.2 and F7 partially blocked the yellow and white enhancers with comparable efficiency (Figure 1B). Deletion of the insulators resulted in recovery of yellow and white expression in most of transgenic lines, confirming the role of these elements in insulation. These results suggest that the 0.86-kb subfragment contains the sequences required for the Fab-7 insulator activity in adult flies.
Functional interaction between two copies of Fab-7 insulator leads to neutralization of their enhancer-blocking activity:
Considering that two consecutive Mcp insulators (Kyrchanova et al. 2007) or gypsy insulators (Cai and Shen 2001; Muravyova et al. 2001; Kuhn et al. 2003) between an enhancer and the target gene promoter fail to block gene activation, we decided to find out whether two Fab-7 insulators would neutralize each other's enhancer-blocking activity. In all further experiments, we used the minimal 0.86-kb Fab-7 insulator (F7).
In the (Eyw)(F7)Y(F7R)W construct (Figure 2), one Fab-7 insulator (F7) flanked by frt sites was inserted at −893 and the second Fab-7 copy flanked by lox sites was inserted between the yellow and white genes in the inverted orientation (designated F7R). To check the contributions of the enhancers to yellow and white expression in the presence or absence of Fab-7 insulators, the fragment containing the enhancers was flanked by 126-bp direct repeats and sites for the rare-cleaving I-SceI endonuclease that permits excision of enhancers as described in materials and methods (Rodin and Georgiev 2005).
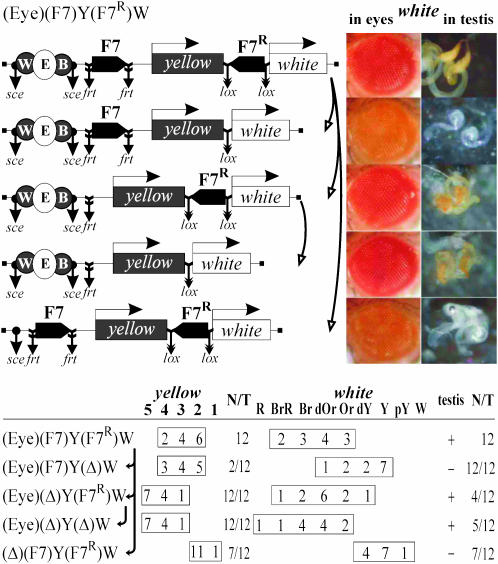
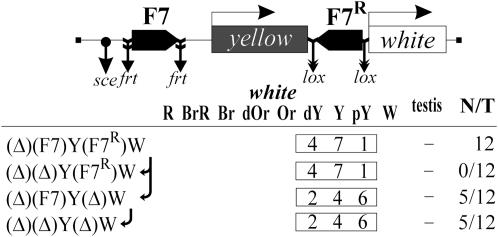
Figure 2.—
Tests for the functional interaction between Fab-7 insulators based on the ability of the eye and testis enhancers to stimulate transcription across the pair of the Fab-7 elements. In schemes of constructs, downward arrows indicate the target sites of Flp recombinase (frt), or Cre recombinase (lox), or I-SceI endonuclease (sce). Arrows near the names of the construct show the order in which the N/T values were calculated. The “testis” column shows white expression in testis: +, extensive pigmentation; −, weak pigmentation or its absence. Other designations are as in Figure 1. Photographs show the eyes and testes of males heterozygous for the construct (P/+) from one transgenic line and its derivatives.
Throughout the text, parentheses in construct designations enclose the elements flanked by the frt (Golic and Lindquist 1989), lox (Siegal and Hartl 2000), or I-SceI sites at which those elements can be excised in crosses with flies expressing Flp or Cre recombinase or I-SceI endonuclease (as noted in materials and methods). By comparing yellow and white expression in transgenic lines and their derivatives with two, one, or no Fab-7 elements, and in the presence or absence of the enhancers, it is possible to estimate the levels of enhancer blocking by different combinations of the elements at the same genomic site.
Twelve transgenic lines carrying a single insertions of the (Eyw)(F7)Y(F7R)W were obtained (Figure 2). By comparing yellow phenotypes in the original and derivative transgenic lines with one Fab-7 element inserted at −893 or no Fab-7 insulators, we found that Fab-7 blocked the yellow enhancers effectively, while the second Fab-7 copy inserted downstream of the yellow gene had no notable effect on insulation. Deletion of the yellow enhancers resulted in decreasing of pigmentation to y2-like level in all transgenic lines, suggesting that the yellow enhancers partially stimulate yellow expression across the Fab-7 insulator in half of the transgenic lines tested.
Comparing eye and testis pigmentation in the (Eyw)(F7)Y(F7R)W lines and their derivatives with the deleted enhancers showed that the eye and testis enhancers can effectively stimulate white expression across two Fab-7 insulators (Figure 2). At the same time, the deletion of the Fab-7 insulator located downstream of the yellow gene led to a strong reduction of eye and testis pigmentation, indicating that one Fab-7 copy at −893 effectively blocked the white enhancers. Unexpectedly, we found that a single Fab-7 insulator located close to the white promoter failed to effectively block the eye and testis enhancers (Figure 2). Thus, the Fab-7 insulator located close to the white promoter proved to lose its enhancer-blocking activity.
Orientation of Fab-7 insulators relative to genes and to each other is not crucial for their insulating activity and the outcome of their functional interaction:
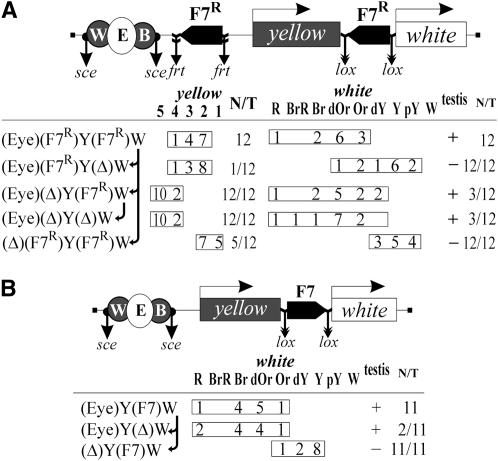
The proximal and distal Fab-7 copies in the (Eyw)(F7)Y(F7R)W construct were in opposite orientations, and this factor might have an effect on the observed enhancer-blocking activity. To test the enhancer-blocking activity of the proximal (relative to the enhancers) Fab-7 insulator inserted in the opposite orientation and to reveal the outcome of pairing between collinear Fab-7 insulators, we made the (Eyw)(F7R)Y(F7R)W construct in which both Fab-7 elements were in the same orientation, opposite to the direction of the yellow and white genes (Figure 3A).
Figure 3.—
Tests for orientation dependence of the Fab-7 insulator. (A) The (Eyw)(F7R)Y(F7R)W and (B) (Eyw)Y(F7)W constructs and phenotypic analysis of the corresponding transgenic lines carrying these constructs and their derivatives. Other designations are as in Figures 1 and 2.
The levels of yellow pigmentation in flies of all 12 transgenic (Eyw)(F7R)Y(F7R)W lines (Figure 3A) were similar to those in flies of the initial (Eyw)(F7)Y(F7R)W lines (Figure 2). Thus, the orientation of the proximal Fab-7 insulator is not crucial for enhancer blocking. Again, deletion of the distal (relative to the enhancers) Fab-7 insulator had no influence on yellow pigmentation.
Comparing eye and testis pigmentation in the (Eyw)(F7)Y(F7R)W (Figure 2) and (Eyw)(F7R)Y(F7R)W (Figure 3A) transgenic lines, we concluded that the insulator bypass by the white enhancers in heterozygous flies did not depend on the relative orientation of Fab-7 insulators. One Fab-7 copy (F7R) inserted at −893 in reverse orientation [in (Eyw)(F7R)Y(Δ)W derivative lines, Figure 3] strongly blocked the white enhancers, confirming that the orientation of the proximal Fab-7 copy is not crucial for insulating activity.
Thereafter, we tested if the activity of Fab-7 located near by the white promoter was also orientation independent. In the (Eyw)Y(F7)W construct, we inserted the Fab-7 insulator flanked by lox sites between the yellow and white genes in the direct orientation (Figure 3B). In all 11 (Eyw)Y(F7)W lines tested, the enhancers effectively stimulated white expression, with the deletion of Fab-7 having no significant effect on it. Thus, the Fab-7 insulator inserted in the direct orientation also failed to block the white enhancers. Taken together, these results suggest that the orientation of Fab-7 relative to the enhancer–promoter pair is not crucial for its enhancer-blocking activity.
Distance between Fab-7 and the white promoter is crucial for enhancer blocking:
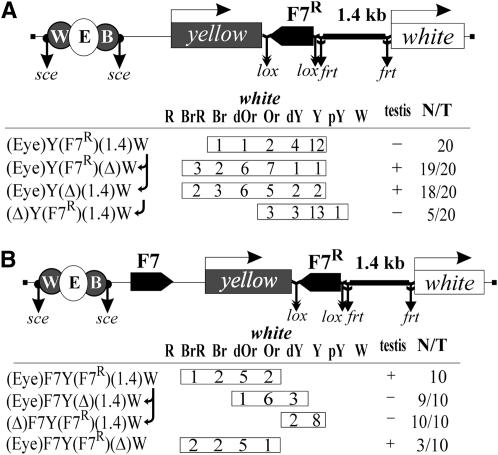
The insulator bypass by the enhancers in the case of Fab-7 placed close to the white promoter suggested that the distance between the promoter and the insulator could have a role in enhancer blocking. To test this possibility, we inserted a 1.4-kb spacer flanked by frt sites between white and Fab-7 in the (Eyw)Y(F7R)(1.4)W construct (Figure 4A). As in the previous constructs, Fab-7 flanked by lox sites was inserted in the reverse orientation on the 3′ side of the yellow gene. Flies in 16 of 20 transgenic (Eyw)Y(F7R)(1.4)W lines had low levels of eye and testis pigmentation, which indicated that the white enhancers were blocked (Figure 4A). Deletion of either Fab-7 insulator or the 1.4-kb spacer restored activity of the enhancers. Thus, the 1.4-kb DNA fragment did not block the enhancers but increased the distance between Fab-7 and the white promoter, which allowed Fab-7 to insulate the enhancers.
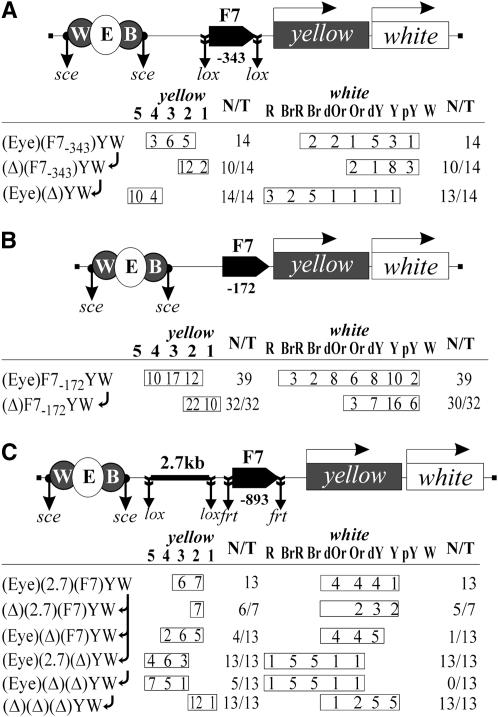
Figure 4.—
Tests for role of the distance between Fab-7 and the white promoter in insulator bypass by the enhancers: (A) (Eye)Y(F7R)(1.4)W and (B) (Eye)(F7)Y(F7R)(1.4)W transgenes and analysis of yellow and white expression in original transgenic lines and their derivatives obtained by deleting either Fab-7, or the enhancers, or the DNA spacer (1.4 kb) inserted between Fab-7 and the white promoter. Other designations are as in Figures 1 and 2.
Next, we checked whether Fab-7 pairing allows enhancer–promoter communication when the distal Fab-7 and the white promoter are separated by the 1.4-kb spacer. In the (Eyw)F7Y(F7R)(1.4)W construct, both Fab-7 insulators were inserted in the same positions as in the original (Eyw)(F7)Y(F7R)W construct, but the white promoter was separated from the distal Fab-7 insulator by the 1.4-kb spacer (Figure 4B). In 10 transgenic (Eyw)F7Y(F7R)(1.4)W lines, the eye and testis enhancers stimulated white expression. Deletion of either the enhancers or the distal Fab-7 insulator led to strong reduction of eye pigmentation in flies, while deletion of the 1.4-kb spacer did not significantly change white expression (Figure 4B). Thus, an increase in the distance between the Fab-7 insulator and the white promoter does not affect the ability of the enhancers to stimulate white expression across the pair of Fab-7 insulators.
The near-by Fab-7 insulator improves the basal activity of the white promoter:
To explain the inability of the Fab-7 insulator to block enhancers in position near the white promoter, we suggest that Fab-7 can strengthen the relatively weak promoter. To test this assumption, we compared eye pigmentation in the enhancerless (Δ)(F7)Y(F7R)W derivatives in the presence of Fab-7 insulators and after their deletion (Figure 5). The deletion of the Fab-7 insulator at −893 had no influence on white expression, while that of the distal Fab-7 insulator markedly decreased eye pigmentation in 5 of 12 transgenic lines, suggesting that the Fab-7 insulator located near the white promoter improves its activity.
Figure 5.—
Tests for the influence of Fab-7 on the activity of white promoter: analysis of eye pigmentation in flies from enhancerless derivatives of (Eyw)(F7)Y(F7R)W lines. Other designations are as in Figures 1 and 2.
Fab-7 displays insulating activity in different positions near the yellow promoter:
The next question was whether Fab-7 fails to block the yellow enhancers when it is located close to the promoter. To study this position dependence, we inserted the Fab-7 insulator flanked by lox sites at −343 bp relative to the yellow transcription start site [(Eye)(F7−343)YW, Figure 6A]. The distance between Fab-7 and the yellow promoter was approximately the same as between Fab-7 and the white promoter in the (Eyw)Y(F7R)W construct. In all 14 (Eye)(F7−343)YW transgenic lines, the yellow and white enhancers were blocked with similar efficiency as in the transgenic lines in which the Fab-7 insulator was inserted at −893 (Figures 1B and 2).
Figure 6.—
Tests for the insulating activity of Fab-7 located at different distances from the yellow promoter or enhancers: (A) (Eye)(F7−343)YW, (B) (Eye)F7−172YW, and (C) (Eye)(2.7)(F7)YW transgenes and analysis of yellow and white expression in original transgenic lines and their derivatives obtained by deletion of Fab-7, or the enhancers, or the DNA spacer (2.7 kb) inserted between the enhancers and Fab-7. Other designations are as in Figures 1 and 2.
In the next (Eye)F7−172YW construct, we did not flank the Fab-7 insulator by lox sites, thereby reducing the distance between Fab-7 and the transcription start site to 172 bp (Figure 6B). Once again, the Fab-7 insulator effectively blocked the yellow and white enhancers. These results suggest that the distance between Fab-7 and the yellow promoter is not crucial for insulation.
Finally, we checked whether the distance between the enhancers and the Fab-7 insulator is crucial for enhancer blocking. In the (Eyw)(2.7)(F7)YW construct, the 2.7-kb spacer flanked by lox sites was inserted between the enhancers and Fab-7 at −893 (Figure 6C). Comparing yellow pigmentation in transgenic lines with or without Fab-7 before and after the deletion of the spacer showed that the presence of the 2.7-kb spacer decreased the activity of the yellow enhancers but had no influence on the insulating properties of Fab-7.
DISCUSSION
Previously, the minimal Fab-7 insulator was mapped with the aid of the eye enhancer–mini-white gene model (Hagstrom et al. 1996). The test fragments of the Fab-7 insulator were inserted in close proximity to the white promoter. As a result, it was found that only the 1.2-kb fragment corresponding to our Fab-71.2 (rather than its 0.86-kb subfragment) blocked the eye enhancer in ∼50% of transgenic lines. Here, we found that both 0.86- and 1.2-kb fragments insulated the enhancers with similar efficiency when placed at a distance from the white promoter. The simplest explanation of these contradictory results is that the 1.2-kb Fab-7 insulator contains the regulatory element that inhibits the activity of the white promoter, which is deleted from the smaller 0.86-kb Fab-7 element. Indeed, our preliminary data show that the 0.4-kb fragment overlapping the proximal part of the 1.2-kb Fab-7 insulator often recruits the transposon in genome regions that negatively influence white expression (O. Kyrchanova, unpublished data). Interestingly, the 1.2-kb Fab-7 insulator contains separable regions that function at different stages of development (Schweinsberg and Schedl 2004). The elements responsible for the insulating activity in embryos and adult flies (the eye enhancer–white promoter) are present in the 0.86-kb Fab-7 element. Thus, these results are in agreement with our finding that the 0.86-kb fragment contains all sequences required for blocking the adult enhancers.
As the 0.86-kb Fab-7 insulator inserted close to the white but not the yellow promoter fails to block the enhancers, we suggest that the Fab-7 insulator stimulates binding of the basic transcription factors to the relatively weak white promoter. The 0.86-kb Fab-7 insulator contains nine consensus-binding sites for the GAGA factor (Schweinsberg et al. 2004). The GAGA factor functions to antagonize the repressive effects of chromatin by promoting the formation of nucleosome-free regions over promoters and other regulatory elements (Leibovitch et al. 2002; Lehmann 2004). It seems likely that GAGA helps in the binding of proteins to the white promoter that helps to overcome the insulating activity of Fab-7. While further study is required to prove this model, it is apparent that the DNA fragment tested for the insulating activity should be placed at a distance from an enhancer and a promoter to avoid effects complicating interpretation of the results.
Recently, we found that relative orientation of the Mcp elements defines the mode of loop formation that either allows or blocks stimulation of the white promoter by the eye enhancer (Kyrchanova et al. 2007). In contrast to previous observations (Zhou et al. 1996; Majumder and Cai 2003), we demonstrated here that the Fab-7 insulators can functionally interact with each other. In contrast to Mcp, however, the relative orientation of Fab-7 does not affect communication between the eye enhancer and the white promoter across the pair of Fab-7 insulators. The Mcp insulator located downstream of the yellow gene significantly improves the enhancer-blocking activity of the insulator located between the enhancers and promoter of the yellow gene (Kyrchanova et al. 2007). It appears that the interaction between the Mcp insulators results in the formation of a loop that restricts communication between the enhancers located outside the loop and the promoters located inside it. We found that Fab-7 inserted downstream of the yellow gene did not contribute to insulation by Fab-7 located between the enhancers and the promoters. If the Fab-7 insulators interact in all tissues, their presence on both sides of a gene does not ultimately improve the blocking of enhancers located outside the chromatin loop formed due to their interaction.
The role of Fab-7 and other boundary elements in transcriptional regulation of Abd-B is as yet uncertain. According to the accepted model, the insulator/boundary element functions as a barrier separating the iab domains differing in the status of chromatin (Mihaly et al. 1998; Sipos and Gyurkovics 2005; Maeda and Karch 2006). Recently, Cleard et al. (2006) directly demonstrated the interaction between Fab-7 and the Abd-B promoter, concluding that Fab-7 and other boundary elements appear to be involved in regulating long-distance interactions between the iab enhancers and the Abd-B promoter. It is noteworthy that the interaction of Fab-7 with the promoter was effective in the tissues where Abd-B is not expressed, e.g., in the eyes. These results are in agreement with our observation that the Fab-7 insulators functionally interact in supporting the interaction between the enhancers and promoters of the white gene. An important task now is to find out whether the Fab-7 insulators are capable of interaction in all tissues and at all developmental stages.
Acknowledgments
This study was supported by the Russian Foundation for Basic Research (project no. 06-04-48360) and by the International Research Scholar Award from the Howard Hughes Medical Institute (to P.G.).
References
- Barges, S., J. Mihaly, M. Galloni, K. Hagstrom, M. Muller et al., 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127: 779–790. [DOI] [PubMed] [Google Scholar]
- Belozerov, V. E., P. Majumder, P. Shen and H. N. Cai, 2003. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 22: 3113–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko, V. A., Y. V. Liu, Y. I. Jiang and V. M. Studitsky, 2003. Communication over a large distance: Enhancers and insulators. Biochem. Cell. Biol. 81: 241–251. [DOI] [PubMed] [Google Scholar]
- Bonifer, C., M. Vidal, F. Grosveld and A. E. Sippel, 1990. Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J. 9: 2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet, A. M., A. Lloyd and S. Sakonju, 1991. Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Development 111: 393–405. [DOI] [PubMed] [Google Scholar]
- Brasset, E., and C. Vaury, 2005. Insulators are fundamental components of the eukaryotic genomes. Heredity 94: 571–576. [DOI] [PubMed] [Google Scholar]
- Burgess-Beusse, B., C. Farrell, M. Gaszner, M. Litt, V. Mutskov et al., 2002. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl. Acad. Sci. USA 99: 16433–16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, H., and M. Levine, 1995. Modulation of enhancer−promoter interactions by insulators in the Drosophila embryo. Nature 376: 533–536. [DOI] [PubMed] [Google Scholar]
- Cai, H. N., and P. Shen, 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291: 493–495. [DOI] [PubMed] [Google Scholar]
- Capelson, M., and V. G. Corces, 2004. Boundary elements and nuclear organization. Biol. Cell 96: 617–629. [DOI] [PubMed] [Google Scholar]
- Celniker, S. E., S. Sharma, D. J. Keelan and E. B. Lewis, 1990. The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 9: 4227–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard, F., Y. Moshkin, F. Karch and R. K. Maeda, 2006. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat. Genet. 38: 931–935. [DOI] [PubMed] [Google Scholar]
- Comet, I., E. Savitskaya, B. Schuettengruber, N. Negre, S. Lavrov et al., 2006. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev. Cell 11: 1–8. [DOI] [PubMed] [Google Scholar]
- Conte, C., B. Dastugue and C. Vaury, 2002. Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol. Cell. Biol. 22: 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett, D., 1999. Distant liaisons: Long range enhancer–promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 9: 505–514. [DOI] [PubMed] [Google Scholar]
- Duncan, I., 1987. The bithorax complex. Annu. Rev. Genet. 21: 285–319. [DOI] [PubMed] [Google Scholar]
- Fraser, P., 2006. Transcriptional control thrown for a loop. Curr. Opin. Genet. Dev. 16: 490–495. [DOI] [PubMed] [Google Scholar]
- Galloni, M., H. Gyurkovics, P. Schedl and F. Karch, 1993. The bluetail transposon: Evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 12: 1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner, M., and G. Felsenfeld, 2006. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7: 703–713. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., and V. G. Corces, 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1: 996–1004. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., and V. G. Corces, 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6: 1865–1873. [DOI] [PubMed] [Google Scholar]
- Golic, K. G., and S. Lindquist, 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59: 499–509. [DOI] [PubMed] [Google Scholar]
- Gruzdeva, N., O. Kyrchanova, A. Parshikov, A. Kullyev and P. Georgiev, 2005. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer–promoter communication. Mol. Cell. Biol. 25: 3682–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurkovics, H., J. Gausz, J. Kummer and F. Karch, 1990. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9: 2579–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom, K., M. Muller and P. Schedl, 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10: 3202–3215. [DOI] [PubMed] [Google Scholar]
- Holdridge, C., and D. Dorsett, 1991. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol. 11: 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, K., G. Arib, C. Lin, G. Van Houwe and U. K. Laemmli, 2002. Chromatin boundaries in budding yeast: The nuclear pore connection. Cell 109: 551–562. [DOI] [PubMed] [Google Scholar]
- Karch, F., B. Weiffenbach, M. Peifer, W. Bender, I. Duncan et al., 1985. The abdominal region of the bithorax complex. Cell 43: 81–96. [DOI] [PubMed] [Google Scholar]
- Karch, F., M. Galloni, L. Sipos, J. Gausz, H. Gyurkovics et al., 1994. Mcp and Fab-7: Molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 22: 3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess, R. E., and G. M. Rubin, 1984. Analysis of P transposable element functions in Drosophila. Cell 38: 135–146. [DOI] [PubMed] [Google Scholar]
- Kellum, R., and P. Schedl, 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950. [DOI] [PubMed] [Google Scholar]
- Kellum, R., and P. Schedl, 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12: 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, E. J., and P. K. Geyer, 2003. Genomic insulators: Connecting properties to mechanism. Curr. Opin. Cell Biol. 15: 259–265. [DOI] [PubMed] [Google Scholar]
- Kuhn, E. J., M. M. Viering, K. M. Rhodes and P. K. Geyer, 2003. A test of insulator interactions in Drosophila. EMBO J. 22: 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova, O., S. Toshchakov, A. Parshikov and P. Georgiev, 2007. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on the enhancer–promoter communication. Mol. Cell. Biol. 27: 3035–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, M., 2004. Anything else but GAGA, a nonhistone protein complex reshapes chromatin structure. Trends Genet. 20: 15–22. [DOI] [PubMed] [Google Scholar]
- Leibovitch, B. A., Q. Lu, L. R. Benjamin, Y. Liu, D. C. Gilmour et al., 2002. GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol. Cell. Biol. 22: 6148–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The genome of Drosophila melanogaster. Academic Press, New York.
- Maeda, R. K., and F. Karch, 2006. The ABC of the BX-C: the bithorax complex explained. Development 133: 1413–1422. [DOI] [PubMed] [Google Scholar]
- Majumder, P., and H. N. Cai, 2003. The functional analysis of insulator interactions in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 100: 5223–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallin, D. R., J. S. Myung, J. S. Patton and P. K. Geyer, 1998. Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics 148: 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M., Y. B. Meng and W. Chia, 1989. Regulatory elements involved in the tissue-specific expression of the yellow gene of Drosophila. Mol. Gen. Genet. 218: 118–126. [DOI] [PubMed] [Google Scholar]
- Mihaly, J., I. Hogga, J. Gausz, H. Gyurkovics and F. Karch, 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124: 1809–1820. [DOI] [PubMed] [Google Scholar]
- Mihaly, J., I. Hogga, S. Barges, M. Galloni, R. K. Mishra et al., 1998. Chromatin domain boundaries in the bithorax complex. Cell. Mol. Life Sci. 54: 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova, E., A. Golovnin, E. Gracheva, A. Parshikov, T. Belenkaya et al., 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291: 495–498. [DOI] [PubMed] [Google Scholar]
- Mutskov, V. J., C. M. Farrell, P. A. Wade, A. P. Wolffe and G. Felsenfeld, 2002. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 16: 1540–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, S., B. Varjavand and V. Pirrotta, 1992. Molecular analysis of the zeste–white interaction reveals a promoter-proximal element essential for distant enhancer–promoter communication. Genetics 131: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin, S., and P. Georgiev, 2005. Handling three regulatory elements in one transgene: Combined use of cre-lox, FLP-FRT and I-SceI recombination systems. Biotechniques 39: 871–875. [DOI] [PubMed] [Google Scholar]
- Roseman, R. R., V. Pirrotta and P. K. Geyer, 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12: 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman, R. R., E. A. Johnson, C. K. Rodesch, M. Bjerke, R. N. Nagoshi et al., 1995. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero, E., 1991. Control of the expression of the bithorax complex abdominal-A and Abdominal-B by cis-regulatory regions in Drosophila embryos. Development 111: 437–448. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero, E., I. Vernos, R. Marco and G. Morato, 1985. Genetic organization of Drosophila bithorax complex. Nature 313: 108–113. [DOI] [PubMed] [Google Scholar]
- Savitskaya, E., L. Melnikova, M. Kostuchenko, E. Kravchenko, E. Pomerantseva et al., 2006. Study of Long-Distance Functional Interactions between Su(Hw) Insulators that Can Regulate Enhancer−Promoter Communication in Drosophila melanogaster. Mol. Cell. Biol. 26: 754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg, S., and P. Schedl, 2004. Developmental modulation of Fab-7 boundary function. Development 131: 4743–4749. [DOI] [PubMed] [Google Scholar]
- Schweinsberg, S., K. Hagstrom, D. Gohl, P. Schedl, R. P. Kumar et al., 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168: 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, K. S., and P. K. Geyer, 1995. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 14: 6258–6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal, M. L., and D. L. Hartl, 2000. Application of Cre/loxP in Drosophila. Site-specific recombination and transgene co-placement. Methods Mol. Biol. 136: 487–495. [DOI] [PubMed] [Google Scholar]
- Sigrist, C. J., and V. Pirrotta, 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos, L., and H. Gyurkovics, 2005. Long-distance interactions between enhancers and promoters. The case of the Abd-B domain of the Drosophila bithorax complex. FEBS J. 272: 3253–3259. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., and G. M. Rubin, 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]
- Sun, F. L., and S. C. Elgin, 1999. Putting boundaries on silence. Cell 99: 459–462. [DOI] [PubMed] [Google Scholar]
- Valenzuela, L., and R. T. Kamakaka, 2006. Chromatin insulators. Annu. Rev. Genet. 40: 107–138. [DOI] [PubMed] [Google Scholar]
- West, A. G., and P. Fraser, 2005. Remote control of gene transcription. Hum. Mol. Genet. 14: R101–R111. [DOI] [PubMed] [Google Scholar]
- Zhou, J., and M. Levine, 1999. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell 99: 567–575. [DOI] [PubMed] [Google Scholar]
- Zhou, J., S. Barolo, P. Szymanski and M. Levine, 1996. The Fab-7 element of the bithorax complex attenuates enhancer–promoter interactions in the Drosophila embryo. Genes Dev. 10: 3195–3201. [DOI] [PubMed] [Google Scholar]
- Zhou, J., H. Ashe, C. Burks and M. Levine, 1999. Characterization of the transvection mediating region of the Abdominal-B locus in Drosophila. Development 126: 3057–3065. [DOI] [PubMed] [Google Scholar]