Abstract
The bacterium Myxococcus xanthus glides over surfaces using two different locomotive mechanisms, called S (social) and A (adventurous) motility that enable cells to move both as groups and as individuals. Neither mechanism involves flagella. The functions of these two motors are coordinated by the activity of a small Ras-like protein, encoded by the mglA gene. The results of previous studies of a second-site suppressor of the mglA-8 missense mutation masK-815 indicate that MglA interacts with a protein tyrosine kinase, MasK, to control social motility. Sequence analysis of the sites of 12 independent insertions of the transposon magellan-4 that result in the loss of motility in an M. xanthus mglA-8 masK-815 double mutant shows that nine of these 12 insertions are in genes known to be required for S gliding motility. This result confirms that the masK-815 suppressor restores S but not A motility. Three of the 12 insertions define three new genes required for S motility and show that the attachment of heptose to the lipopolysaccharide inner core, an ortholog of the CheR methyltransferase, and a large protein with YD repeat motifs, are required for S motility. When these three insertions are backcrossed into an otherwise wild-type genetic background, their recombinants are found to have defects in S, but not, A motility. The spectrum of magellan-4 insertions that lead to the loss of S motility in the mglA-8 masK-815 double mutant background is different than that resulting from a previous mutant hunt starting with a different (A mutant) genetic background, suggesting that the number of genes required for S motility in M. xanthus is quite large.
EUBACTERIAL genomes are extraordinarily diverse and range in size from <1 Mbp to >10 Mbp. In general, the bacteria with genomes at the larger extreme of genome size, such as species of Anabena, Myxococcus, and Streptomyces, display more complex behaviors in response to environmental stresses, including programs of multicellular development that involve the differentiation of cells into specialized forms. For example, Myxococcus xanthus, with its 9-Mb genome, responds to starvation by aggregating large groups of individual cells into fruiting bodies, in which a subset of cells differentiate into heat-resistant, diploid myxospores.
This response to starvation by M. xanthus requires the functions of a large set of genes, many of which do not have homologs in other bacteria. Within the set of genes required for the multicellular development of M. xanthus are two subsets of genes involved in its two different mechanisms of gliding motility. M. xanthus can glide over solid surfaces without the use of flagella, both as individual cells (called A, or “adventurous” motility) and as groups of cells (called S, or “social” motility). These two mechanisms can be separated genetically. Most mutations induced by chemical mutagens or transposons that impair motility in M. xanthus affect its ability to glide as single cells, or as groups of cells, but not both (Hodgkin and Kaiser 1979a,b; Macneil et al. 1994a,b; Wu and Kaiser 1995; Youderian et al. 2003; Youderian and Hartzell 2005).
Wild-type strains of M. xanthus form large, spreading colonies on agar plates. Most single mutants of M. xanthus with defects in either A or S motility form colonies of intermediate size. Double mutants of M. xanthus with pairs of mutations, one in an A motility gene plus one in an S motility gene, form smaller colonies than either single A or S mutants. These colonies have sharp edges, and the colonies formed by double mutants with both A and S defects can be distinguished readily from those made by either a wild-type strain or single A or S mutants, both on the basis of their relative size and their morphology. In the past, we have used these phenotypic differences to screen for double mutants with additional defects in A and S motility (Macneil et al. 1994a,b; Youderian et al. 2003; Youderian and Hartzell 2005).
Our most fruitful screens for mutants defective in the two motility systems have involved making double mutants using the transposons Tn5 and magellan-4. Because transposons are both genetic and physical markers, genes disrupted by transposon insertions can be subcloned rapidly, and the sequence junctions between transposons and their target genes can be determined to identify these target genes. Previously, we have employed a simple technique to screen for double mutants defective in both A and S motility with Tn5 and magellan-4 insertions and identify their target A and S genes. Starting with mutants defective in either A or S motility, we mutagenized these single mutants with transposons and screened for double mutants that form smaller, nonmotile colonies, because they carry second mutations (insertions) in S or A genes, respectively. Using this strategy, we have shown that the functions of at least 34 genes are required for A motility and have identified 45 of the 113 genes known to be required for S motility (Youderian et al. 2003; Youderian and Hartzell 2005; Hartzell et al. 2007).
The results of our studies, and those from other laboratories, have identified only three genes: mglA, (Stephens and Kaiser 1987; Stephens et al. 1989; Hartzell and Kaiser 1991a,b; Hartzell 1997); agmA, predicted to encode an amidase involved in the biogenesis of the cell wall (Youderian et al. 2003); and epsI/nla24, predicted to encode a positive activator of transcription (Caberoy et al. 2003; Lancero et al. 2004; Lu et al. 2005) required for both the S and A motility mechanisms.
The first of these three genes, mglA, encodes a 22-kD protein in the Ras family of GTPases, which behaves like the other small “G proteins” in this family, by coupling the hydrolysis of its GTP substrate with multiple protein–protein interactions that trigger signal transduction cascades (Hartzell and Kaiser 1991b; Hartzell 1997). MglA interacts with two different proteins, AglZ, a myosin-like coiled-coil protein involved in A motility (Yang et al. 2004; Mignot et al. 2007), and MasK, a protein kinase involved in S motility (Thomasson et al. 2002). The interactions between MglA and each of these proteins likely regulate the simultaneous operation of the S and A gliding motors of M. xanthus, to coordinate the actions of both motors simultaneously so that they function in the same direction.
The interaction between MglA and the protein tyrosine kinase, MasK, was discovered by using a classical genetic approach. A missense mutation in the mglA gene, mglA-8, results in a loss of mglA function, and impairs both A and S motility. An allele-specific suppressor of mglA-8, masK-815, also is a missense mutation. This extragenic second-site suppressor mutation maps to the 3′ end of the essential masK gene. Cells of the mglA-8 masK-815 double mutant can move as groups, but not as individuals, and appear to have regained S, but not A, motility. In addition, when a plasmid subclone of the masK gene was used in the yeast two-hybrid selection as bait against a library carrying plasmid subclones of M. xanthus chromosomal DNA, subclones carrying fusions of mglA with the GAL4 activation domain were recovered, confirming the interaction between MglA and MasK. MasK, when expressed in Escherichia coli, has tyrosine kinase activity (Thomasson et al. 2002). Together, these results argue that the interaction between MglA and MasK likely mirrors those between eukaryotic GTPases and MAP kinases and controls a signal transduction cascade in M. xanthus that, in turn, controls S motility.
MATERIALS AND METHODS
Bacterial strains and growth conditions:
The M. xanthus strains generated in this study are derivatives of the wild-type strain DK1622 and its mglA-8 masK-815 double mutant derivative MxH1104 (Thomasson et al. 2002) and are listed in Table 1 . M. xanthus was grown at 32° in CTPM liquid medium (1% casitone, 10 mm Tris pH 7.6, 1 mm potassium phosphate pH 7.5, 5 mm MgSO4) and CTPM agar (1.5%) plates; CTPM was supplemented with kanamycin (Kan; 40 μg/ml). Plasmids were introduced into M. xanthus by electroporation (Kashefi and Hartzell 1995; Youderian et al. 2003). E. coli strain DH5α(λ pir) was used for the recovery and propagation of plasmids and the preparation of plasmid DNA. Plasmids were introduced into this strain by electroporation, and derivatives with plasmids were grown in LB medium supplemented with kanamycin (Kan; 40 μg/ml). Plasmid pMycoMar, donor of the mini-mariner element magellan-4, has been described (Rubin et al. 1999). Restriction endonucleases and DNA modifying enzymes were from New England Biolabs (Ipswich, MA).
TABLE 1.
Insertions of magellan-4 that impair social gliding motility
| Coordinates | Locus | Insertion | Straina | Strainb | Gene | Isoallelesc |
|---|---|---|---|---|---|---|
| 5901872–5901873 | MXAN_4707 | mis-185 | MxH1185 | sgmU; rfaF | mis-55 | |
| 5903725–5903726 | MXAN_4710 | mis-198 | MxH1198 | MxH1298 | sgnG; rfaE | |
| 7149933–7149934 | MXAN_5774 | mis-178 | MxH1178 | pilO | mis-78, mis-79 | |
| 7151612–7151613 | MXAN_5776 | mis-168 | MxH1168 | pilM | ||
| 7151612–7151613 | MXAN_5776 | mis-182 | MxH1182 | pilM | ||
| 8154610–8154611 | MXAN_6627 | mis-181 | MxH1181 | sgnC | mis-52 | |
| 8200360–8200361 | MXAN_6671 | mis-196 | MxH1196 | sglK | mis-45 | |
| 8208665–8208666 | MXAN_6679 | mis-195 | MxH1195 | MxH1295 | sgnH | |
| 8671663–8671664 | MXAN_7103 | mis-189 | MxH1189 | MxH1289 | sgnI | |
| 9064019–9064020 | MXAN_7441 | mis-190 | MxH1190 | epsH | ||
| 9072529–9072530 | MXAN_7448 | mis-179 | MxH1179 | epsD | ||
| 9072529–9072530 | MXAN_7448 | mis-180 | MxH1180 | epsD |
The 12 insertions of magellan-4 in the M. xanthus genome described in this report are listed in order of their sites within the genome sequence, which can be found at: http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gmx. Given are the gene numbers in which the insertions are situated, the allele numbers of the insertions, the strain numbers of the derivatives of strain MxH1104 with each insertion, and the gene names. The new genes identified by magellan-4 insertions in this study, MXAN_4710, MXAN_6679, and MXAN_7103, have been designated sgnG, sgnH, and sgnI, respectively.
Strain name of the original isolate containing the mariner insertion in M. xanthus MxH1104 (mglA8 masK-815).
Strain name of the corresponding mariner insertion in the wild-type M. xanthus (DK1622) background.
The allele numbers of insertions that were also isolated in an ΔaglU are listed as isoalleles (Youderian and Hartzell 2005).
Isolation and phenotypic screening of potential social motility mutants:
The electroporation of MxH1104 cells with plasmid pMycoMar was performed as described (Youderian et al. 2003). Electroporation was used to backcross transposon insertions from the MxH1104 background into the wild-type (DK1622) background. Chromosomal DNA was prepared from strains MxH1189, 1195, and 1198 using the Easy DNA method (Invitrogen, Carlsbad, CA). Electroporation of DK1622 cells with purified chromosomal DNA (1–3 μg) was performed as described (Youderian et al. 2003). Strains with the mis-189, mis-195, and mis-198 insertions in an otherwise wild-type genetic background were designated MxH1289, 1295, and 1298, respectively. In all cases, the KanR determinant was found to be 100% linked with a defect in S motility (see results). Electroporation mixes were plated on CTPM Kan agar and incubated for 5 days at 32°. After incubation, plates were screened visually to identify small colonies with smooth edges. Mutants were purified twice, and the phenotypes of single colonies formed by each mutant were compared after each purification step.
Cloning and sequence analysis of M. xanthus genomic DNA flanking magellan-4 insertions in S genes:
To subclone magellan-4 insertions in S genes, M. xanthus genomic DNA was isolated from vegetative cultures of MxH1104 S∷magellan-4 strains, cleaved with BssHI, ligated, and electroporated into E. coli DH5α (λ pir) as described (Youderian et al. 2003; Youderian and Hartzell 2005). Plasmid DNAs with subcloned BssHII fragments were isolated from KanR electroporants and sequenced with primers Mar1 and Mar2 (Biosource/Invitrogen), complementary to the ends of magellan-4 (Youderian et al. 2003); sequencing was performed by Commonwealth Biotechnologies (Richmond, VA). BLASTn searches (Altschul et al. 1990) against the DK1622 genome sequence were used to identify the TA target site for each magellan-4 insertion, given as the coordinates of the M. xanthus sequence available from TIGR (http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org_search=&org=gmx) (Table 1). In all cases, these searches yielded a unique dinucleotide target site of insertion without accompanying deletion or rearrangement. The probable functions of proteins encoded by target genes inactivated by magellan-4 insertions were deduced using the CD-search program (Marchler-Bauer and Bryant 2004) available at http://www.ncbi.nlm.nih.gov/BLAST/.
Analysis of spreading motility and single cell gliding:
Motility phenotypes of mutants were compared with that of the wild-type strain using spreading assays on 0.3 and 1.5% CTPM agar as described (Shi and Zusman 1993) and by microscopic examinations of colony edges. Individual cells were tracked by time-lapse videomicroscopy and analyzed using Metamorph tracking software. Cells were grown in CTPM medium, diluted to 5 × 105 cells/ml and spotted on agar pads as described elsewhere (Mignot et al. 2005). The agar pad containing 1% agar in CTPM was poured on a cover slip containing a 0.5-mm silicon gasket and allowed to dry for 30 min. Cells were placed on the agar pad, inverted on a glass slide, and incubated at 32° for 30 min. The cells were viewed with a Nikon FXA microscope at 20×. Images were captured using a CCD camera at 30 sec intervals for 30 min. Stacks (consecutive series of images) were created with Metamorph, and individual cells (minimum 30 cells selected at random) were tracked to quantify the rate of cell movement and cell reversal frequency.
Developmental assays:
Fruiting body formation and spore production were monitored on TPM starvation medium as described elsewhere (Yang et al. 2004). In each of these experiments, mutants were assayed in triplicate alongside control strains DK1622 (wild-type), DK4135 (mglA8), and MxH1104 (mglA8 masK815).
RESULTS
Insertions of magellan-4 that impair the mobility of an mglA-8 masK-815 double mutant map within old and define new genes required for S motility:
To confirm the result that the masK-815 restores S but not A motility, we mutagenized the double mglA-8 masK-815 mutant strain, MxH1104, with transposon magellan-4, which confers kanamycin resistance (KanR). We electroporated MxH1104 with the donor, suicide plasmid pMycoMar (Rubin et al. 1999) and screened for triple mutants among 2000 independent KanR mutants of MxH1104 that form nonmotile colonies. Among these 2000 mutants, we recovered 22 that form nonmotile colonies. Chromosomal DNA was purified from these mutants, cleaved with restriction endonucleases that do not have recognition sites within the magellan-4 element, ligated, and used to electroporate an E. coli host expressing the Pir protein. Because the defective magellan-4 transposon carries the plasmid R6Kγ origin, its replication is conditionally dependent on the Pir protein. Thus, KanR recombinants of this E. coli host arising after electroporation carry plasmids with the entire magellan-4 element and flanking M. xanthus chromosomal DNA (Youderian et al. 2003; Youderian and Hartzell 2005). KanR plasmids with magellan-4 insertions were recovered from 12 of the nonmotile M. xanthus mglA-8 masK-815 magellan-4 triple mutants, and the sequences of both of the junctions between magellan-4 and the M. xanthus chromosome were determined for each insertion by using primers complementary to, and pointing outward from, the ends of magellan-4. These sequences were aligned with the sequence of the M. xanthus genome to determine the identities of the genes inactivated by magellan-4 insertions.
Table 1 summarizes the results of this analysis. Insertions of magellan-4 in all 12 nonmotile triple mutants are flanked by repeats of the dinucleotide TA, the preferred target for magellan-4 insertion. Nine of the 12 insertions are found to lie within the pilM, pilO, sglK, epsD, epsH, sgmU (MXAN_4707), and sgnC (MXAN_6627) genes, previously shown to be required for S motility. Three of the 12 insertions lie within three different genes, MXAN_4710, MXAN_6679, and MXAN_7103, previously not known to be required for S motility (Figures 1 and 2). Many M. xanthus strains that carry a mutation in an A motility gene and an S motility gene are completely devoid of motility (Spormann and Kaiser 1999).
Figure 1.—
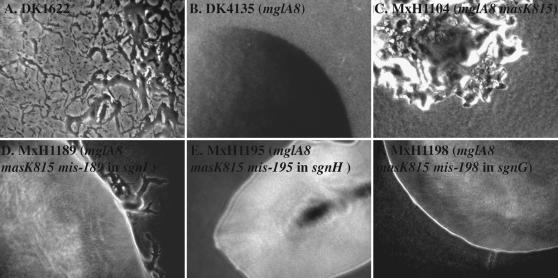
Colony morphologies of mutants with magellan-4 insertions that abolish S motility in the mglA-8 masK-815 genetic background. Colonies formed by strains (A) DK1622 (wild type), (B) DK4135 (mglA-8), (C), MxH1104 (mglA-8 masK-815), (D) MxH1189 (mglA-8 masK-815 mis-189), (E) MxH1195 (mglA-8 masK-815 mis-195), and (F) MxH1195 (mglA-8 masK-815 mis-198) are shown. The mis-189, mis-195, and mis-189 alleles are magellan-4 insertions in the MXAN_7103, MXAN_6679, and MXAN_4710 genes, respectively (Table 1), predicted to encode a homolog of the CheR methyltransferase named SgnI, a large protein with YD repeat motifs named SgnH and an enzyme that is involved in the LPS biosynthesis pathway named SgnG. Photographs were taken using a Nikon FXA microscope, and are shown at 15× magnification.
Figure 2.—
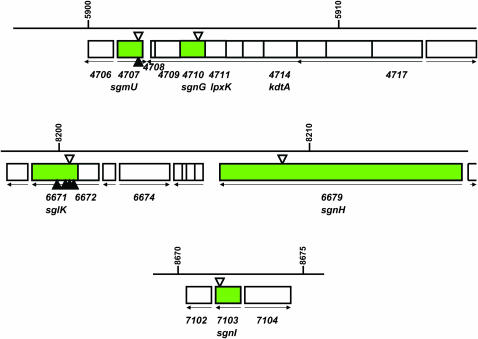
Magellan-4 insertions in the mglA-8 masK-815 genetic background identify three additional genes required for S motility. The regions of the M. xanthus genome with the MXAN_4710, MXAN_6679, and MXAN_7103 genes are shown. Genes are depicted as boxes, with arrows below the boxes indicating their directions of transcription. Coordinates and gene numbers are those of the M. xanthus genome sequence (http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org_search=&org=gmx). The positions of magellan-4 insertions obtained in the current mutant hunt are indicated by open triangles above the genes; those of magellan-4 insertions obtained in our previous mutant hunt (Youderian and Hartzell 2005) as filled triangles below the genes. Adjacent genes are considered to be in the same transcription unit if they are separated by <20 bp. (Top) Genes MXAN_4711 (lpxK) and MXAN_4714 (kdtA), predicted to be upstream of MXAN_4710 in the same operon, are predicted to encode essential functions required for the synthesis of the lipid A-KDO moiety of O-antigen to which heptoses are then attached. MXAN_4710 and MXAN_4709 together encode the two functional domains of the enzyme HldE (RfaE); MXAN_4710 is predicted to encode d-β-d-heptose 7-phosphate kinase, and MXAN4709 is predicted to encodes d-β-d-heptose 1-phosphate adenosyltransferase. MXAN_4708 is predicted to encode a 55 amino acid hypothetical protein. Gene MXAN_4717, which may be the first gene in this operon, is predicted to encode a protein with both DnaJ and response regulator domains. (Middle) The gene MXAN_6672 is predicted to be upstream of sglK (MXAN_6671) (Weimer et al. 1998) in the same operon. It likely encodes a homolog of GrpE, which participates with DnaK in the HSP70 chaperone complex (Szabo et al. 1994). We have yet to obtain magellan-4 insertions in this gene. If this gene is not essential (the M. xanthus MXAN_4331 gene likely encodes a second homolog of GrpE), then we predict that these insertions will also result in an S motility defect, because such insertions should, at the least, be polar on the expression of sglK. The sgnH gene (MXAN_6679) appears to be the only gene in its operon. (Bottom) The MXAN_7103 gene is predicted to encode a homolog of CheR; it also appears to be the only gene in its operon.
When the triple mutants (MxH1189, MxH1195, and MxH1198) were examined by time-lapse videomicroscopy, subtle differences in their motility were identified. More than 80% of wild-type (DK1622) cells move during the 20 min video with a typical speed of 2.7 μm/min (range 0.8–3.2 μm/min) on CTPM agar pads. In contrast, <5% of the cells of strain MxH1104 displayed any movement at all. Those that do move, move at the much slower rate of ∼0.1 μm/min. Isolated cells of MxH1104 do not show any movement. The majority of cells of strains MxH1189 and MxH1198 show no movement on CTPM agar pads, but a subset (5%) of MxH1195 cells show a rapid back-and-forth movement similar to that described by Spormann and Kaiser (1999) for mglA mutants.
Methylcellulose has been found to restore the motility of a subset of S mutants, because it can stimulate the retraction of type IV pili (Sun et al. 2000; Li et al. 2003). Therefore, we compared the motility of triple mutants with the motility of the wild-type and parental MxH1104 strains in the presence of 1% methylcellulose. In contrast with their movement on CTPM agar pads, most (>90%) MxH1104 cells, including isolated cells, were found to be motile in the presence of methylcellulose. Although their rate of movement (2.3 μm/min) is similar to that of the wild type, MxH1104 cells reverse direction every 1–2 min, whereas wild-type cells reverse direction approximately every 6 min. About one-third (28/80) of MxH1189 cells actively rotate in the presence of methylcellulose, whereas no MxH1189 cells show movement on a CTPM surface. The motility of MxH1195 is nearly identical in the presence of methylcellulose as it is on CTPM agar pads. Whereas MxH1198 cells are nonmotile on CTPM agar, >60% of these cells are motile in the presence of methylcellulose. However, these cells display erratic, jerky movements in contrast with the characteristic, smooth movement of wild-type or MxH1104 parent cells under the same conditions.
The fact that most of these insertions lie within genes required for S motility confirms our conclusion that the masK-815 suppressor restores S, but not A, motility in an mglA-8 genetic background. The majority of genes known to be required for S motility are required for the biogenesis of polar type IV pili, the exopolysaccharide (EPS) component of peritrichous fibrils, or lipopolysaccharide (LPS). In a previous mutant hunt for genes required for S motility, we mutagenized an A (ΔaglU) mutant of M. xanthus with magellan-4 and defined the sites of 128 independent insertions resulting in a nonmotile phenotype in this different genetic background. We found that the majority of magellan-4 insertions resulting in an S mutant phenotype inactivate genes in two large, contiguous regions of the M. xanthus genome, the pil and eps gene clusters, required for pili and EPS biogenesis, respectively. Four of the 128 insertions were found within sglK, only one insertion was found in the sgmU gene, predicted to encode a heptosyl transferase, and only one insertion was found in the sgnC gene, predicted to encode one of six different response regulators required for S motility (Youderian and Hartzell 2005).
Half (6/12) of the insertions resulting from our current mutant hunt lie within the pil and eps gene clusters, and 3 of the 12 insertions lie within the sglK, sgmU, and sgnC genes. The fact that we obtained independent insertions in our current hunt in the sgmU and sgnC genes, predicted to be the only genes in their transcription units (Figure 2), confirms that these genes are required for S motility. Among the 12 independent magellan-4 insertions that disrupt S genes, only 4 of these are found at the same sites in the genes we identified in our previous mutant hunt. This result shows that the spectrum of magellan-4 insertions that we have obtained in the mglA masK double mutant background is very different than the spectrum we obtained in the ΔaglU genetic background.
The addition of heptose to the lipid A-KDO component of LPS is required for S motility:
Three of the magellan-4 insertions define new S motility genes. One of these is in MXAN_4710, which appears to be the third to last gene in a large operon of at least seven genes predicted to encode proteins involved in the biosynthesis of the lipid A, KDO (3-deoxy-d-manno-octulosonic acid), and additional inner core components of LPS (Figure 2). The MXAN_4710 gene, identified by the mis-198 allele, is predicted to encode a protein with the N-terminal domain of the bifunctional enzyme RfaE, required for the synthesis of d-glycero-d-manno-heptose-1-phosphate. The C-terminal domain of RfaE required for the subsequent transfer of this substrate to ADP to form ADP-d-glycero-d-manno-heptose is predicted to be encoded by MXAN_4709, immediately downstream of MXAN_4710. It may be the case that this insertion not only inactivates MXAN_4710, but also is polar on the expression of MXAN_4709. The nearby convergently transcribed MXAN_4707 gene is predicted to encode RfaF, ADP-heptose:LPS heptosyltransferase, which uses the product of the reaction catalyzed by RfaE to add heptose to initiate the formation of the inner core of LPS. The result that we obtain insertions in both MXAN_4710 and MXAN_4707 and the fact that MXAN_4710, MXAN_4709, and MXAN_4707 are the only three genes present in the M. xanthus genome sequence predicted to encode activities required for the addition of heptose to LPS, argue that the addition of heptose to the inner core of LPS is required for S motility.
These results confirm the finding that the biosynthesis of O-antigen is required for S motility (Bowden and Kaplan 1998; Yang et al. 2000a,b; Youderian and Hartzell 2005). We do not as yet know whether LPS plays a direct role in the mechanism of S motility by participating in cell–cell recognition, or an indirect role in S motility, because mutations that result in defects in LPS biosynthesis have pleiotropic effects on the biogenesis of EPS, as well as on the proportionation of reduced carbon into biopolymers, since more than half of the reduced carbon in many Gram-negative bacteria is invested into O-antigen.
Using generalized transduction with phage Mx8, we were unable to backcross the mis-198 allele into a wild-type genetic background to confirm its role in S motility. This is because mutants with this mutation, like other “rough” mutants defective in O-antigen biosynthesis, are resistant to phage Mx8, which requires O-antigen for adsorption (Fink et al. 1989). Therefore, to determine if disruption of mis-198 affects S motility in a wild-type genetic background, chromosomal DNA from M. xanthus MxH1198 was transferred into M. xanthus DK1622 by electroporation. As shown in Figure 3, MxH1298 (which carries the sgnG∷magellan-4 insertion mis-198, in an otherwise wild-type genetic background) displays reduced spreading on 0.3% CTPM agar compared with that of the wild-type strain, but shows a near normal level of spreading on 1.5% agar, characteristic of mutants defective in S motility. As shown in Figure 4, isolated cells are present at the edges of colonies formed by the MxH1298 mutant strain.
Figure 3.—
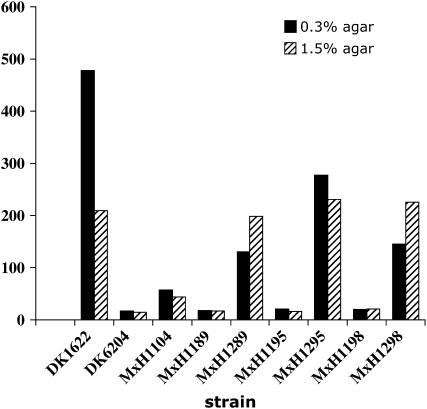
Magellan-4 insertions in the MXAN_4710 (sgnG), MXAN_6679 (sgnH), and MXAN_7103 (sgnI) genes result in defects in S motility but not A motility. The spreading areas of single (MxH1289, MxH1295, and MxH1298), double (DK6204 and MxH1104), and triple mutants (MxH1189, MxH1195, and MxH1198) are compared with the spreading of the wild-type strain (DK1622) on CTPM medium containing 0.3 and 1.5% agar. The final spreading area is the difference between the colony area at T = 0 and T = 120 hr after incubation at 32°.
Figure 4.—
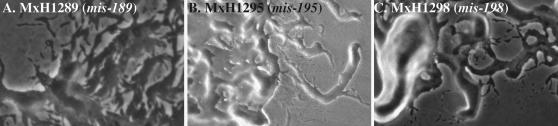
Gliding motility of isolated cells is evident in the MXAN_4710 (sgnG), MXAN_6679 (sgnH), and MXAN_7103 (sgnI) mutants. The colony edge morphology of M. xanthus strains (A) MxH1289, (B) MxH1295, and (C) MxH1298 are shown. The 15× (final magnification) images were taken with a Nikon FXA microscope.
A homolog of the CheR methyltransferase is required for S motility:
The second new S gene MXAN_7103 is predicted to encode a homolog of CheR methyltransferase and appears to be expressed in its transcription unit as a single gene (Figure 2). M. xanthus is predicted to encode nine paralogs of CheR, one of which is encoded by the frzF gene. FrzF methyltransferase is required for the sensory adaptation of M. xanthus in response to repellent chemotactic stimuli (Shi and Zusman 1993), and the multicellular development of M. xanthus at lower cell densities (Kashefi and Hartzell 1995). Although FrzF participates in the Frz chemotaxis system of M. xanthus, it is not required for social motility.
To confirm that the mis-189 insertion confers an S motility defect, we backcrossed this magellan-4 insertion into a wild-type genetic background by electroporation to produce M. xanthus MxH1289. All KanR electroporants (34/34) formed smaller colonies than their wild-type (DK1622) parent. As shown in Figures 3 and 4, the MxH1289 mutant is defective in spreading on 0.3% agar. MxH1289 is delayed in fruiting body formation and forms mounds after 4 days whereas the wild-type forms mounds within one day on TPM starvation agar (P. L. Hartzell, unpublished data). The production of heat-resistant spores is reduced 10-fold compared with the wild type.
The result that MXAN_7103, identified by the mis-189 allele, is essential for S motility suggests that a methyl-accepting chemotaxis protein must also play an essential role in S motility. This substrate of the MXAN_7103-encoded methyltransferase may be the product of the difA gene, predicted to encode a methyl-accepting chemotaxis protein, which is required for the production of extracellular fibrils, and therefore for S motility (Yang et al. 1998). However, because there is no evidence that methylation of DifA occurs (Bonner et al. 2005), the product of MXAN_7103 may participate in a different chemotaxis pathway required for S motility. We are currently testing whether a mutation in this gene affects EPS production. Although the dif gene cluster encodes other homologs of chemotaxis proteins as well as DifA, it does not include a gene encoding a paralog of CheR (Yang et al. 1998). Pseudomonas aeruginosa also has cluster of chemotaxis genes required for S (or “twitching” motility). This cluster includes a gene encoding a required methyl-accepting chemotaxis protein, but the gene encoding its partner methyltransferase is not linked with this cluster (Kato et al. 1999).
The MXAN_6679 gene, predicted to encode a large YD repeat protein, is required for S motility:
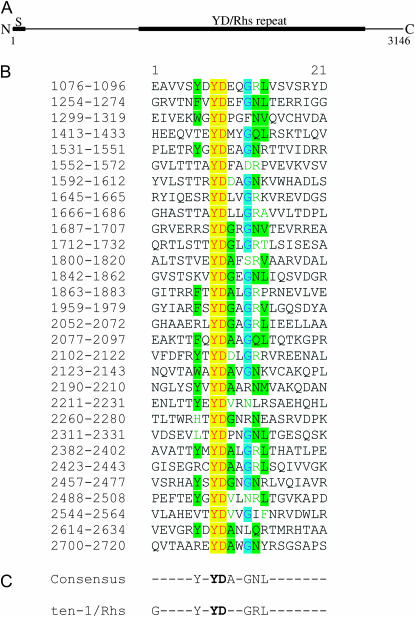
The third new S gene MXAN_6679 also appears to be expressed in its transcription unit as a single gene. It is predicted to encode a product with a N-terminal signal sequence, which is cleaved after residue 30, to yield a protein with a predicted molecular mass of ∼340,000 Da. The 3146 amino acid protein product of MXAN_6679 has an unusual domain structure that includes at least 30 repeats that are typically 21 amino acids in length and include the dipeptide Tyr–Asp (YD, Figure 5). These repeats are found in very few proteins, including the predicted protein products of the rhs (rearrangement hot spot) elements of E. coli (Hill et al. 1994), the cell-wall associated WapA protein of Bacillus subtilis (Foster 1993), toxin A of Clostridium difficile (Dove et al. 1990), the first pair-rule protein Ten-m of Drosophila melanogaster (Baumgartner et al. 1994; Levine et al. 1994), and the vertebrate teneurin-1 protein (Minet et al. 1999). The four latter proteins are secreted extracellular proteins; among these, the vertebrate protein is critical for neuronal development (Minet et al. 1999). Although little is known about the function of the YD repeat motifs in WapA and Ten-m, the YD repeat motifs of both C. difficile toxin A and of teneurin-1 have been shown to bind carbohydrate (Krivan et al. 1986; Wren 1991; Minet et al. 1999), and the E. coli RhsA protein has been shown to be involved in the export of EPS (Mcnulty et al. 2006).
Figure 5.—
MXAN_6679 is predicted to encode a protein product with 36 YD repeats. The product of MXAN_6679 (sgnH) is predicted to be a 340-kDa protein. The presence of an N-terminal signal sequence, shown in A, suggests that SgnH is secreted. Over half of the protein includes a series of YD/Rhs repeats (solid bar from 1076 to 2720). Rhs proteins contain extended repeat regions that are thought to be involved in ligand binding. The extracellular repeat contains the dipeptide YD. Potential YD repeats in the predicted product of the MXAN_6679 (sgnH) gene were aligned by visual inspection and by SMART (Simple Modular Architecture Research Tool) LeTunic et al. 2002. Repeat regions, shown in B, are numbered by the residue numbers of the predicted protein product of the sgnH gene. The consensus sequence of the YD repeats shared by the vertebrate teneurin-1 protein and the predicted products of the E. coli rhs genes (Minet et al. 1999) is shown in C. Amino acids occurring in the consensus sequence are shaded.
To confirm that the mis-195 insertion in MXAN_6679 confers an S motility defect, we backcrossed this magellan-4 insertion into a wild-type genetic background to produce M. xanthus MxH1295. The KanR determinant in MxH1195 (mglA-8 masK-815 mis-195) is 100% linked to a defect in S motility. All KanR electroporants form smaller colonies than their wild-type parent (DK1622) that are defective in spreading on 0.3% agar (Figure 3). Their ability to spread on 1.5% agar is similar to that of the wild type. Isolated cells are detected at the edge of vegetative colonies formed by MxH1295, characteristic of A motility (Figure 4). MxH1295 is able to produce fruiting bodies that are similar in size and color with those of the wild type, but the production of heat-resistant spores in these fruit is reduced 90-fold (P. L. Hartzell, unpublished data).
What role might this YD repeat protein play in the mechanism of S motility of M. xanthus? In D. melanogaster, the Ten-m protein was identified using an antibody specific for phosphotyrosine (Levine et al. 1994). Thus, YD repeats may represent targets for tyrosine phosphorylation, perhaps by the essential MasK tyrosine kinase of M. xanthus (Thomasson et al. 2002). If the product of MXAN_6679 is secreted, as we predict, then it may play a role for sensing the presence of the carbohydrate polymers that are a component of EPS. Additional biochemical studies of this unusual protein product, and a more careful analysis of its roles in S motility and multicellular development, may help us understand why the Rhs elements, predicted to encode similar YD repeat proteins, have been acquired recently by the Gram-negative enteric pathogens (Hill et al. 1994).
DISCUSSION
When we compare the results of a previous mutagenesis to obtain magellan-4 insertions in S genes starting with an A mutant genetic background, we find that the spectrum of magellan-4 insertions in S motility genes is different from that when we screen for nonmotile mutants starting with the mglA-8 masK-815 genetic background. Although many of the insertions we obtained in the latter double mutant background are in the same genes “hit” with 128 independent insertions starting with an A mutant, 3 of 12 are not. This is not surprising. The differences in genetic background may favor the recovery of a different subset of mutations in S motility genes. Such differences also help to distinguish components that function in the signaling pathways needed for S motility from structural components. We did not recover magellan-4 insertions in mglA or masK although disruption of either gene in the M. xanthus MxH1104 background would be expected to yield nonmotile colonies. This result is consistent with our previous results showing that masK is an essential gene. Disruption of mglA in a wild-type background does not affect cell growth and hence mglA is not essential. However the pairing of the mglA8 mutation with the masK-815 suppressor of mglA might make it difficult to identify strains with insertions in mglA.
We find that a mutation in MXAN_4707 (sgnG; M. xanthus MxH1198), which is predicted to encode an enzyme required for addition of heptose to the inner core of LPS, abolishes motility on CTPM agar. However, the fact that methylcellulose stimulates the movement (albeit jerky) of this mutant suggests that its type IV pili are capable of retraction. Hence, SgnG may play a role in the signaling pathway that stimulates the retraction of type IV pili, required for S motility. The motility of cells carrying a disruption of the sgnH gene (MXAN_6679), predicted to encode a 340-kDa extracellular YD-repeat protein, is not stimulated by the addition of methylcellulose. This result suggests that SgnH may be a structural component for the S motor. Finally, cells carrying a disruption of the sgnI gene (MXAN_7103), predicted to encode a protein with methyltransferase activity, exhibit rotational movement that occurs only in the presence of methylcellulose. We speculate that this rotational movement indicates that the mutant can produce and retract type IV pili, yet is blocked in the ability to switch the extension of pili from one cell pole to the other. Hence, these cells, which adhere by their polar pili to the surface of a microscope slide, can retract their pili when methylcellulose is present. If SgnI is involved in polar switching, it may be an integral part of the Mgl signal transduction pathway that coordinates reversals of the A and S motors. Specifically, SgnI may an S-gliding specific component that is downstream of the Mgl coordination signal, so a mutation in sgnI would not be expected to affect A gliding.
The 140 magellan-4 insertions, which we have characterized in a recent and in this current study, identify 36 new genes required for S motility; when added to the 77 genes characterized previously, we now know the identities of >113 different genes required for S motility. The result that 3 among 12 independent insertions of magellan-4 in an mglA-8 masK-815 genetic background that impair motility identify three new genes involved in S motility argues that we have not saturated the genes required for S motility. This idea is supported by the fact that we have found magellan-4 insertions in only three-quarters of the genes shown to be involved in S motility by all genetic methods employed to date. The mechanism of S motility is extraordinarily complex and will likely involve two to four times the number of genes required for the mechanism of flagellar-dependent motility in the Gram-negative enteric bacteria.
The choice of different starting genetic backgrounds for screening S mutants made by transposon insertions will likely improve our chances of finding additional S motility genes. Thus, we expect that the characterization of additional magellan-4 insertions in the double mutant mglA masK genetic background will reveal additional genes required for S motility. Second, because many S genes are clustered in the genome sequence, detailed analyses of mutations in nearby genes likely will identify additional genes required for S motility. This has certainly proven to be the case for the genes required for the biogenesis of EPS. For example, the result that both the sglK (MXAN_6671) and sgnH (MXAN_6679) genes are required for S motility suggests that the seven genes between these two also may be required for S motility. Third, because many of the genes known to be involved in S motility are predicted to encode products of related function, genome-based approaches involving the systematic inactivation of genes predicted to encode functions similar to those of known S motility genes will certainly complement our more classical genetic methods, including transposon mutagenesis, to identify additional S motility genes.
Acknowledgments
This work was supported by a grant MCB0242191 from the National Science Foundation to P.L.H. and grants GM59336 and GM075242 from the National Institutes of Health to P.Y. and P.L.H.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Baumgartner, S., D. Martin, C. Hagios and R. Chiquet-Ehrismann 1994. Tenm, a Drosophila gene related to tenascin, is a new pair-rule gene. EMBO J. 13: 3728–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner, P. J., Q. Xu, W. P. Black, Z. Li, Z. Yang and L. J. Shimkets 2005. The Dif chemosensory pathway is directly involved in phosphatidylethanolamine sensory transduction in Myxococcus xanthus. Mol. Microbiol. 57: 1499–1508. [DOI] [PubMed] [Google Scholar]
- Bowden, M. G., and H. B. Kaplan, 1998. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 30: 275–284. [DOI] [PubMed] [Google Scholar]
- Caberoy, N. B., R. D. Welch, J. S. Jakobsen, S. C. Slater and A. G. Garza, 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185: 6083–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove, C. H., S. Z. Wang, S. B. Price, C. J. Phelps, D. M. Lyerly et al., 1990. Molecular characterization of the Clostridium difficile toxin A gene. Infect. Immun. 58: 480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink, J. M., M. Kalos and J. F. Zissler 1989. Isolation of cell surface antigen mutants of Myxococcus xanthus by use of monoclonal antibodies. J. Bacteriol. 171: 2033–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, S. J., 1993. Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two-domain ligand-binding protein. Mol. Microbiol. 8: 299–310. [DOI] [PubMed] [Google Scholar]
- Hartzell, P. L., 1997. Complementation of Myxococcus xanthus sporulation and motility defects by a eukaryotic RAS homolog. Proc. Natl. Acad. Sci. USA 97: 9881–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell, P. L., and D. Kaiser, 1991. a Function of MglA, a 22-Kilodalton Protein Essential for Gliding in Myxococcus xanthus. J. Bacteriol. 173: 7615–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell, P. L., and D. Kaiser, 1991. b Upstream gene of the mgl operon controls the level of MglA protein in Myxococcus xanthus. J. Bacteriol. 173: 7625–7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell, P. L., W. Shi and P. Youderian, 2007. Gliding motility of Myxococcus xanthus, pp. 103–122 in Myxobacteria: Multicellularity and Differentiation, edited by D. Warick. ASM Press, Washington, DC.
- Hill, C. W., C. H. Sandt and D. A. Vlazny, 1994. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol. Microbiol. 12: 865–871. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., and D. Kaiser, 1979. a Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171: 167–171. [Google Scholar]
- Hodgkin, J., and D. Kaiser, 1979. b Genetics of gliding motility in Myxococcus xanthus (Myxobactererales): two gene systems control movement. Mol. Gen. Genet. 171: 177–191. [Google Scholar]
- Kashefi, K., and P. L. Hartzell, 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF- defect. Mol. Microbiol. 15: 483–494. [DOI] [PubMed] [Google Scholar]
- Kato, J., T. Nakamura, A. Kuroda and H. Ohtake, 1999. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 63: 155–161. [DOI] [PubMed] [Google Scholar]
- Krivan, H. C., G. F. Clark, D. F. Smith and T. D. Wilkins, 1986. Cell surface binding site for Clostridium difficile enterotoxin: evidence for a glycoconjugate containing the sequence Gal alpha 1–3Gal beta 1–4GlcNAc. Infect. Immun. 53: 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancero, H., N. B. Caberoy, S. Castaneda, Y. Li, A. Lu et al., 2004. Characterization of a Myxococcus xanthus mutant that is defective for adventurous motility and social motility. Microbiology 150: 4085–4093. [DOI] [PubMed] [Google Scholar]
- Letunic, I., L. Goodstadt, N. J. Dickens, T. Doerks, J. Schultz et al., 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30: 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, A., A. Bashan-Ahrend, O. Budai-Hadrian, D. Gartenberg, S. Menasherow et al., 1994. Odd Oz: a novel Drosophila pair rule gene. Cell 77: 587–598. [DOI] [PubMed] [Google Scholar]
- Li, Y., H. Sun, X. Ma, A. Lu, R. Lux et al., 2003. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100: 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, A., K. Cho, W. P. Black, X. Y. Duan, R. Lux et al., 2005. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol. Microbiol. 55: 206–220. [DOI] [PubMed] [Google Scholar]
- Macneil, S. D., F. Calara and P. L. Hartzell, 1994. a New clusters of genes required for gliding motility in Myxococcus xanthus. Mol. Microbiol. 14: 61–71. [DOI] [PubMed] [Google Scholar]
- Macneil, S. D., A. Mouzeyan and P. L. Hartzell, 1994. b Genes required for both gliding motility and development in Myxococcus xanthus. Mol. Microbiol. 14: 785–795. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., and S. H. Bryant, 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32: W327–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcnulty, C., J. Thompson, B. Barrett, L. Lord, C. Andersen et al., 2006. The cell surface expression of group 2 capsular polysaccharides in Escherichia coli: the role of KpsD, RhsA and a multi-protein complex at the pole of the cell. Mol. Microbiol. 59: 907–922. [DOI] [PubMed] [Google Scholar]
- Mignot, T., J. P. Merlie, Jr. and D. Zusman, 2005. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science 310: 855–857. [DOI] [PubMed] [Google Scholar]
- Mignot, T., J. W. Shaevitz, P. Hartzell and D. Zusman, 2007. Evidence that focal adhesion complexes power bacterial gliding motility. Science 315: 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet, A. D., B. P. Rubin, R. P. Tucker, S. Baumgartner and R. Chiquet-Ehrismann, 1999. Teneurin-1, a vertebrate homologue of the Drosophila pair-rule gene ten-m, is a neuronal protein with a novel type of heparin-binding domain. J. Cell Sci. 112: 2019–2032. [DOI] [PubMed] [Google Scholar]
- Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson et al., 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96: 1645–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, W., and D. R. Zusman, 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. USA 90: 3378–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spormann, A. M., and D. Kaiser, 1999. Gliding mutants of Myxococcus xanthus with high reversal frequencies and small displacements. J. Bacteriol. 181: 2593–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, K., and D. Kaiser, 1987. Genetics of gliding in Myxococcus xanthus: molecular cloning of the mgl locus. Mol. Gen. Genet. 207: 256–266. [Google Scholar]
- Stephens, K., P. L. Hartzell and D. Kaiser, 1989. Gliding motility in Myxococcus xanthus: mgl locus, RNA, and predicted protein products. J. Bacteriol. 171: 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., D. R. Zusman and W. Shi, 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10: 1143–1146. [DOI] [PubMed] [Google Scholar]
- Szabo, A., T. Langer, H. Schroder, J. Flanagan, B. Bukau et al., 1994. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl. Acad. Sci. USA 91: 10345–10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasson, B., J. Link, A. G. Stassinopoulos, N. Burke, L. Plamann et al., 2002. The GTPase, MglA, interacts with a tyrosine kinase to control type-IV pili-mediated motility of Myxococcus xanthus. Mol. Microbiol. 46: 1399–1413. [DOI] [PubMed] [Google Scholar]
- Weimer, R.M., C. Creighton, A. Stassinopoulos, P. Youderian and P. L. Hartzell, 1998. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J. Bacteriol. 180: 5357–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren, B. W., 1991. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol. Microbiol. 5: 797–803. [DOI] [PubMed] [Google Scholar]
- Wu, S. S., and D. Kaiser, 1995. Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18: 547–558. [DOI] [PubMed] [Google Scholar]
- Yang, R., S. Bartle, R. Otto, A. Stassinopoulos, M. Rogers et al., 2004. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J. Bacteriol. 186: 6168–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Y. Geng and W. Shi, 1998. A DnaK homolog in Myxococcus xanthus is involved in social motility and fruiting body formation. J. Bacteriol. 180: 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., D. Guo, M. G. Bowden, H. Sun, L. Tong et al., 2000. a The Myxococcus xanthus wgbB gene encodes a glycosyltransferase homologue required for lipopolysaccharide O-antigen biogenesis. Arch. Microbiol. 174: 399–405. [DOI] [PubMed] [Google Scholar]
- Yang, Z., X. Ma, L. Tong, H. B. Kaplan, L. J. Shimkets et al., 2000. b Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182: 5793–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian, P., and P. L. Hartzell, 2005. Transposon insertions of magellan-4 that impair social gliding motility in Myxococcus xanthus. Genetics 172: 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian, P., N. Burke, D. J. White and P. L. Hartzell, 2003. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol. Microbiol. 49: 555–570. [DOI] [PubMed] [Google Scholar]