Abstract
Female infertility syndromes are among the most prevalent chronic health disorders in women, but their genetic basis remains unknown because of uncertainty regarding the number and identity of ovarian factors controlling the assembly, preservation, and maturation of ovarian follicles. To systematically discover ovarian fertility genes en masse, we employed a mouse model (Foxo3) in which follicles are assembled normally but then undergo synchronous activation. We developed a microarray-based approach for the systematic discovery of tissue-specific genes and, by applying it to Foxo3 ovaries and other samples, defined a surprisingly large set of ovarian factors (n = 348, ∼1% of the mouse genome). This set included the vast majority of known ovarian factors, 44% of which when mutated produce female sterility phenotypes, but most were novel. Comparative profiling of other tissues, including microdissected oocytes and somatic cells, revealed distinct gene classes and provided new insights into oogenesis and ovarian function, demonstrating the utility of our approach for tissue-specific gene discovery. This study will thus facilitate comprehensive analyses of follicle development, ovarian function, and female infertility.
DISORDERS of female infertility and early menopause due to depletion of ovarian follicles, such as premature ovarian failure, are among the most common chronic medical conditions affecting women. The incidence of female infertility in the United States is ∼13%, and ovarian dysfunction is the most common underlying cause (Kumar et al. 2007). Premature ovarian failure (menopause prior to the age of 40) has multiple systemic consequences due to sex steroid deficiency (such as osteoporosis) and affects 1% of women (Kalantaridou and Nelson 2000). Substantial progress has recently been made in studies of male infertility, as exemplified by the discovery of Y chromosome microdeletions resulting in azoospermia (Pryor et al. 1997), but common causes of female infertility syndromes remain elusive. Although rare metabolic and genetic causes of female infertility are known, such as galactosemia (Kaufman et al. 1979) or mutations in the follicle stimulating hormone receptor (Aittomaki et al. 1995), the etiologies of female infertility syndromes including primary amenorrhea, premature ovarian failure, and polycystic ovarian disease are largely unknown. These disorders, however, are believed to have an ovarian basis with a major hereditary component (Kumar et al. 2007).
Female infertility likely results from defects at various stages of follicle assembly and development. In mammals, the postnatal period (birth to 2 weeks in the mouse) is an active phase of ovarian development when primordial follicles (PFs) are formed and the first wave of follicle activation begins at around postnatal day (PND) 3. Individual PFs remain quiescent until they resume growth via a poorly understood process known as PF activation (PFA). PFA begins immediately after follicle assembly is complete at PND3 and continues until follicle depletion at menopause. Because all activated follicles ultimately undergo ovulation or atresia, PFA represents an irreversible commitment to follicle growth (Mcgee and Hsueh 2000). Thus, in the normal premenopausal ovary, the vast majority of follicles are primordial and quiescent (Figure 1C), and the small percentage of follicles that are actively growing do so in an asynchronous manner. Furthermore, large preovulatory follicles comprise a disproportionate share of the ovarian mass. These aspects of ovarian physiology have presented major hurdles to the identification and study of factors that function during early follicle growth.
Figure 1.—
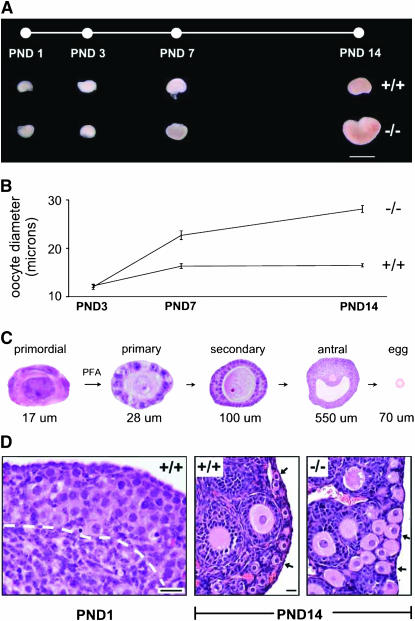
Experimental approach and timepoints selected for expression profiling. PND1 and PND3 precede the earliest morphologic manifestations of the Foxo3 phenotype, whereas PND7–14 allow for accumulation of transcripts induced during early follicle growth. (A) Foxo3 +/+ and −/− ovaries at PND1–14. Bar, 2 mm. (B) Oocyte diameters at PND3–14 in Foxo3 +/+ and −/− ovaries. At PND3, just after follicle individualization is complete, oocyte diameter is unchanged, but by PND7, oocyte diameters are significantly increased. Measurements made on H&E-stained sections; error bars, SEM. (C) Schematic of follicle maturation. PFs are small and quiescent, with a single layer of flattened granulosa cells. Primary follicles have initiated growth and have increased oocyte diameter and a single layer of granulosa cells, whereas secondary follicles have two layers. (D) Ovarian histology at PND1 and -14. At PND1, Foxo3 +/+ ovaries contain large numbers of germ cells (oogonia) (−/− ovaries are indistinguishable, not shown). White dashed line demarcates boundary between oogonial clusters and ovarian stroma. Bottom left corner of image corresponds to approximate center of ovary. By PND14, global PFA is evidenced in Foxo3 −/− ovaries by increased oocyte diameter (approximately two times normal). Arrows point to PFs (+/+) and early growing follicles (−/−). Bar, 20 μm; two right panels at same magnification.
The forkhead transcription factor Foxo3 is a master regulator of PFA (Castrillon et al. 2003; Hosaka et al. 2004). PF assembly is normal in female mice bearing a null Foxo3 mutation (John et al. 2007). Immediately thereafter, however, PFs undergo global activation and grow in an essentially synchronized manner, resulting in ovarian hyperplasia by PND14. Of particular relevance for this study, Foxo3 females are fertile until follicles are depleted at ∼4 months of age, demonstrating that subsequent steps of follicle maturation (ovulation, fertilization, zygotic development, etc.) are unaffected (Castrillon et al. 2003).
Gene discovery efforts with purified oocytes obtained following superovulation (Rajkovic et al. 2001), for example, have led to the discovery of essential oocyte-specific factors such as Gdf9, Bmp15, and Factor in the germline α (Fig1α) (Dong et al. 1996; Dube et al. 1998; Soyal et al. 2000). However, oocyte genes required for early oocyte development and growth are not necessarily expressed in mature eggs, given the prolonged time interval required to complete follicle maturation—at least 3–5 weeks in mice (Hirshfield 1991) and 280 days in women (Gougeon 1986). Furthermore, relatively little is known about factors unique to the somatic cells of the ovary (granulosa cells and stroma) that play specialized roles in ovarian development, particularly in the early stages of follicle growth. Other studies have documented temporal gene expression changes in embryonic gonads (Small et al. 2005), neonatal to adult ovaries (Herrera et al. 2005), primordial to secondary follicles (Yoon et al. 2006), and the egg to embryo transition (Evsikov et al. 2006), providing a wealth of information pertaining to global changes in gene expression during various phases of ovarian development and oogenesis. However, these studies were not designed to systematically discover individual genes specifically expressed in the ovary.
We hypothesized that many ovarian fertility genes remain unidentified, limiting efforts to understand ovarian function and define the hereditary factors that influence female fertility. To identify such genes in a high-throughput manner, we developed a microarray approach generally applicable toward the discovery of genes with tissue-specific expression. Through this approach and by profiling Foxo3 ovaries (which should be enriched for genes induced during early follicle growth) and other samples, we obtained the most comprehensive view of ovarian genes to date, yielding several new insights into ovarian function and gene expression. Furthermore, these genes represent an inclusive set of functional and genetic entry points into studies of female infertility and ovarian function in mammals.
MATERIALS AND METHODS
Sample procurement and RNA amplification:
Total RNA was prepared from single pairs of ovaries at PND1, -3, -7, and -14 using 1 ml of Tripure reagent (Roche) and 2 μl (20 mg/ml) glycogen (Invitrogen, Carlsbad, CA) and then subjected to two rounds of RNA amplification using the Eberwine method of amplification (Phillips and Eberwine 1996). Heterozygous Foxo3 mice backcrossed to FVB mice for six generations (Castrillon et al. 2003) were interbred to generate matched −/− and +/+ sibling pairs; animal use was approved by an institutional committee. Adult mouse tissues were dissected from six week FVB mice and labeled per manufacturer's specifications without amplification. KitlSl/KitlSl-d male mice were purchased from the Jackson Laboratories. The embryonic stem (ES) cell data sets were downloaded from the Gene Expression Omnibus (GEO) repository (unamplified samples 1–8; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4308) (Kurimoto et al. 2006). For the first round of amplification, RNA was resuspended in 11 μl RNAse-free H2O and combined with 1 μg oligodT24-T7 primer (Proligo), denatured at 70° for 10 min then chilled on ice. The remaining synthesis reagents [4 μl first-strand buffer, 2 μl 0.1 m DTT, 1 μl 10 mm dNTPs, 1 μl Superscript II (Invitrogen)] were added and incubated at 42° for 1 hr. Second-strand synthesis was performed by adding 91 μl H2O, 30 μl second-strand buffer, 3 μl 10 mm dNTPs, 1 μl Escherichia coli ligase, 4 μl DNA polymerase, 1 μl RNaseH (Invitrogen), and incubated at 16° for 2 hr. The samples were blunt ended by addition of 2 μl of T4 DNA polymerase (Invitrogen) and incubation at 16° for 15 min. Ethanol precipitation was performed (70 μl 5 m NH4OAc, 650 μl chilled 100% ethanol, 2 μl glycogen) and the sample was resuspended in 8 μl of RNase-free water. In vitro transcription was performed with the MEGAscript high-yield transcription kit (Ambion) and incubated at 37° for 4 hr. Samples were purified using RNeasy mini kit (QIAGEN), eluted in 50 μl H2O, quantitated by OD260, and 400 ng cRNA in 12 μl H2O was subjected to a second round of amplification.
For the second round of amplification, the samples were combined with 1 μg random hexamers (Invitrogen), mixed and denatured at 70° for 10 min, and then chilled on ice. The remaining first-strand reagents were combined and incubated as above. Once the first-strand synthesis was complete, the reaction was incubated with 1 μl RNaseH at 37° for 20 min then heat inactivated at 95° for 5 min. OligodT24-T7 primer was added to samples and incubated at 70° for 10 min then chilled on ice. Second-strand synthesis was completed by adding 91 μl H2O, 30 μl second-strand buffer, 3 μl dNTPs, and 4 μl DNA polymerase and incubated for 2 hr at 16°. Blunt ending was performed as above. Sample was ethanol precipitated and resuspended in 22 μl H2O. In vitro transcription was performed this time using BioArray RNA transcription labeling kit (Enzo) per the manufacturer's recommendation and incubated at 37° for 4 hr. The cRNA was again purified with the RNeasy kit and concentration was measured using 5 μl.
Laser capture microdissection:
Ovaries from FVB mice (3 weeks of age) were sectioned onto neutral glass slides and lightly counterstained using the HistoGene LCM Frozen Section kit (Arcturus). Laser capture microdissection (LCM; Arcturus PixCell IIe) was performed on primary and secondary oocytes and somatic cells to isolate the two cell populations. RNA was purified using the PicoPure RNA isolation kit (Arcturus) and then subjected to two rounds of amplification as described above. Unfertilized egg and cumulus complexes were collected from FVB females (3 weeks of age) by first injecting with pregnant mare serum (5 IU/mouse) followed by a single dose 42 hr later of human chorionic gonadotropin (5 IU/mouse). Complexes were collected 12 hr later, washed with PBS, and then digested for 5 min with hyaluronidase (Type IV-S, Sigma) and HEPES solution. Eggs were removed from the cumulus cells and placed into 1× HEPES; cumulus cells were collected and placed into a separate tube. Microscopic inspection confirmed the purity of both preparations and the absence of any cross-contaminating cells. These cell populations were washed two times with 1× HEPES, spun down, and layered with 1 ml of Tripure.
Affymetrix microarray hybridization:
Microarray hybridization was performed per the manufacturer's specifications with 15 μg of labeled cRNA. Hybridization, washing, and scanning of GeneChip Mouse Genome 430 2.0 arrays was performed by the University of Texas Southwestern DNA Microarray Core Facility. Analysis was performed using Affymetrix GeneChip Operating Software v1.4.0.036 and GeneSpring GX 7.3. Scaling was set to 250, normalization to 1, and probability calls to 0.5. Signal strengths were calculated on the basis of the unadjusted data's mean. Signals for PND1, -3, -7, and -14 ovaries were averaged since these were performed in triplicate. Quality control analysis of all microarray data sets indicated an average percentage present call of 42.68% (± 4.6 SD). Acceptable 3′:5′ ratios were obtained for unamplified samples in the range of 1 and found to be slightly higher (4–5) for all the amplified samples. The computed signal strengths from each array were imported into Excel. An average somatic signal strength was calculated for each probe set from all adult tissues except for reproductive organs and adrenal. This average somatic signal was then compared to the corresponding signal strengths from the neonatal ovaries, unfertilized eggs, etc. Probe sets that had a signal >100 and were ≥20-fold higher than the average somatic signal were tabulated for further analysis. Probe sets were annotated using NetAffx, Ensembl (v42), and Entrez Gene ID. Graphs were generated in Excel by importing signal strengths from each array for a particular probe set and calculating error (SEM) where applicable.
Data access:
All array data sets were deposited into the GEO, a curated gene expression repository supporting MIAME compliant data submissions. GEO can be accessed through the National Center for Biotechnology Information web page (http://www.ncbi.nlm.nih.gov/geo/). Our project, including 24 arrays for the PD1–PD14 timepoints plus 24 additional gonadal, normal adult tissues, and other related data sets, can be accessed through project number GSE8249.
RNA in situ hybridization:
cDNA plasmid clones with suitable T7, T3, or SP6 sites were purchased from Open Biosystems and confirmed by end sequencing. Plasmid DNA was linearized with restriction enzymes and RNA polymerase reactions were performed with 1 μg of plasmid DNA using the Maxiscript transcription kit (Ambion) and DIG RNA labeling mix (Roche). RNA probes were purified with NucAway spin column (Ambion). Ovaries were frozen at −80° in OCT compound (Tissue-Tek) and 16-μm sections were placed on SuperFrost Plus slides (Fisher). Slides were air dried for 30 min and then immediately fixed in 4% paraformaldehyde/PBS (Electron Microscopy Sciences) for 30 min. Slides were washed in 1× DEPC-treated PBS for 15 min. Sections were treated with proteinase K/PBS (20 μg/ml; Invitrogen, Carlsbad, CA) for 15 min and washed in 1× PBS. Slides were then treated with 0.1% active DEPC-PBS for 15 min two times and hybridized with 40 ng DIG-labeled probe per 100 μl hybridization buffer (50% formamide, 5× SSC, 40 μg/ml salmon sperm DNA. A total of 300 μl of hybridization solution was used per slide and covered with Hybrislips (Research Products International). Slides were incubated at 58° in a humid chamber containing 50% formamide and 5× SSC for 24 hr. Cover slips were removed in 2× SSC; slides were rinsed in 2× SSC then washed in 0.1× SSC at 65° for 1 hr each. Slides were equilibrated in buffer 1 (100 mm Tris, pH 7.5, 150 mm NaCl) for 5 min at room temperature; subsequent washes were at room temperature. Anti-DIG antibody (AP-conjugated) was diluted 1:5000 in buffer 1 plus 0.5% blocking reagent (Roche). The slides were blocked for 2 hr, rinsed two times for 15 min in buffer 1 and equilibrated in buffer 2 (100 mm Tris, pH 9.5, 100 mm NaCl, 50 mm MgCl2 for 5 min). BM purple chromogenic substrate (Roche) was generously applied to the slides and incubated 3–48 hr. Slides were then washed with water and mounted with Crystalmount (Electron Microscopy Sciences) without air drying.
RT–PCR:
RNA was isolated using Tripure reagent and resuspended in 11 μl H2O. Samples were mixed with 1 μg random hexamers, heat denatured at 70° for 10 min, and then chilled on ice. cDNA was synthesized using the Superscript II enzyme in 20 μl at 42° for 1 hr and then treated with 1 μl RNaseH and incubated at 42° for another 20 min. Samples were first normalized to relative amounts using Gapdh. All PCRs were performed using HotStart Taq (Perkin Elmer) for 28 cycles. PCR conditions and primer sequences are available upon request.
Northern hybridization:
RNA isolation was performed using Tripure reagent and resuspended in RNAse-free H2O. Five micrograms total RNA was loaded onto 2.2 m formaldehyde gels. Transfer was completed overnight in 20× SSC to Hybond-N+ (Amersham, Piscataway, NJ). Radioactive probes were prepared using cDNA inserts purified from human clones purchased from Open Biosystems (clone identifiers available upon request) with the Radprime DNA labeling system (Invitrogen). Overnight hybridizations were performed in Church-Gilbert hybridization buffer (0.5 m sodium phosphate, pH 7.0, 1% BSA, 7% SDS, and 1 mm EDTA) with 1 × 106 counts/ml at 65° and exposed to BioMax MS film (Kodak, Rochester, NY). Human fetal samples were obtained following Institutional Review Board approval.
RESULTS
Systematic identification of candidate ovarian fertility genes:
We selected four timepoints (PND1, -3, -7, and -14) spanning follicle assembly and early growth. Foxo3−/− ovaries are enlarged by PND14 due to global PFA beginning after PND3, leading to greatly increased numbers of growing follicles (Castrillon et al. 2003) (Figure 1, A–C). Neonatal ovaries, unlike adult ovaries, contain a high proportion of germ cells (∼30% at PND1–3) (Figure 1D) and thus should permit the detection of genes expressed in germline and somatic compartments. Total RNA from +/+ and −/− ovaries (n = 3 replicates per timepoint and genotype, a total of 24 microarrays) was subjected to linear RNA amplification and hybridized to Affymetrix 430 2.0 mouse whole-genome microarrays, which interrogate >39,000 transcripts including the vast majority of protein-coding genes. We also profiled 14 somatic tissues and, to provide more refined views of gene expression, adult ovaries, adult testes, KitlSl/KitlSl-d testes (devoid of germ cells except for rare spermatogonia) (Shinohara et al. 2000), ES cells, LCM primary oocytes, LCM somatic cells (granulosa and stromal cells), superovulated unfertilized eggs, cumulus granulosa (CG) cells, and E11 Foxo3 +/+ and −/− embryos. Each array data set was independently normalized by global median scaling, with signal strengths averaged for those samples with replicates (PND1–14).
For each array probe set, the signal ratio at each ovarian timepoint and genotype relative to the average of the 14 somatic tissues was calculated. A cutoff ratio of 20× for any one of the PND1–14 sample sets (Foxo3 +/+ or −/−) was used to select putative ovarian fertility genes highly expressed in the ovary relative to other tissues—and thus likely to serve discrete ovarian functions. This ratio was chosen because, in gene set enrichment analyses, it yielded the most significant P-values for gene ontology (GO) terms relating to reproduction and ovarian function (below and data not shown). These criteria were stringent but permitted the identification of ovarian genes expressed to a limited extent in other tissues, such as adrenal. The data were internally consistent, with similar patterns of expression in +/+ and −/− ovaries and many genes induced during early follicle growth were more highly expressed in −/− ovaries, as expected. Multiple probe sets corresponding to the same targets gave virtually identical results (e.g., no. 47 Dazl) (supplemental Figure 1 at http://www.genetics.org/supplemental/). This analysis of PD1–14 ovaries led to the identification of 208 distinct ovarian genes (supplemental Table 1).
Profiling wild-type and Foxo3 null ovaries identified ovarian factors likely to participate in early follicle development but might have missed genes uniquely induced in late antral follicles or corpora lutea, structures abundant in sexually mature ovaries but not present by PD14. To identify such genes, we profiled wild-type adult ovaries and employed the same selection criteria as for PD1–PD14, leading to the identification of an additional 22 genes. Finally, we also sought to identify genes expressed in mature eggs and CG cells, which may be required for fertilization, support of the mature egg, or as maternal factors needed during early embryonic development. To complete our efforts to generate an inclusive list of ovarian genes, we compared the expression profiles of superovulated eggs and CG cells again with the panel of somatic tissues and identified an additional 110 egg and 8 CG genes (supplemental Table 1 at http://www.genetics.org/supplemental/).
Success in the identification of known and novel ovarian genes:
The vast majority of previously identified ovarian genes were reidentified, including Zp1–3, Bmp15, Gdf9, Mos, Vasa, Inhibin-α/β, Wt1, Fig1α, Obox1, Oosp1, Stella/Dppa3, Dazl, Dax1, FK binding protein 6 (Fkbp6), LIM homeobox protein 8 (Lhx8), and Amh, among many others (supplemental Table 1). Of the total set of 348 genes, ∼218 (∼63%) were not previously documented as ovarian factors, as determined by extensive literature review and cross-checking with online databases (Pubmed, Entrez, MGI). Some of these factors have received little previous scrutiny, identified only as ESTs or by gene prediction algorithms, while others (such as the monoamine transporter VMAT2/Slc18a2) have been researched extensively in other physiologic contexts but not appreciated to also be highly expressed in the ovary (Uhl et al. 2000). Still others have not been recognized as ovarian specific per se. A substantial number have been described as testis specific (supplemental Table 1). That so many well-studied genes were overlooked as ovarian factors we attribute to the fact that ovarian gene expression is often evaluated only in adult ovaries, where many of these genes are not highly expressed or readily detectable (e.g., oocytes comprise <1% of cells in the adult ovary). Our approach was thus reliable and led to the identification of a large number of ovarian genes. Furthermore, the set presented here is expected to represent the great majority of ovarian genes, at least as defined by our stringent criteria.
Classification of ovarian factors:
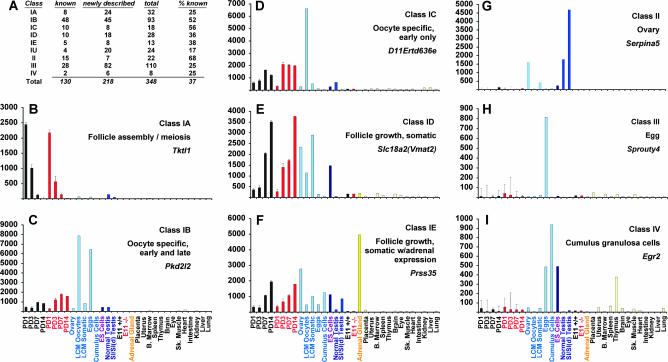
Graphs were generated for the 348 genes to provide detailed views of their relative expression across all samples (supplemental Figure 1 at http://www.genetics.org/supplemental/). These “digital Northerns” revealed distinctive, recurring spatiotemporal patterns of expression, permitting grouping of the PD1–14 ovarian genes (termed class I) into distinct subclasses (IA–IE) (Figure 2). As discussed below, the functions of known ovarian genes in each of these subclasses (Figure 2A) argue that the subclasses are biologically meaningful. In the following sections, we provide representative examples of genes in each class I subclass, taking the opportunity to present interpretations of the expression data. This is followed by descriptions of the genes specifically expressed in the adult ovary (class II), eggs (class III), and CG cells (class IV).
Figure 2.—
Systematic classification of ovarian factors. (A) Total ovarian genes identified in this study and percent documented in previous studies (B–I). Graphs show relative expression levels of single probe set for indicated gene across multiple samples; error bars represent SEM. Samples are (left to right) Foxo3 +/+ PND1, -3, -7, -14 (black), Foxo3 −/− PND1, -3, -7, -14 (red), adult ovary, LCM primary and secondary oocytes, LCM somatic cells (primary plus secondary granulosa cells and surrounding stroma), eggs, cumulus cells, ES cells, normal adult testis, Sl/Sld (germ cell-depleted) adult testis, Foxo3 +/+ E11 embryos, Foxo3 −/− E11 embryos, adrenal gland, placenta, uterus, bone marrow, spleen, thymus, brain, eye, skeletal muscle, heart, intestine, kidney, liver, and lung.
Class IA—follicle assembly/meiosis:
This class of ovarian genes is distinguished by high expression at PD1 followed by a rapid decline, implying a specific functional requirement in those processes (meiotic entry, follicle assembly) taking place at PD1 or before. As a representative example, Transketolase1 (Tktl1) is highly expressed at PD1 but rapidly declines by PD14, a pattern seen in both wild-type and Foxo3−/− ovaries. Expression is virtually undetectable in adult ovaries and all somatic tissues, although there is low but above-background expression in testis (Figure 2B). Higher signal in the LCM oocyte vs. somatic samples implies that Tktl1 is specifically expressed in oocytes. Thirty-two genes were assigned to this class; of these, we could find documentation for only eight as ovarian specific (Figure 2A). Of these eight genes, several are canonical meiotic regulators, including Fkbp6, Synaptonemal complex protein 3 (Sycp3), and synaptonemal complex central element protein 1 (Syce1), while others are well-known regulators of PF assembly and early survival—Fig1α, Lhx8, Lhx9, and Spermatogenesis and oogenesis specific basic helix-loop-helix 1 (Sohlh1) (Birk et al. 2000; Soyal et al. 2000; Yuan et al. 2000; Crackower et al. 2003; Costa et al. 2005; Pangas et al. 2006). Finally, piwi like homolog 2 (piwil2) was recently shown to bind a novel class of microRNAs (piRNAs) unique to the germline (Kim 2006). By extension, the 24 novel members of this class are also likely to serve as yet unknown roles in meiotic entry, follicle assembly, and related biological processes taking place in the ovary at and prior to PND1.
Class IB—oocyte-specific, early, and late maturation:
These genes are induced during early follicle growth and are specifically expressed in early growing oocytes and also in mature eggs. They exhibit a characteristic pattern of increasing expression from PD1 to PD14, with relative expression levels often significantly higher in Foxo3−/− ovaries relative to wild type, consistent with increased follicle activation and early oocyte growth in mutant ovaries.
For example, Polycystic kidney disease 2-like 2 (Pkd2l2) is specifically expressed in PD1–PD14 ovaries, with very low levels of expression in all somatic tissues and low (albeit significant) expression in adult ovaries (Figure 2C). The low level of signal in adult ovary relative to perinatal ovaries is consistent with oocyte-specific expression, since oocytes comprise a small proportion of cells in the adult ovary. Indeed, the LCM samples show much higher signal in oocytes relative to somatic cells, demonstrating that this gene is specifically expressed in oocytes. Although the LCM samples tend to have higher background likely due to the additional round of RNA amplification, they permit qualitative determinations as to germline vs. somatic expression. This was the largest class I subclass with 93 members. Known factors include members of oocyte-specific multicopy gene clusters such as the NACHTs, oogenesins, oocyte maturation α and β, the Fbox and WD40 domain cluster (Paillisson et al. 2005), as well as essential oocyte maturation factors such as Gdf9, Bmp15, Mos, and Dazl (Ruggiu et al. 1997). Members of this class are thus likely to function either during early or late follicle growth, or perhaps at multiple stages of follicular development. For example, Gdf9 is essential for the primordial to primary follicle transition but also functions during later stages of follicle maturation, whereas Mos is first needed in mature, unfertilized mouse eggs for their maintenance in meiosis II arrest (Colledge et al. 1994; Dong et al. 1996).
Class IC—oocyte-specific, early maturation only:
These ovarian genes are specifically expressed in oocytes and induced during early growth but, unlike the previous class, are defined by the absence of expression in mature eggs (Figure 2D). Since the maturation of an oocyte from the primordial stage to a fully mature egg is a complex, prolonged process requiring at least 3–5 weeks in the mouse (Hirshfield 1991), this previously undescribed class of oocyte gene likely serves essential roles specifically during early follicle growth. Nonetheless, members of this class can also function later in follicle maturation, e.g., in eggs, given that (1) mechanisms for translational control temporally uncouple mRNA synthesis from protein translation in oocytes, and (2) some proteins synthesized early may be stable and persist until ovulation (Seydoux 1996). This is exemplified by the zona pellucida genes Zp1–3, all of which are members of this class. Mouse knockouts for the Zp genes exhibit early defects in follicle growth, consistent with the need for oocyte extracellular matrix (ECM) reorganization during early follicle growth, but the zona pellucida is also an essential component of the mature egg and required for fertilization (Rankin et al. 1996).
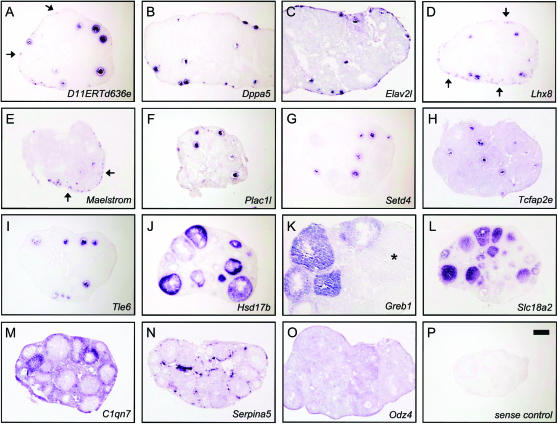
Of the 18 IC genes, 7 have been previously documented as ovarian genes. Among these are Nobox and Igf2bp2. Nobox encodes a homeobox protein that regulates the transcription of genes required for PF survival and early follicle growth (Rajkovic et al. 2004), whereas Igf2bp2 is an RNA-binding protein that represses Igf2 mRNA translation. Igf2bp2 is expressed in the ovary, although its ovarian function remains poorly understood (Hammer et al. 2005). Among the novel ovarian factors in this class is Maelstrom, the mouse homolog of the Drosophila gene maelstrom required for oogenesis (Clegg et al. 1997). maelstrom is expressed in the early Drosophila female germline, where the protein is a component of nuage. Maelstrom is also a component of nuage in mouse spermatogenesis (Costa et al. 2006); however, its expression in the female germline had not been noted prior to this study. RNA in situ hybridization (RISH) shows that expression of Maelstrom is high in primordial oocytes but rapidly disappears at the initiation of follicle growth (Figure 4E), in agreement with its classification, and suggesting a possible role in PF maintenance. Other notable mammalian ovarian factors with Drosophila homologs are presented below.
Figure 4.—
Patterns of expression for ovarian factors closely correspond to in silico analysis. Mouse ovaries at 3–6 weeks of age were embedded, cryosectioned, and subjected to RISH with digoxigenin-labeled antisense probes. (A–I) Genes predicted to be expressed in oocytes (J–O) and genes predicted to be expressed in somatic compartments. A brief description of follicle stages where expression was detected is provided. (A) D11ERTd636e, primordial to preantral; (B) Dppa5, primordial to preantral; (C) Elav2l, primary to antral; (D) Lhx8, primordial to antral; (E) Maelstrom, primordial, weak in primary and secondary, undetectable in subsequent stages; (F) Plac1l, primary to antral; (G) Setd4, primary to antral; (H) Tcfap2e, primary to antral; (I) Tle6, primary to preantral; (J) Hsd17b, primary to antral; (K) Greb1, strongest in preantral and antral, absent in corpora lutea (*); (L) Slc18a2, primary to antral; (M) C1qn7, thecal and stromal cells; (N) Serpina5, interfollicular stroma; (O) Odz4, broad expression in all somatic compartments; (P) sense negative control. Bar, 0.2 mm; A–P at same magnification.
Class ID—follicle growth, somatic:
These genes are induced during follicle growth but are expressed predominantly in a somatic compartment. Most are expressed in granulosa cells, with some expressed in the stroma (supplemental Table 1 at http://www.genetics.org/supplemental/; see also Figure 4, J–O). A representative member of this class is Slc18a2 (Vmat2), which is strongly induced from PD1–PD14 and is abundantly expressed in the adult ovary but is expressed at relatively low levels in other tissues (Figure 2E). Slc18a2 serves to transport cytosolic monoamines into synaptic vesicles and has been extensively studied since its discovery in 1993 for its role in neurotransmitter release (Liu et al. 1992), with over 250 published studies. However, we could find no previous report of its ovarian expression. Further studies will be required to assess its ovarian function, but the high level of expression of Slc18a2 in the developing and adult ovary, as well as its specific pattern of expression in granulosa cells (Figure 4L), raise the possibility of an unexpected role for monoamine vesicular transport in ovarian follicle maturation.
Class IE—follicle growth, somatic, with high adrenal expression:
A small number (n = 13) of somatic follicle growth genes were distinguished by high adrenal expression. Given that steroidogenesis is the central physiologic function shared by the gonads and adrenal, this pattern of expression strongly suggests a role in steroidogenesis. Consistent with this hypothesis, four of the five members of this family are known ovarian factors with established roles in sterol sensing and/or regulation (Cyp11a1, Hsd3b1, inhibin α, and Dax1) and have been shown to be expressed in granulosa/theca cells (supplemental Table 1 at http://www.genetics.org/supplemental/). We also note that per our profiling data (supplemental Figure 1), 10 of the 13 genes are also expressed in the testis, all predominantly somatic. The other previously identified ovarian factor in this class was Prss35, which encodes an extracellular protease recently shown to be expressed in the theca and granulosa cells of the ovary (Miyakoshi et al. 2006). We show here that among somatic tissues, Prss35 is also highly expressed only in the adrenal (Figure 2F). Another member of this class, Greb1, was not known as a predominantly ovarian factor but was discovered on the basis of its strong induction by estrogen in mammary epithelium (Ghosh et al. 2000) and is also induced by androgens in the prostate (Rae et al. 2006). The remaining seven genes in this class are rather poorly characterized ESTs, although at least two encode proteins with known domains (supplemental Table 1).
Class IU—early follicle growth not further classified:
The remaining 24 class I genes were not readily subclassified by the above criteria; many appear to be strongly expressed in both germline and somatic compartments (supplemental Table 1).
Class II—adult ovary:
This class had the highest proportion of previously known genes, consistent with the fact that late follicle maturation and ovulation have been more comprehensively studied than early follicle development (Figure 2A). The proteinase inhibitor Serpina5, known to regulate the degradation of ECM components in the testis (Odet et al. 2006) is shown as a representative example; it is highly expressed in the ovary but not in other adult tissues. LCM samples show it is expressed in somatic cells but not oocytes, and it is also expressed in testis, here again in the soma (Figure 2G and 4N). All 16 of the previously described adult ovarian factors are known to be specifically expressed in granulosa cells, corpora lutea, or thecal cells; none are oocyte specific (supplemental Table 1 at http://www.genetics.org/supplemental/). Consistent with their expression in late follicles or corpora lutea, many play fundamental roles in steroid metabolism, including Akr1c18 (involved in progesterone catabolism) (Vergnes et al. 2003), Cyp11a1 (Hu et al. 2002), Cyp17a1 (Gray et al. 1996), LH receptor (Mcfarland et al. 1989), prostaglandin F receptor (Sugatani et al. 1996), prolactin receptor (Clarke and Linzer 1993), steroidogenic acute regulatory protein (Star) (Clark et al. 1995), and endothelial lipase (Ishida et al. 2003; Ma et al. 2003). Other previously described factors include anti-müllerian hormone (Durlinger et al. 1999) and secreted frizzled-related sequence protein 4 (Hsieh et al. 2005).
Class III—eggs:
We identified a large number (n = 110) of factors specifically expressed in eggs, the majority of which have not been documented as ovarian or egg-specific factors (Figure 2A). As an example of this class, Sprouty4, one of the four mammalian homologs of Drosophila Sprouty (see below) that dampen receptor tyrosine kinase signaling, is expressed in eggs but not in any other adult tissue we examined (Figure 2H). These genes may be required for fertilization or serve as maternal factors that sustain early embryonic development.
Class IV—CG cells:
Last, we identified eight genes highly expressed in the CG cells that remain attached to the egg following ovulation. Early growth response 2 (Egr2) is highly expressed in CGs, but transcripts are also abundant in eggs (Figure 2I). Remarkably, transcripts for all eight genes are also abundant in eggs, albeit at lower levels than in CGs (supplemental Figure 1 at http://www.genetics.org/supplemental/, graphs 341–348). This does not appear to be due to cross-contamination, as the CG and egg preparations (assessed by microscopy) revealed no contaminating cells in either preparation, and many profiles (e.g., Sprouty4; Figure 2H) do not show evidence of such cross contamination. Although we cannot exclude the possibility that these genes are all simultaneously co-expressed in both egg and GC cells, it is perhaps more likely that the genes are expressed in only one compartment but that the mRNAs are transported via the rich network of gap junctions that functionally couple both the CGs and the egg (Kidder and Mhawi 2002). If so, the expression patterns of this gene class raises the possibility that such transport may be a surprisingly general phenomenon.
Validation of ovarian factors by RT–PCR and RISH:
In the above studies, we identified a total of 348 ovarian factors, the majority of them novel. To validate our microarray-based gene discovery methodology and to further document the ovarian expression of the factors identified, RT–PCR was performed on PD1–14 ovaries and a panel of 12 adult tissues including testis and ovary. The expression patterns of 22 representative genes closely matched the global expression profiling and digital Northern analysis, showing strong expression in PD1–14. The majority were also expressed in adult gonads but were undetectable in most somatic tissues (Figure 3). A few previously known ovarian genes, such as Fkpb6, inhibin β, Lhx8, and Nrf2, were included in this analysis for comparative purposes; RT–PCR results for these genes closely match their previously documented expression patterns.
Figure 3.—
Validation of ovarian gene expression (n = 22). cDNA was prepared for the tissues shown and subjected to RT–PCR using intron-spanning primers and reactions loaded onto ethidium-bromide stained agarose gels. The first four samples on the left correspond to wild-type postnatal ovary at day 1, 3, 7, and 14; remaining tissues are from adult mice. Bottom, positive control for housekeeping gene GAPDH.
Next, we performed RISH on ovarian tissue sections. Although a minority of probes failed to give detectable signal (data not shown), most probes revealed discrete patterns of expression in either oocytes or soma, closely matching our in silico analysis. Representative examples of genes predicted to be expressed in oocytes are shown in Figure 4, A–I; RISH for these genes revealed distinct temporal patterns of expression in primordial and maturing follicles. For example, Maelstrom (class IC: oocyte specific, early only) is readily detectable in primordial oocytes but rapidly disappears following PFA (Figure 4E), whereas the similarly classified D11ERTd636e is expressed in primordial oocytes and also in early growing (primary and secondary) follicles (Figure 4A). In contrast, class IB genes were not detectable in PFs but were strongly induced in more advanced stages following PFA (e.g., Elav2l, Tcfap2E, Tie6; Figure 4, C, H, and I). RISH also confirmed predicted ovarian somatic genes, revealing diverse expression patterns within distinct ovarian somatic compartments. Hsd17b, Greb1, and Slc18a2 were strongly induced in granulosa cells in growing follicles (Figure 4, J–L), whereas C1qn7 was expressed in theca and stromal cells (Figure 4M). Serpina5 was expressed in the interfollicular stromal cells (Figure 4N), and Odz4 was broadly expressed in all somatic compartments of the ovary (Figure 4O). Taken together, RT–PCR and RISH validated our in silico approach to systematically identify ovarian factors, provide detailed views of their expression by digital Northern analysis, and classify them on the basis of spatio-temporal patterns of expression.
General properties of ovarian factors:
Of the 348 ovarian factors, 131 (38%) are also significantly expressed in the testis (defined as more than two times higher signal strength in testis than in the average of all somatic tissues), consistent with the biological functions (such as meiosis) shared by oogenesis and spermatogenesis (supplemental Table 1 at http://www.genetics.org/supplemental/). Interestingly, when analyzed by class, the percentage of ovarian genes also expressed in testis was highest (69%) in class I genes and lowest (26 and 25%) in class III (egg) and IV (CG cells), arguing that a higher proportion of early acting genes have such shared functions in spermatogenesis and oogenesis (P = 1.7 × 10−5, Fisher exact test). Most ovarian factors are expressed predominantly in the germline; the ratio (germline vs. soma) for all 348 factors is ∼3:1.
Although systematic programs to generate mutant alleles of all mouse genes are underway, knockout alleles have already been generated for ∼10% of mouse genes (Austin et al. 2004). Of the 348 class I–IV genes, 103 (30%) have been previously knocked out, and of these, 45 (44%) result in female subfertility/ovarian phenotypes (supplemental Table 1 at http://www.genetics.org/supplemental/). Thus, a very high proportion of the set of ovarian factors we have identified are mutable to produce female subfertility phenotypes, proving that these genes are highly relevant for future studies of oogenesis and female infertility. This is especially so considering that many of the 103 knockouts were associated with embryonic/perinatal lethality that precluded an assessment of potential functions in oogenesis.
Preloading of PFs:
The profiles presented here provide opportunities to gain new insights into gene expression during early follicle growth. One unanticipated observation is that the vast majority (>98%) of genes induced during early follicle growth (i.e., class IB–E) are already turned on by PD1. Since PD1 precedes the initiation of follicle growth by several days (indeed, by PD1, PF formation is not complete), this demonstrates that PFs, even as they become individualized and quiescent at around PD3, are already engaged in the expression of genes they will require during activation and early growth. Presumably, this phenomenon, which we term preloading, reflects the establishment of stable oocyte and granulosa chromatin states and patterns of gene expression that permit PFs to quickly resume growth following their reactivation. Even genes classically described as induced following the primordial to primary follicle transition, such as Zp1–3, Gdf9, and Inhibin-β, or expressed later in follicle maturation, such as Mos or Mater, are in fact already significantly expressed by PD1 (Figure 2, C–F, and Figure 3; supplemental Figure 1 at http://www.genetics.org/supplemental/).
GO data mining highlights ECM remodeling during oogenesis:
To derive insights into important biological processes during follicle growth, we searched for enrichment of GO terms. There was significant enrichment of multiple GO terms for each class (Table 1). Many of these enriched terms were not unanticipated but confirmed the validity of our screening methodology and classification scheme. For example, class IA: follicle assembly/meiosis genes were significantly enriched for the GO terms meiosis and synaptonemal complex (P = 0.003 and 0.0001), whereas other classes were enriched for genes involved in reproduction and fertilization (P = 6 × 10−5 and 3 × 10−6) or C21-steroid biosynthesis (P = 8 × 10−9).
TABLE 1.
Statistically significant overrepresented GO terms per ovarian fertility factor class
| Class IA: follicle assembly/meiosis |
| Biological process |
| Meiotic cell cycle (P = 0.003) |
| M phase of meiotic cell cycle (P = 0.003) |
| Meiosis (P = 0.003) |
| Synapsis (P = 0.003) |
| Molecular function |
| Transcriptional regulator activity (P = 0.008) |
| Nucleic acid binding* (P = 0.0009) |
| DNA binding* (P = 0.0004) |
| Cellular component |
| Condensed nuclear chromosome* (P = 0.0003) |
| Synaptonemal complex* (P = 0.0001) |
| Transcription elongation factor complex* (P = 7 × 10−6) |
| Class IB: oocyte-specific, early and late maturation |
| Biological process |
| Embryonic pattern specification (P = 0.009) |
| Reproduction* (P = 0.0001) |
| Fertilization* (P = 0.0003) |
| Sperm egg recognition (P = 0.002) |
| Binding of sperm to zona pellucida (P = 0.002) |
| Molecular function |
| Nucleic acid binding (P = 0.007) |
| RNA binding (P = 0.003) |
| mRNA binding (P = 0.007) |
| Molecular function unknown (P = 0.003) |
| Cellular component |
| Pronucleus (P = 0.001) |
| Class IC: oocyte-specific, early maturation only |
| Biological process |
| Reproduction* (P = 6 × 10−5) |
| Fertilization* (P = 3 × 10−6) |
| Sperm-egg recognition* (P = 8 × 10−5) |
| Binding of sperm to zona pellucida* (P = 8 × 10−5) |
| Cellular component |
| ECM (P = 0.001) |
| Class ID: follicle growth, somatic |
| Biological process |
| Cell adhesion (P = 0.009) |
| Development* (P = 0.0006) |
| Sex differentiation (P = 0.003) |
| Molecular function |
| ECM structural constituent (P = 0.004) |
| Metalloendopeptidase activity (P = 0.009) |
| Transmembrane receptor protein kinase activity (P = 0.004) |
| Cellular component |
| ECM* (P = 0.0005) |
| Class IE: follicle growth, somatic, with high adrenal expression |
| Biological process |
| Steroid metabolism (P = 0.001) |
| Steroid biosynthesis* (P = 0.0003) |
| C21-steroid hormone biosynthesis* (P = 1 × 10−5) |
| Class II: ovary |
| Biological process |
| Reproductive physiological process (P = 0.002) |
| Cellular lipid metabolism* (P = 0.0002) |
| Steroid metabolism* (P = 3 × 10−5) |
| C21-steroid biosynthesis* (P = 8 × 10−9) |
| Molecular function |
| Lipid binding (P = 0.004) |
| Steroid binding (P = 0.002) |
| Steroid hydroxylase activity* (P = 6 × 10−5) |
| Endopeptidase inhibitor activity* (P = 0.0009) |
| Class III: egg |
| Biological process |
| Development (P = 0.007) |
| Organ morphogenesis (P = 0.008) |
| Reproduction (P = 0.002) |
| BMP signaling pathway (P = 0.009) |
| Retinoic acid metabolism* (P = 0.0006) |
| Molecular function |
| Polysaccharide binding (P = 0.009) |
| Hyaluronic acid binding (P = 0.004) |
Parent–child relationships are shown by indented lines. All terms for which P < 0.01 are shown; *P < 0.001. GO analysis was performed using the WebGestalt gene set analysis tool kit; Affymetrix identifiers were used for gene set retrieval. Gene set analysis and statistical comparisons were done against the 430 2.0 reference set, with P-values calculated by the hypergeometric test. Criteria for enriched GO categories were a minimum of two genes at a 0.01 significance level.
Strikingly, however, this analysis also identified multiple GO terms relating to ECM function, including extracellular matrix (P = 0.0005), metalloendopeptidase activity (P = 0.009), cell adhesion (P = 0.009), endopeptidase inhibitor activity (P = 0.0009), and hyaluronic acid binding (P = 0.004), across multiple gene classes (Table 1). A manually curated list of 30 ovarian factors with likely roles in ECM remodeling in the ovary is provided (Table 2). These factors all encode bona fide secreted or transmembrane proteins with N-terminal signal sequences (data not shown). That nearly 10% of all ovarian factors are well-described ECM factors (and other ovarian factors encoding secreted/transmembrane proteins may serve as yet unknown ECM functions) highlights the importance of ECM remodeling in ovarian biology, not only during oocyte/zona pellucida and follicle growth but also ovulation and early embryonic development. Furthermore, this analysis provides many new entry points for the systematic analysis of these poorly understood biological processes, particularly as nearly one-half (13/30) of these ECM factors were not previously recognized as ovarian factors. In this regard, it is notable that expression of several prominent structural components of the ECM or remodeling proteases, such as Col9a1, Col9a2, integrin α9, and adamts2, had not been previously documented in the ovary (supplemental Table 1 at http://www.genetics.org/supplemental/), although several other members of this class, including Timp1, protein kinase C mu, and tissue plasminogen activator (tPA), have been extensively studied for their roles in ECM remodeling in oogenesis.
TABLE 2.
List of ovarian factors that are known structural components of or involved in remodeling of the ECM (n = 30 for all classes)
| Gene | Class |
|---|---|
| adamts2 | ID |
| astacin-like metalloendopeptidase (M12 family) | III |
| carboxypeptidase A1 | IB |
| chondroitin sulfate proteoglycan 2 (versican) | III |
| C1q and tumor necrosis factor related protein 7 | IU |
| cystatin 8 | II |
| cystatin 12 | IU |
| dipeptidase 3 | IC |
| elastin microfibril interfacer 3 (emilin 3) | IA |
| heparan sulfate 6-O-sulfotransferase 2 | IU |
| fibrillin 2 | ID |
| fras1 related extracellular matrix protein 1 (frem1) | ID |
| interalpha (globulin) trypsin inhibitor H5 | IU |
| integrin α 9 | III |
| laminin a1 | IU |
| lysyl oxidase | III |
| matrix metallopeptidase 23 | ID |
| pentraxin 3 | III |
| procollagen, type IX, α 1 (Col9a1) | IU |
| procollagen, type IX, α 2 (Col9a2) | ID |
| procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 | III |
| protease, serine, 35 | IE |
| protein kinase C, mu | IB |
| serpin a5 | II |
| tissue inhibitor of metalloproteinase 1 (Timp1) | II |
| tPA | III |
| tumor necrosis factor α-induced protein 6 | III |
| zona pellucida glycoprotein 1 | IC |
| zona pellucida glycoprotein 2 | IC |
| zona pellucida glycoprotein 3 | IC |
Chromosomal localization reveals ovarian gene “miniclusters” and a novel member of the Rhox gene family:
A number of oocyte-specific genes are part of multiple gene-copy tandem repeat clusters, including the oogenesin (Oog), oligoadenylate synthetase (Oas), Nalp (Mater-like), F-box and WD-40 repeat (Fbxw), Tcl1-related, and the sperm-associated glutamate (E)-rich protein (Speer) gene families (Paillisson et al. 2005). Although most of the individual genes are nominally represented on Affymetrix arrays, the probe sets for some are identical or highly similar, leading to cross-hybridization and inability to reliably distinguish among all members of these families (see comments, supplemental Table 1). Nonetheless, our analysis did identify at least some representative members for all of the above gene families. To reveal previously undefined gene clusters, all 346 genes for which chromosome location could be determined were ordered on the most recent genome assembly (NCBI Build 36), and any clusters of two or more genes were noted. No new large cluster of tandemly repeated loci were identified (supplemental Figure 2A at http://www.genetics.org/supplemental/), demonstrating that all such clusters have been previously found. However, we did identify four new ovarian gene miniclusters consisting of two immediately adjacent loci, a finding highly unlikely due to chance. Interestingly, in three of the four miniclusters, both genes encode proteins with no detectable homology (e.g., Zp3 and Deltex2), arguing that the functional significance of these clusters relates to the existence of shared cis-regulatory elements directing coexpression. Consistent with this possibility, we note that in all four cases, both genes have strikingly similar patterns of gene expression (supplemental Figure 2B).
We also identified a new member of the recently described X-linked Rhox homeobox gene cluster whose members are expressed during gonadal development (Maclean et al. 2006). This gene, 1700123J19Rik, is immediately distal to Rhox12 (NCBI mouse genome Build 37.1) and encodes a homeodomain protein with all of the signature features of Rhox family members, including the placement of two introns within the homeodomain, similar overall protein length, and conserved C-terminal location of the homeodomain (data not shown). The predicted amino acid sequence, when compared against the entire mouse proteome by BLASTP (Altschul et al. 1990), was most similar to Rhox11. Thus, 1700123J19Rik is a bona fide Rhox family gene and not a pseudogene, and its physical proximity and sequence similarity support its categorization as a member of Rhox γ subcluster. Interestingly, 1700123J19Rik is a class I gene highly expressed in PD1 ovaries (no. 18, supplemental Table 1 and supplemental Figure 1 at http://www.genetics.org/supplemental/), and thus appears to be uniquely ovarian specific among subcluster γ genes. In following the convention of numbering Rhox genes according to their order on the X chromosome (Maclean et al. 2005), we propose that 1700123J19Rik be renamed Rhox13.
Homologs of Drosophila fertility or patterning factors:
We identified 21 homologs of Drosophila gametogenesis or patterning genes, most not previously recognized as mammalian ovarian factors. For example, we identified two homologs of kelch, required in Drosophila for oogenesis and intracytoplasmic transport via ring canals (Xue and Cooley 1993). These mammalian kelch homologs may serve a conserved role in intracytoplasmic transport, particularly since intercellular transport between oocytes and their somatic support cells are shared features of Drosophila and mammalian oogenesis (Anderson and Albertini 1976; Kidder and Mhawi 2002). Other Drosophila homologs include Sprouty4, Maelstrom, Seven-in-absentia 2, Transducin-like enhancer of split 6, Tudor domain containing 9, and Sex comb on midleg-like 2, among others (supplemental Table 2 at http://www.genetics.org/supplemental/; see also Figures 3–5).
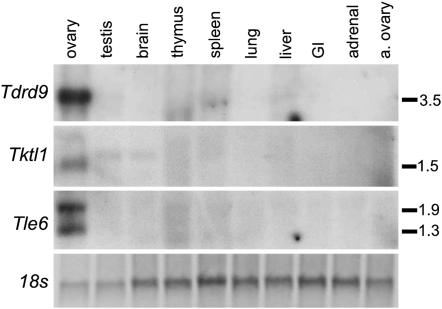
Figure 5.—
Northern analysis of human homologs shows ovary-specific expression. cDNAs for human homologs were end-sequenced for verification and cDNA inserts used for probe generation. Tissue samples were derived from the same female fetus at 20 weeks gestation except a. ovary, adult postmenopausal ovary. Genes are Tudor domain containing 9 (Tdrd9), Tktl1, and Transducin-like enhancer of split 6 (Tle6); 18s rRNA subunit as loading control (ethidium bromide stain of post-transfer membrane, negative image). Transcript sizes shown on right in kilobases. Note that the lack of detectable expression in adult postmenopausal ovary is expected given that all are predicted oocyte genes.
Conservation of ovarian factors among mice and humans:
Known ovarian factors are, almost without exception, conserved among mice and humans (Matzuk and Lamb 2002). Most mouse ovarian factors we identified have recognizable protein domains, and virtually all of the protein-coding genes (and several putative RNA genes; see discussion) have readily identifiable human homologs (supplemental Table 1). To confirm that our newly described genes are similarly ovarian specific in humans, we analyzed three genes, chosen at random, whose expression had not been previously evaluated in humans (tudor domain containing 9, transketolase-like 1, and transducin-like enhancer of split 6). Northern analysis showed that these genes were specifically expressed in the fetal human ovary (Figure 5), suggesting that most of the ovarian genes we identified are indeed conserved and have common functions in oogenesis in mammals.
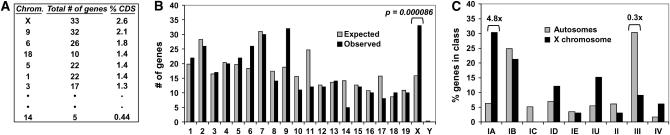
Ovarian factors are significantly overrepresented on the X chromosome:
To determine if ovarian genes were differentially represented on chromosomes, we studied the 346 genes for which chromosomal location is known. The X chromosome bears the highest number of ovarian genes and, more importantly, is also the top-ranked chromosome following normalization to genes per chromosome (Figure 6A). The number of ovarian genes on the X is more than two times higher than expected (P = 8.6 × 10−5, Fisher exact test) (Figure 6B). Chromosome 9 is enriched for ovarian genes, but this correlates with two gene clusters comprising 10 genes; the dearth of clustered genes on the X (supplemental Figure 2 at http://www.genetics.org/supplemental/) underscores the significance of ovarian factor overrepresentation on the X. Surprisingly, genes that act early in oogenesis (class IA: follicle assembly/meiosis) account for the overrepresentation of ovarian factors on the X, while genes that act late (class III: egg) are underrepresented (only 3/110 genes) (Figure 6C). These findings are reminiscent of observations that among testis genes, only those expressed prior to meiosis are enriched on the X (Wang et al. 2001; Khil et al. 2004), although the underlying mechanisms likely differ.
Figure 6.—
Enrichment of ovarian genes on X chromosome. (A) Total gene count per chromosome, rank order, and percent coding sequences (CDS) per chromosome that are ovarian genes. (B) Number of expected vs. observed ovarian genes on each chromosome based on the latest genome build (NCBI 36). Ovarian genes are significantly enriched on chromosomes X and 9; P-value by Fisher's exact test. Chromosome 9 contains multiple ovarian gene clusters, whereas the X chromosome does not. (C) Differences in X chromosome enrichment by class. Ovarian genes localizing to autosomes or X chromosomes is shown as percent of each class. The sum of each series is 100%.
DISCUSSION
The restricted expression of a gene in a unique tissue or cell type implies a specific functional role maintained by natural selection. While most developmental or physiologic processes depend on interactions between both tissue-specific and ubiquitous factors, tissue-specific factors often represent especially useful molecular and genetic entry points for a variety of investigations. Consequently, much effort has been invested to develop techniques (such as differential display and serial analysis of gene expression/SAGE) to identify genes with restricted expression patterns, and the deployment of such techniques has provided a wealth of information and useful targets. The success of our efforts to identify ovarian genes demonstrates that our general approach employing whole-genome arrays is useful and is much more efficient and less labor intensive than available methodologies. For example, in this study, we more than doubled the number of ovarian/oocyte factors identified previously. One key requirement for this approach is a panel of expression data sets for normal tissues. However, once such a panel is assembled, as we have done in this study, it can be easily shared with other investigators. In addition, analyses can be easily refined and expanded by inclusion of additional expression profiles, e.g., of specific tissues, developmental stages, or cell types purified by LCM. While our approach and digital Northern analysis is similar in some respects to that of a web based-resource (Su et al. 2004), our use of a noncustomized and widely available microarray platform greatly facilitates such integration of additional data sets.
Several observations demonstrate that the set of 348 ovarian genes we identified is biologically meaningful and a rich resource for functional and genetic analyses of oogenesis and female fertility. First, we identified the great majority of previously studied ovarian factors. Second, validation by RT–PCR and RISH proved that our analyses and screening criteria were reliable. Third, other observations, including the enrichment of ovarian factors on the X chromosome and that engineered null mutations in close to one-half of the mouse loci result in female sterility without affecting viability, strongly argue that this set of genes is biologically meaningful. Finally, the human orthologs of these mouse genes are also ovary specific. We also note that there can be no absolute cutoff to determine whether a gene evolved to serve some ovarian function on the basis of its expression pattern alone, and many genes not highly expressed in the ovary relative to other tissues can nonetheless serve discrete ovarian functions, such as Foxo3 itself (Castrillon et al. 2003).
The high yield of our approach, combined with the detailed views of gene expression afforded by digital Northern analysis, permitted several new insights into ovarian gene expression, such as the finding that CG-specific genes transcripts are also present in eggs, raising the possibility that transcripts may be actively transported. Furthermore, we found that PFs transcribe follicle growth genes even before they initiate growth, indeed, even before their assembly is complete (preloading). While these phenomena merit further investigation, they illustrate the ability of combined high-throughput gene discovery and digital Northern approaches to reveal unexpected properties of gene expression in a particular developmental process.
This screen was not designed to identify small RNAs or RNA-coding genes but nonetheless did succeed in identifying ∼20 loci that do not appear to encode proteins; many are evolutionary conserved and thus may encode functional RNAs. One of these genes, forkhead box L2 opposite strand transcript (Foxl2os) (no. 153, supplemental Table 1 at http://www.genetics.org/supplemental/), encodes an antisense RNA believed to play a role in the regulation of FoxL2, a forkhead transcription factor required for normal ovarian follicle assembly and maturation (Crisponi et al. 2001; Cocquet et al. 2005). A poorly annotated EST (1447352_at; Mm.412326) identified as an ovarian factor contains no obvious ORF (no. 145, supplemental Table 1) but is highly conserved in humans and spans microRNA (miRNA) mmu-mir-202, suggesting that this EST may represent a pre-miRNA. A unique class of small RNAs (Piwi-interacting RNAs, or piRNAs) larger (26–31 nt) than most small RNAs was recently discovered in mammalian testes. It is notable, however, that piRNAs have not been identified in oogenesis, and murine piwi homologs are required for spermatogenesis but apparently not oogenesis. Knockout mice for these genes are male sterile with a block in spermatogenesis, while females have normal fertility (Kim 2006). This suggests that piRNAs play more important roles in spermatogenesis (where they are believed to function in the repression of transposons) (Carmell et al. 2007) but leaves open the possibility that some miRNAs or other RNAs serve important functions in oogenesis (Murchison et al. 2007).
The oocyte is enveloped in a highly specialized ECM—the zona pellucida—and the three major zona pellucida glycoproteins Zp1–3 were the first identified ovary-specific ECM components. Their study has led to many insights into oocyte gene expression, follicle growth, and fertilization (Epifano et al. 1995). However, there is growing interest in ECM remodeling throughout oogenesis (Rodgers 2006), the importance of which is further emphasized by our identification of a large number of ovarian-specific ECM factors. For example, in response to the ovulatory luteinizing hormone surge, a unique matrix envelops the cumulus–oocyte complex (cumulus expansion), creating a microenvironment required for fertilization. Three factors required for this process—pentraxin 3, Tnfip-6, and versican—were all among the ECM factors we identified (Table 2), and still other factors known to be absolutely required for cumulus matrix assembly, such as Cox-2, were also among our list of ovarian factors. Knockouts of three of these four factors (except versican, associated with embryonic lethality) are female subfertile due to defects in ovulation/cumulus matrix expansion (Russell and Salustri 2006). Our findings should help fill some important remaining gaps in our understanding of cumulus expansion and other ECM-related processes in the ovary. For example, inter-α-trypsin inhibitor (IαI) is also required for cumulus expansion and is believed to be transported into the follicular space from serum (IαI is synthesized by the liver). Although a serum-derived IαI activity can serve this cumulus expansion function in vitro (Chen et al. 1992; Nagyova et al. 2004), our identification of an ovarian-specific inter-α-trypsin gene, Itih5 (no. 191, supplemental Table 1 at http://www.genetics.org/supplemental/), raises the possibility that Itih5 represents a physiologic source of ovarian IαI activity.
In species ranging from Drosophila to mammals, the gene content of sex chromosomes differs substantially from that of the autosomes, due to a complex interplay of opposing evolutionary forces (Vallender and Lahn 2004). For example, the X chromosome spends most of its time (two-thirds) in females, and this may lead genes that benefit females, such as those required for fertility, to migrate to the X (sexual antagonism). On the other hand, genes present on the X are subject to hemizygous exposure in males, permitting selection to operate more effectively in males, a phenomenon postulated to lead (somewhat counterintuitively) to the accumulation of male genes on the X (Wang et al. 2001), although for testis genes this may be limited to those expressed prior to the onset of meiosis (Khil et al. 2004). Since most ovarian genes we identified on the X were class IA and also highly expressed in testis probably prior to meiosis, their expression in males may thus have contributed to their enrichment on the X.
The ovarian genes presented here are attractive candidates as causal hereditary factors in female infertility and early menopause (Matzuk and Lamb 2002) because these conditions have an ovarian basis (Kumar et al. 2007). Given that the number of ovarian factors is large (the 348 genes described here represent >1% of the genome), it is not surprising that previous small-scale efforts to identify mutations in candidate genes have not yet yielded common causal mutations. Such mutations are likely to exist, but it is now clear that systematic, large-scale gene resequencing studies will be required to identify them. The set of ovarian factors presented here is a starting point for such an effort.
Acknowledgments
We thank Keith Wharton, Andrew Zinn, Dennis McKearin, Scott Armstrong, Luigi Marchionni, and David Berman for helpful discussions and comments on the manuscript, and Anwu Zhou and the UT Southwestern Microarray Core Facility for technical assistance. Requests for materials should be addressed to D.H.C. This work was supported by a grant from the N.I.H. (HD048690). C.M.C. is supported by an N.I.H. N.R.S.A. fellowship.
References
- Aittomaki, K., J. L. Lucena, P. Pakarinen, P. Sistonen, J. Tapanainen et al., 1995. Mutation in the follicle-stimulating hormone receptor gene causes hereditary hypergonadotropic ovarian failure. Cell 82: 959–968. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson, E., and D. F. Albertini, 1976. Gap junctions between the oocyte and companion follicle cells in the mammalian ovary. J. Cell. Biol. 71: 680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, C. P., J. F. Battey, A. Bradley, M. Bucan, M. Capecchi et al., 2004. The knockout mouse project. Nat. Genet. 36: 921–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birk, O. S., D. E. Casiano, C. A. Wassif, T. Cogliati, L. Zhao et al., 2000. The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature 403: 909–913. [DOI] [PubMed] [Google Scholar]
- Carmell, M. A., A. Girard, H. J. van de Kant, D. Bourc'his, T. H. Bestor et al., 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12: 503–514. [DOI] [PubMed] [Google Scholar]
- Castrillon, D. H., L. Miao, R. Kollipara, J. W. Horner and R. A. DePinho, 2003. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science 301: 215–218. [DOI] [PubMed] [Google Scholar]
- Chen, L., S. J. Mao and W. J. Larsen, 1992. Identification of a factor in fetal bovine serum that stabilizes the cumulus extracellular matrix. A role for a member of the inter-alpha-trypsin inhibitor family. J. Biol. Chem. 267: 12380–12386. [PubMed] [Google Scholar]
- Clark, B. J., S. C. Soo, K. M. Caron, Y. Ikeda, K. L. Parker et al., 1995. Hormonal and developmental regulation of the steroidogenic acute regulatory protein. Mol. Endocrinol. 9: 1346–1355. [DOI] [PubMed] [Google Scholar]
- Clarke, D. L., and D. I. Linzer, 1993. Changes in prolactin receptor expression during pregnancy in the mouse ovary. Endocrinology 133: 224–232. [DOI] [PubMed] [Google Scholar]
- Clegg, N. J., D. M. Frost, M. K. Larkin, L. Subrahmanyan, Z. Bryant et al., 1997. maelstrom is required for an early step in the establishment of Drosophila oocyte polarity: posterior localization of grk mRNA. Development 124: 4661–4671. [DOI] [PubMed] [Google Scholar]
- Cocquet, J., M. Pannetier, M. Fellous and R. A. Veitia, 2005. Sense and antisense Foxl2 transcripts in mouse. Genomics 85: 531–541. [DOI] [PubMed] [Google Scholar]
- Colledge, W. H., M. B. Carlton, G. B. Udy and M. J. Evans, 1994. Disruption of c-mos causes parthenogenetic development of unfertilized mouse eggs. Nature 370: 65–68. [DOI] [PubMed] [Google Scholar]
- Costa, Y., R. Speed, R. Ollinger, M. Alsheimer, C. A. Semple et al., 2005. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J. Cell Sci. 118: 2755–2762. [DOI] [PubMed] [Google Scholar]
- Costa, Y., R. M. Speed, P. Gautier, C. A. Semple, K. Maratou et al., 2006. Mouse MAELSTROM: The link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum. Mol. Genet. 15: 2324–2334. [DOI] [PubMed] [Google Scholar]
- Crackower, M. A., N. K. Kolas, J. Noguchi, R. Sarao, K. Kikuchi et al., 2003. Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science 300: 1291–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisponi, L., M. Deiana, A. Loi, F. Chiappe, M. Uda et al., 2001. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat. Genet. 27: 159–166. [DOI] [PubMed] [Google Scholar]
- Dong, J., D. F. Albertini, K. Nishimori, T. R. Kumar, N. Lu et al., 1996. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature 383: 531–535. [DOI] [PubMed] [Google Scholar]
- Dube, J. L., P. Wang, J. Elvin, K. M. Lyons, A. J. Celeste et al., 1998. The bone morphogenetic protein 15 gene is X-linked and expressed in oocytes. Mol. Endocrinol. 12: 1809–1817. [DOI] [PubMed] [Google Scholar]
- Durlinger, A. L., P. Kramer, B. Karels, F. H. de Jong, J. T. Uilenbroek et al., 1999. Control of primordial follicle recruitment by anti-Mullerian hormone in the mouse ovary. Endocrinology 140: 5789–5796. [DOI] [PubMed] [Google Scholar]
- Epifano, O., L. F. Liang, M. Familari, M. C. Moos, Jr. and J. Dean, 1995. Coordinate expression of the three zona pellucida genes during mouse oogenesis. Development 121: 1947–1956. [DOI] [PubMed] [Google Scholar]
- Evsikov, A. V., J. H. Graber, J. M. Brockman, A. Hampl, A. E. Holbrook et al., 2006. Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev. 20: 2713–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, M. G., D. A. Thompson and R. J. Weigel, 2000. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 60: 6367–6375. [PubMed] [Google Scholar]
- Gougeon, A., 1986. Dynamics of follicular growth in the human: a model from preliminary results. Hum. Reprod. 1: 81–87. [DOI] [PubMed] [Google Scholar]
- Gray, S. A., M. A. Mannan and P. J. O'Shaughnessy, 1996. Development of cytochrome P450 17 alpha-hydroxylase (P450c17) mRNA and enzyme activity in neonatal ovaries of normal and hypogonadal (hpg) mice. J. Mol. Endocrinol. 17: 55–60. [DOI] [PubMed] [Google Scholar]
- Hammer, N. A., T. O. Hansen, A. G. Byskov, E. Rajpert-De Meyts, M. L. Grondahl et al., 2005. Expression of IGF-II mRNA-binding proteins (IMPs) in gonads and testicular cancer. Reproduction 130: 203–212. [DOI] [PubMed] [Google Scholar]
- Herrera, L., C. Ottolenghi, J. E. Garcia-Ortiz, M. Pellegrini, F. Manini et al., 2005. Mouse ovary developmental RNA and protein markers from gene expression profiling. Dev. Biol. 279: 271–290. [DOI] [PubMed] [Google Scholar]
- Hirshfield, A. N., 1991. Development of follicles in the mammalian ovary. Int. Rev. Cytol. 124: 43–101. [DOI] [PubMed] [Google Scholar]
- Hosaka, T., W. H. Biggs, III, D. Tieu, A. D. Boyer, N. M. Varki et al., 2004. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. USA 101: 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, M., D. Boerboom, M. Shimada, Y. Lo, A. F. Parlow et al., 2005. Mice null for Frizzled4 (Fzd4−/−) are infertile and exhibit impaired corpora lutea formation and function. Biol. Reprod. 73: 1135–1146. [DOI] [PubMed] [Google Scholar]
- Hu, M. C., N. C. Hsu, N. B. El Hadj, C. I. Pai, H. P. Chu et al., 2002. Steroid deficiency syndromes in mice with targeted disruption of Cyp11a1. Mol. Endocrinol. 16: 1943–1950. [DOI] [PubMed] [Google Scholar]
- Ishida, T., S. Choi, R. K. Kundu, K. Hirata, E. M. Rubin et al., 2003. Endothelial lipase is a major determinant of HDL level. J. Clin. Invest. 111: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, G. B., L. J. Shirley, T. D. Gallardo and D. H. Castrillon, 2007. Specificity of the requirement for Foxo3 in primordial follicle activation. Reproduction 133: 855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantaridou, S. N., and L. M. Nelson, 2000. Premature ovarian failure is not premature menopause. Ann. NY Acad. Sci. 900: 393–402. [DOI] [PubMed] [Google Scholar]
- Kaufman, F., M. D. Kogut, G. N. Donnell, H. Koch and U. Goebelsmann, 1979. Ovarian failure in galactosaemia. Lancet 2: 737–738. [DOI] [PubMed] [Google Scholar]
- Khil, P. P., N. A. Smirnova, P. J. Romanienko and R. D. Camerini-Otero, 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 36: 642–646. [DOI] [PubMed] [Google Scholar]
- Kidder, G. M., and A. A. Mhawi, 2002. Gap junctions and ovarian folliculogenesis. Reproduction 123: 613–620. [DOI] [PubMed] [Google Scholar]
- Kim, V. N., 2006. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 20: 1993–1997. [DOI] [PubMed] [Google Scholar]
- Kumar, A., S. Ghadir, N. Eskandari and A. H. DeCherney, 2007. Infertility, pp. 917–925 in Current Diagnosis & Treatment Obstetrics & Gynecology, edited by A. H. DeCherney and L. Nathan. McGraw-Hill, New York
- Kurimoto, K., Y. Yabuta, Y. Ohinata, Y. Ono, K. D. Uno et al., 2006. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 34: e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., D. Peter, A. Roghani, S. Schuldiner, G. G. Prive et al., 1992. A cDNA that suppresses MPP+ toxicity encodes a vesicular amine transporter. Cell 70: 539–551. [DOI] [PubMed] [Google Scholar]
- Ma, K., M. Cilingiroglu, J. D. Otvos, C. M. Ballantyne, A. J. Marian et al., 2003. Endothelial lipase is a major genetic determinant for high-density lipoprotein concentration, structure, and metabolism. Proc. Natl. Acad. Sci. USA 100: 2748–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean, II, J. A., M. A. Chen, C. M. Wayne, S. R. Bruce, M. Rao et al., 2005. Rhox: a new homeobox gene cluster. Cell 120: 369–382. [DOI] [PubMed] [Google Scholar]
- Maclean, II, J. A., D. Lorenzetti, Z. Hu, W. J. Salerno, J. Miller et al., 2006. Rhox homeobox gene cluster: recent duplication of three family members. Genesis 44: 122–129. [DOI] [PubMed] [Google Scholar]
- Matzuk, M. M., and D. J. Lamb, 2002. Genetic dissection of mammalian fertility pathways. Nat. Cell Biol. 4(Suppl.): s41–49. [DOI] [PubMed] [Google Scholar]
- McFarland, K. C., R. Sprengel, H. S. Phillips, M. Kohler, N. Rosemblit et al., 1989. Lutropin-choriogonadotropin receptor: an unusual member of the G protein-coupled receptor family. Science 245: 494–499. [DOI] [PubMed] [Google Scholar]
- McGee, E. A., and A. J. Hsueh, 2000. Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 21: 200–214. [DOI] [PubMed] [Google Scholar]
- Miyakoshi, K., M. J. Murphy, R. R. Yeoman, S. Mitra, C. J. Dubay et al., 2006. The identification of novel ovarian proteases through the use of genomic and bioinformatic methodologies. Biol. Reprod. 75: 823–835. [DOI] [PubMed] [Google Scholar]
- Murchison, E. P., P. Stein, Z. Xuan, H. Pan, M. Q. Zhang et al., 2007. Critical roles for Dicer in the female germline. Genes Dev. 21: 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagyova, E., A. Camaioni, R. Prochazka and A. Salustri, 2004. Covalent transfer of heavy chains of inter-alpha-trypsin inhibitor family proteins to hyaluronan in in vivo and in vitro expanded porcine oocyte-cumulus complexes. Biol. Reprod. 71: 1838–1843. [DOI] [PubMed] [Google Scholar]
- Odet, F., A. Verot and B. Le Magueresse-Battistoni, 2006. The mouse testis is the source of various serine proteases and serine proteinase inhibitors (SERPINs): serine proteases and SERPINs identified in Leydig cells are under gonadotropin regulation. Endocrinology 147: 4374–4383. [DOI] [PubMed] [Google Scholar]
- Paillisson, A., S. Dade, I. Callebaut, M. Bontoux, R. Dalbies-Tran et al., 2005. Identification, characterization and metagenome analysis of oocyte-specific genes organized in clusters in the mouse genome. BMC Genomics 6: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangas, S. A., Y. Choi, D. J. Ballow, Y. Zhao, H. Westphal et al., 2006. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc. Natl. Acad. Sci. USA 103: 8090–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, J., and J. H. Eberwine, 1996. Antisense RNA amplification: a linear amplification method for analyzing the mRNA population from single living cells. Methods 10: 283–288. [DOI] [PubMed] [Google Scholar]
- Pryor, J. L., M. Kent-First, A. Muallem, A. H. Van Bergen, W. E. Nolten et al., 1997. Microdeletions in the Y chromosome of infertile men. N. Engl. J. Med. 336: 534–539. [DOI] [PubMed] [Google Scholar]
- Rae, J. M., M. D. Johnson, K. E. Cordero, J. O. Scheys, J. M. Larios et al., 2006. GREB1 is a novel androgen-regulated gene required for prostate cancer growth. Prostate 66: 886–894. [DOI] [PubMed] [Google Scholar]
- Rajkovic, A., M. S. C. Yan, M. Klysik and M. Matzuk, 2001. Discovery of germ cell-specific transcripts by expressed sequence tag database analysis. Fertil. Steril. 76: 550–554. [DOI] [PubMed] [Google Scholar]
- Rajkovic, A., S. A. Pangas, D. Ballow, N. Suzumori and M. M. Matzuk, 2004. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science 305: 1157–1159. [DOI] [PubMed] [Google Scholar]
- Rankin, T., M. Familari, E. Lee, A. Ginsberg, N. Dwyer et al., 1996. Mice homozygous for an insertional mutation in the Zp3 gene lack a zona pellucida and are infertile. Development 122: 2903–2910. [DOI] [PubMed] [Google Scholar]
- Rodgers, R., 2006. Extracellular matrix in the ovary. Semin. Reprod. Med. 24: 193–194. [DOI] [PubMed] [Google Scholar]
- Ruggiu, M., R. Speed, M. Taggart, S. J. McKay, F. Kilanowski et al., 1997. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 389: 73–77. [DOI] [PubMed] [Google Scholar]
- Russell, D. L., and A. Salustri, 2006. Extracellular matrix of the cumulus-oocyte complex. Semin. Reprod. Med. 24: 217–227. [DOI] [PubMed] [Google Scholar]
- Seydoux, G., 1996. Mechanisms of translational control in early development. Curr. Opin. Genet. Dev. 6: 555–561. [DOI] [PubMed] [Google Scholar]
- Shinohara, T., K. E. Orwig, M. R. Avarbock and R. L. Brinster, 2000. Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proc. Natl. Acad. Sci. USA 97: 8346–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, C. L., J. E. Shima, M. Uzumcu, M. K. Skinner and M. D. Griswold, 2005. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol. Reprod. 72: 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyal, S. M., A. Amleh and J. Dean, 2000. FIGalpha, a germ cell-specific transcription factor required for ovarian follicle formation. Development 127: 4645–4654. [DOI] [PubMed] [Google Scholar]
- Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching et al., 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101: 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugatani, J., Y. Masu, M. Nishizawa, K. Sakamoto, T. Houtani et al., 1996. Hormonal regulation of prostaglandin F2 alpha receptor gene expression in mouse ovary. Am. J. Physiol. 271: E686–693. [DOI] [PubMed] [Google Scholar]
- Uhl, G. R., S. Li, N. Takahashi, K. Itokawa, Z. Lin et al., 2000. The VMAT2 gene in mice and humans: amphetamine responses, locomotion, cardiac arrhythmias, aging, and vulnerability to dopaminergic toxins. FASEB J. 14: 2459–2465. [DOI] [PubMed] [Google Scholar]
- Vallender, E. J., and B. T. Lahn, 2004. How mammalian sex chromosomes acquired their peculiar gene content. BioEssays 26: 159–169. [DOI] [PubMed] [Google Scholar]
- Vergnes, L., J. Phan, A. Stolz and K. Reue, 2003. A cluster of eight hydroxysteroid dehydrogenase genes belonging to the aldo-keto reductase supergene family on mouse chromosome 13. J. Lipid Res. 44: 503–511. [DOI] [PubMed] [Google Scholar]
- Wang, P. J., J. R. McCarrey, F. Yang and D. C. Page, 2001. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27: 422–426. [DOI] [PubMed] [Google Scholar]
- Xue, F., and L. Cooley, 1993. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72: 681–693. [DOI] [PubMed] [Google Scholar]
- Yoon, S. J., K. H. Kim, H. M. Chung, D. H. Choi, W. S. Lee et al., 2006. Gene expression profiling of early follicular development in primordial, primary, and secondary follicles. Fertil. Steril. 85: 193–203. [DOI] [PubMed] [Google Scholar]
- Yuan, L., J. G. Liu, J. Zhao, E. Brundell, B. Daneholt et al., 2000. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell 5: 73–83. [DOI] [PubMed] [Google Scholar]