Abstract
Schizosaccharomyces pombe cells can switch between two mating types, plus (P) and minus (M). The change in cell type occurs due to a replication-coupled recombination event that transfers genetic information from one of the silent-donor loci, mat2P or mat3M, into the expressed mating-type determining mat1 locus. The mat1 locus can as a consequence contain DNA encoding either P or M information. A molecular mechanism, known as synthesis-dependent strand annealing, has been proposed for the underlying recombination event. A key feature of this model is that only one DNA strand of the donor locus provides the information that is copied into the mat1. Here we test the model by constructing strains that switch using two different mutant P cassettes introduced at the donor loci, mat2 and mat3. We show that in such strains wild-type P-cassette DNA is efficiently generated at mat1 through heteroduplex DNA formation and repair. The present data provide an in vivo genetic test of the proposed molecular recombination mechanism.
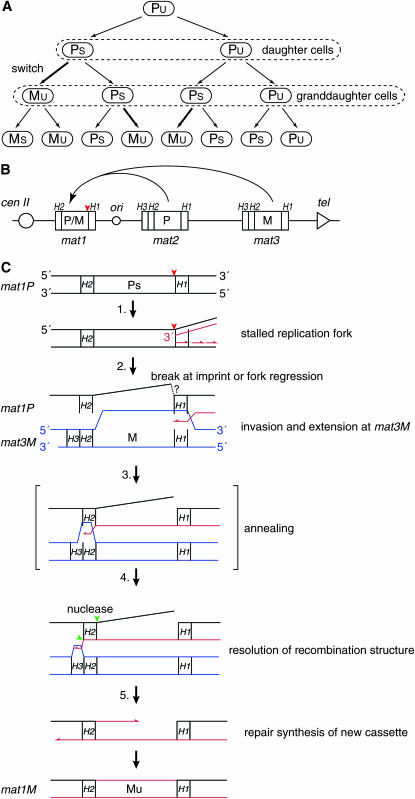
SCHIZOSACCHAROMYCES pombe mating-type switching is the first system identified where a stalled replication fork acts to induce a programmed DNA rearrangement required for cellular differentiation (Egel et al. 1984; Arcangioli 1998; Dalgaard and Klar 1999, 2000; Arcangioli and De Lahondes 2000). Thus, characterization of the underlying recombination event is crucial for gaining a more general understanding not only of the molecular responses to stalled replication forks, but also of cellular differentiation mechanisms. S. pombe is a fission yeast that lives predominantly in a haploid state (reviewed by Egel 1989). Only during nutritional starvation will cells of the two opposite mating types, called plus (P) and minus (M), mate. Generally, the diploid zygote forms and then directly undergoes meiosis followed by sporulation leading to the formation of an ascus containing four haploid spores. Cells of homothallic S. pombe strains are able to highly efficiently switch between the two mating types (Leupold 1950; Miyata and Miyata 1981). The switching occurs by a specific pattern: analysis of S. pombe switching pedigrees has established that cell division of an “unswitchable” cell leads to the formation of a “switchable” and an unswitchable daughter cell, both of the parental mating type, while cell division of a switchable cell gives rise to an unswitchable daughter cell of the opposite mating type and a switchable daughter cell of the parental mating type (a P-to-M pedigree is shown in Figure 1A; Miyata and Miyata 1981; Egel 1984; Egel and Eie 1987; Klar 1987, 1990b). The two cell types, P and M, are genetically different as they possess two different gene cassettes at the mat1 locus located on chromosome II (Egel and Gutz 1981; Beach 1983; Beach and Klar 1984; Kelly et al. 1988; Klar 1990b). Furthermore, switchable cells of both the P- and the M-cell types carry an epigenetic modification of the mat1-cassette DNA (Egel 1984; Egel and Eie 1987; Klar 1987, 1990b; Klar and Bonaduce 1993). This modification, which historically has been referred to as an imprint, is required for induction of the replication-coupled recombination that underlies the mating-type switching process (Egel et al. 1984; Dalgaard and Klar 2000; Holmes et al. 2005). The recombination event acts to copy the genetic information encoded by one of the two transcriptional silenced donor loci mat2P and mat3M into the expressed mat1 locus, thus leading to the change in the cell's mating type (Egel 1977; Egel and Gutz 1981; Beach 1983; Beach and Klar 1984; Kelly et al. 1988, see below). Importantly, pedigree experiments have established that the process of mating-type switching is continuously ongoing where each division propels cells through this program of cellular differentiation (Egel 1984; Egel and Eie 1987; Klar 1987, 1990b).
Figure 1.—
Molecular mechanism of S. pombe mating-type switching. (A) Switching pedigree. Unswitchable cell divides to form an unswitchable (lowercase “u”) and a switchable (lowercase “s”) daughter cell. Cell division of the switchable daughter yields a switchable granddaughter cell and a granddaughter cell that has switched mating type. (B) The mating-type region on chromosome II harbors the three mating-type loci: mat1, mat2P, and mat3M. The mat1 locus encodes either P or M information and is transcriptionally active while the donor loci, mat2P and mat3M, are transcriptionally silent. Homology domains H1, H2, and H3 flank the cassettes. An origin of replication (ori) is located between the mat1 and mat2 cassettes. The relative positions of the centromere and the telomere are given. The position of the imprint is indicated with a red arrowhead. There are interesting consequences of this highly efficient process that are of importance for this study. Theoretically, 50% of the mat1 cassettes will represent switching events that occurred in the last two generations. Also, the individual switching events will have given rise only to a very minor fraction of the mat1 cassettes present in the population. Importantly, pedigree analysis of switching cells has determined switching rates that closely correspond to these theoretical values; in the wild-type homothallic strains both the rate of initial switching and the rate of recurrent switching are ∼90% of that predicted (Klar 1990a, 1991). (C) The proposed model for the underlying recombination mechanism that transfers mating-type cassette information from one of the two donor loci into the mat1 locus. Only a P-to-M switch is shown. (1) The replication fork (red lines) initiated at a cenII-distal origin stalls at the imprint (red arrowhead) present in the template of the leading-strand polymerase. (2) A 3′ single-strand end is formed either by hydrolysis at the imprint followed by 5′ recession mediated by an exonuclease, or by fork regression. The 3′ end invades at the homology of the H1 domain in the donor cassette (mat3M cassette shown; blue lines) and one strand of the new cassette is synthesized using the donor as template. (3) When the replication fork passes through the donor cassette's H2 domain, homology to the mat1-H2 domain is created. The homology allows annealing between the newly synthesized H2 sequence and the older mat1-H2 sequence. (4) Resolution by flap endonucleases leads to the removal of the old outgoing cassette strand as well as newly synthesized nonhomologous sequences cenII-proximal to the donor locus' H2 domain (green arrowheads). (5) The second strand of the new cassette is synthesized, using the newly copied strand as a template. Ligation leads to the establishment of the intact chromatid containing a newly switched mat1 cassette.
The molecular mechanism that establishes the observed asymmetrical pattern of switching relies on the asymmetry of the DNA replication process (Dalgaard and Klar 2001b; Dalgaard and Vengrova 2004). Central to the mechanism is that the mat1 locus is replicated in a unidirectional manner by replication forks initiated at a mat1 centromere-distal origin (Dalgaard and Klar 1999, 2001a). This unidirectional replication dictates that one of the two mat1 DNA strands (the “upper” strand, Figure 1B) always acts as a template for leading-strand replication and the other (“lower”) for lagging-strand replication. Two different molecular events, each associated with one of the two replication processes, act in concert to establish the asymmetrical switching pattern of S. pombe. During each S phase the sister chromatid where the mat1 locus is replicated as lagging strand is imprinted/epigentically modified by introduction of ribonucleotides (Arcangioli 1998; Dalgaard and Klar 1999, 2001a; Kaykov and Arcangioli 2004). Experiments have established that the imprinting process is tightly linked with the replication process. A mutation (swi7-1) that abolishes imprinting has been identified in polymerase-α, which is responsible for Okazaki fragment synthesis (Singh and Klar 1993). In addition, genetic alterations that relax, invert, and, subsequently, restore the strict control of the direction of replication at mat1 lead to corresponding reduction, abolishment, and restoration of mat1 imprinting (Figure 1B) (Dalgaard and Klar 1999, 2000, 2001a). Also characterization of synchronized cultures has shown that imprinted mat1 DNA appears concurrently with mat1 replication (Holmes et al. 2005). Finally, the characterization of replication intermediates has established that mat1 imprinting correlates with replication-fork pausing in the proximity of the H1-mat1 cassette junction (Dalgaard and Klar 2000; Kaykov et al. 2004). Furthermore, the molecular anatomy of the paused replication fork suggests that the mat1 pause signal is read during lagging-strand replication (Vengrova and Dalgaard 2004). It is not known how the mat1 imprint is introduced, but experiments have shown that it is constituted by two ribonucleotides (Vengrova and Dalgaard 2004, 2006). These ribonucleotides are incorporated into the DNA such that a DNA-(RNA)2-DNA hybrid strand is formed at the precise junction between the mat1 cassette and the H1 homology domain. The two ribonucleotides are maintained in the DNA, and the imprinted chromosome is inherited by one daughter cell. Here, the imprint licenses the cell for mating-type switching during the next S phase (Figure 1C) (Egel 1984; Klar and Bonaduce 1993; Arcangioli and De Lahondes 2000; Dalgaard and Klar 2001a). The replication-coupled recombination event underlying mating-type switching is initiated when the leading-strand polymerase encounters the ribonucleotides present in the mat1 template DNA and is stalled in its progression (Kaykov et al. 2004; Vengrova and Dalgaard 2004). The precise 3′ end of the newly synthesized leading strand of the stalled replication fork has been mapped to the nucleotide preceding the imprint. Furthermore, intermediates suggestive of fork regression can be observed at the stalled replication fork when mat1-replication intermediates are analyzed by two-dimensional gels (Vengrova and Dalgaard 2004). However, it is not known whether the stalled replication fork is resolved by hydrolysis of the ribonucleotide imprint, leading to a single-sided double-stranded break (DSB), or that a double-stranded end generated by fork regression acts to induce recombination (Vengrova and Dalgaard 2004). In either case, it has been proposed that the recombination event, resembling synthesis-dependent strand annealing (SDSA), proceeds as follows (Arcangioli 1998; Arcangioli and De Lahondes 2000; Dalgaard and Klar 2001b): (i) the homology domain H1 present at the 3′ end invades at one of the two donor cassettes' H1 domains; (ii) the invading 3′ end is extended across the donor locus until homologous sequence is synthesized at the H2 domain; and (iii) the synthesized H2 DNA anneals to the homologous mat1 H2 sequence, thus allowing resolution by endonucleases (most likely involving the Swi4, Swi8, Swi9, and Swi10 factors, Egel et al. 1984; Schmidt et al. 1989) and repair by polymerase(s) and DNA ligase (Figure 1C). Two different types of evidence support this model; first, intermediates can be detected by PCR that link the cen-distal side of mat1 to the cen-proximal side of either donor loci (Arcangioli and De Lahondes 2000). Second, experiments using shifts from media containing heavy radioisotopes to media that contain light radioisotopes have shown that both DNA strands of a newly switched mat1 cassette are newly synthesized (Arcangioli 2000). However, the precise molecular mechanism still remains unknown and is the subject of this study.
We present here an in vivo test of the proposed recombination mechanism by testing its unique prediction; during every switching event a heteroduplex can potentially form between the outgoing “old” mat1-cassette strand and the incoming “new” cassette strand that has been copied from one of the donor loci (Figures 1C and 2). In wild-type cells, when switching occurs between P and M cassettes, the lack of homology between the cassettes prevents heteroduplex formation. Meanwhile, if cells were switching between two cassettes with related sequences, the formed mat1 heteroduplex would potentially be recognized and repaired by cellular heteroduplex repair enzymes leading to high levels of mitotic gene conversion at mat1. We test this prediction by constructing experimental strains that switch using two different dysfunctional mutant alleles of the P cassette introduced at the donor loci mat2 and mat3 and establish that switching in such strains allows efficient restoration of functional wild-type mat1P cassette DNA through heretoduplex formation and repair.
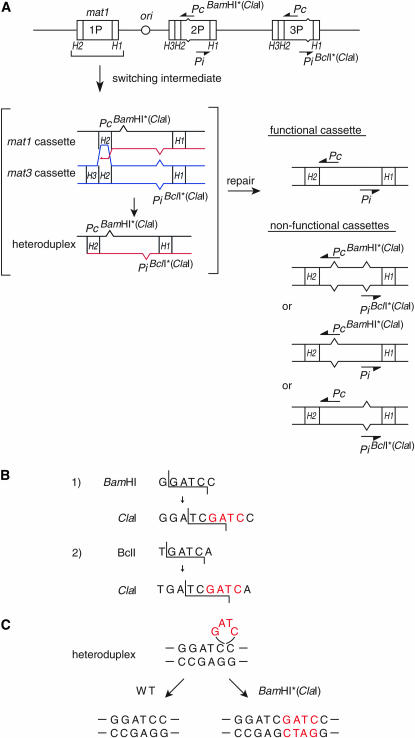
Figure 2.—
Heteroduplex DNA is predicted to form during switching between homologous sequences. (A) Line drawing of the mat2PBamHI* mat3PBclI* mutant mating-type region. The two transcriptional units, Pc and Pi, are indicated, as well as the relative position of the introduced mutations. A diagram, given in brackets, displays the predicted heteroduplex intermediate formed during switching, between the outgoing old strand and the incoming newly synthesized strand. The displayed intermediate corresponds to that enclosed in brackets in Figure 1C. Only the heteroduplex DNA species formed during a switch when a mat1PBamHI* cassette is changed to a mat1PBclI* cassette is shown. DNA mismatch repair (horizontal arrow) can repair this heteroduplex to form either a functional (top) or three nonfunctional cassettes (bottom). (B) Intact and mutated BamHI and BclI restriction sites. The ClaI site was created by filling in the BamHI (1) or BclI (2) restriction sites by introducing four nucleotides (GATC; red letters). (C) Heteroduplex DNA (GATC; red letters) can be repaired by two pathways leading to either the removal or the maintenance of the inserted nucleotides. Only heteroduplex DNA formed at the BamHI site is shown.
MATERIALS AND METHODS
Strain construction:
Mutant alleles were constructed using plasmids pAK67 (A. Klar) and pGT67 (G. Thon). pAK67 contains the mat2P HindIII fragment where the ura4+ gene has been integrated at the XbaI site. pGT67 (Thon and Klar 1992) contains a synthetic mat3P 4.2-kb HindIII fragment of a mat3P cassette where the ura4+ gene has been integrated at the EcoRV site (gift from G. Thon). The mutant mat3P alleles were integrated into the genome of strain pG598 using standard techniques (Moreno et al. 1991). The clr1 mutation allows recombination in the otherwise recombinationally inert K region located between mat2 and mat3 (Thon and Klar 1992). Subsequently, 5-fluoroorotic acid-resistant ura4 colonies were isolated. The obtained strains were transformed with the mat2P mutant alleles, and leucine auxotroph, uracil prototroph colonies were selected. The desired genotypes were verified by Southern analysis. Such strains were crossed with tester strain JZ108 to cross out the clr1 mutation. clr1+ segregants were identified by detection of epigenetic silencing of the ura4+ marker integrated in the mating-type region (Thon and Klar 1992). The strains were reanalyzed by Southern analysis to verify the genotype of the mating-type region (see supplemental data at http://www.genetics.org/supplemental/).
Haploid strains containing M-cassette information at the ade6 locus were constructed as follows. M-cassette DNA was amplified using primers mat1M-forward (5′-AAAGGATCCTTATAATTGTTGTGTCTTTTTT-3′) and mat1M-reverse (5′-AAAACTGCAGATTGAAAATAAATAAAAACG-3′) and cloned into pCR4-Blunt TOPO (Invitrogen, Carlsbad, CA) to obtain plasmid TY22. The ade6+ PCR product obtained using primers test-1 (5′-AAAACTGCAGCCAACGTTGCATTCTAATGAGCAAAG-3′) and test-2 (5′-AAAACTGCAGCCAATATATTTAGAATTAGCAATGA-3′) was digested with PstI and cloned into the PstI site (underlined in primer mat1M-reverse) present in plasmid TY22 to obtain plasmid TY33. Plasmid TY33 was transformed into strains JZ44, JZ149, TY07, and Ade+ transformants were selected. Southern analysis was performed using a probe specific to ade6 to verify the correct integration of the plasmid TY33 into the ade6 locus, as well as with a probe specific to the P cassette to make sure that no rearrangements had occurred in the mating-type region in the process of strain construction.
Quantifications of sporulation:
Mating mix:
Cells grown in liquid YEA were mixed with an equal number of the M-tester cells and patched on PMA+ plates. Plates were incubated at 30°. Cells, zygotic cells, and asci were counted on the third and fourth days. The experiment was repeated three times.
Sporulation assays using diploid cells:
Diploid cells were isolated by selection of ade6 complementation. Such strains were grown as single colonies on PMA+ plates at 30°. Sporulation was determined from five different single colonies of each strain on the fourth day. The experiment was repeated three times, except for TY07 where only two experiments were performed. Thus, the standard error on the measurement of the sporulation frequency is given for the TY07 strain in Figure 5. The strains' genotypes are: JZ1, h90 ade6-M210 leu1-32 ura4-D18; JZ5, mat1M smt-0 ade6-M216 leu1-32; JZ36, mat2PBamHI(BglII)∷LEU2,(XbaI)∷ura4+ ade6-M210 leu1-32 ura4-D18 clr1-5; JZ44, mat2P(BglII)∷LEU2 mat3P(EcoRV)∷ura4+ ade6-M210 leu1-32 ura4-D18; JZ60, h90 ade6-M216 leu1-32; JZ67, mat2PBclI(XbaI)∷ura4+ ade6-M210 leu1-32 ura4-D18 clr1-5; JZ108, mat1M smt-0 Δmat2,3∷LEU2 ade6-M216 leu1-32 his2; JZ109, mat1M smt-0 Δmat2,3∷LEU2 ade6-M210 leu1-32 his2; JZ149, mat2PBclI(XbaI)∷ura4+ mat3PBamHI(EcoRV):ura4FOAr ade6-M210 leu1-32 ura4-D18; TY07, mat2PBamHI(XbaI)∷ura4+ mat3PBclI(EcoRV)∷ura4FOAr ade6-M216 leu1-32 ura4-D18; pG598, h90 mat2(BglII)∷LEU2 ade6-M216 leu1-32 ura4-D18 clr1-5 (gift from G. Thon); TY128, mat2P(BglII)∷LEU2 mat3P(EcoRV)∷ura4+ msh6∷arg3 ade6-M210 leu1-32; TY131, mat2PBclI(XbaI)∷ura4+ mat3PBamHI(EcoRV)∷ura4FOAr msh6∷arg3 leu1-32; TY134, mat2PBamHI(XbaI)∷ura4+ mat3PBclI(EcoRV)∷ura4FOAr msh6∷arg3 leu1-32; OL712, h− msh6∷arg3 arg3-D4 (gift from O. Fleck); TY34, mat2P(BglII)∷LEU2 mat3P (EcoRV)∷ura4+ ade6-M210∷pTy33(Ade6+) leu1-32 ura4-D18; TY36, mat2PBclI(XbaI)∷ura4+ mat3PBamHI(EcoRV):ura4FOAr ade6-M210∷pTy33(Ade6+) leu1-32 ura4-D18; TY88, mat2PBamHI(XbaI)∷ura4+ mat3PBclI(EcoRV)∷ura4FOAr ade6-M216∷pTy33(Ade6+) leu1-32 ura4-D18.
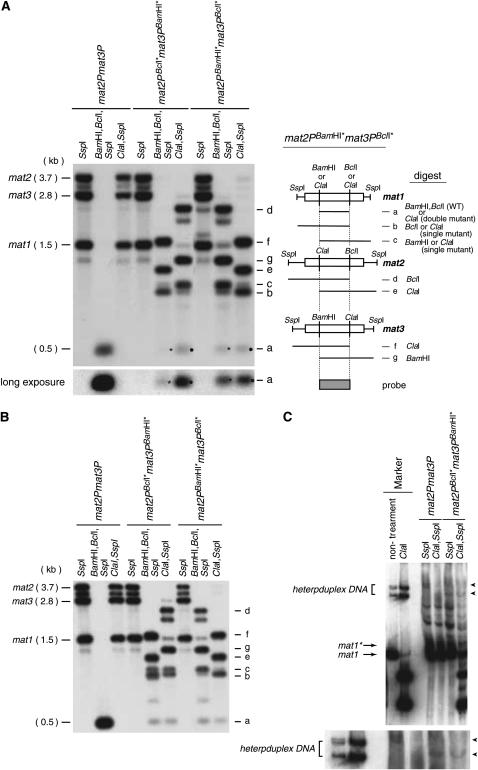
Figure 5.—
(A) Detection of heteroduplex DNA repair by Southern blot analysis. (Left) Chromosomal DNAs from the indicated haploid strains were digested with the given restriction enzymes (top) and analyzed by Southern blot using the central BclI/BamHI P-cassette fragment as a probe. The lowercase letters (right side of autoradiogram) identify the fragments detected by the Southern analysis for the mat2PBamHI* mat3PBclI* (see line drawing to the right). Molecular sizes for the SspI fragments are given on the left. An asterisk marks the signal corresponding to the wild-type cassette digested with BamHI and BclI. A black dot marks the ClaI fragment from mat1P double-mutant containing cassettes. The bottom left displays a long exposure for the detection of the repaired cassettes. The relative intensities of the bands marked “b” and “c” indicate the ratios of mat1PBclI* and mat1PBamHI* alleles for the strains. The ratios between mat1PBclI* and mat1PBamHI* are 0.42 ± 0.00 and 2.4 ± 0.2 for strains JZ149 and TY07, respectively (standard error is given). In both mutant strains the allele encoded by mat3 (which normally encodes M information) is predominantly present at mat1, verifying that neither the Pc nor the Pi transcript is required for establishing directionality of switching (Ruusala 1991; Thon and Klar 1993; Dalgaard and Vengrova 2004). A similar analysis of diploid strains constructed using JZ149 and TY07 (Figure 4A, II; data not shown) gave ratios of 0.77 ± 0.1 and 1.6 ± 0.0, respectively (standard error is given). A lower ratio is expected from a more random donor choice. The line drawing to the right defines bands and their predicted molecular sizes. (B) Southern analysis of msh6 strains detects wild-type mat1P-cassette DNA. Refer to A for the description. (C) Detection of heteroduplex DNA. Chromosomal DNA was purified and digested with either SspI or SspI and ClaI. A Southern analysis of the digested DNA separated on a native 10% polyacrylamide gel is displayed (Nagamine et al. 1989). The mobility of the identical heteroduplex DNA generated in vitro is shown as a marker. Heteroduplex DNA species (indicated by arrowheads), which are resistant to ClaI digestion, are observed for the mat2PBcIl* mat3PBamHI* strain, but not for the reference strain mat2P mat3P. The probe used is specific to the sequence centromere-distal to mat1 and is contained within the SspI fragment. Note that imprinted mat1* and unimprinted mat1 SspI fragments are separated on the acrylamide gel as two bands (Arcangioli 1998).
Analysis of heteroduplex DNA:
Heteroduplex DNA was generated in vitro by first PCR amplification of mutant PBamHI* and PBclI* cassette DNA using the primers mat1P-ssp1-f (5′-ATTGGAAGAGGTAGTATTTTTCTGT-3′) and mat1P-ssp1-R (5′-ATTAGTGAGTA TATTATGGTAGGGA-3′). These primers generate a 1452-bp PCR product with ends corresponding to those formed by SspI digestion. The PCR products were mixed, denatured at 96° for 2 min, and incubated at room temperature for 30 min. An aliquot of the mixture was digested with ClaI using standard procedures and both undigested and digested heteroduplex DNA were separated alongside digested genomic DNA on a native 10% polyacrylamide gel (acrylamide/bis-acrylamide ratio 29:1). After electrophoresis the gel was electroblotted to a nylon membrane using a submarine system (Bio-Rad, Hercules, CA). The Southern analysis was performed using a probe specific to the mat1P SspI fragment.
RESULTS
Efficient heteroduplex formation detected using genetic assay:
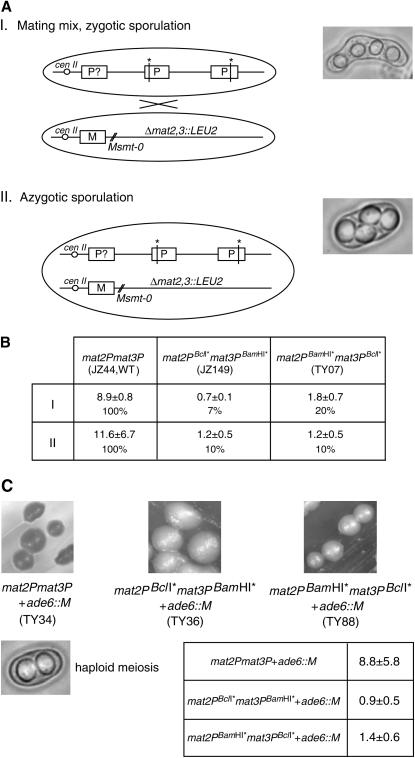
Two mutant P alleles, here referred to as PBamHI* and PBclI*, were constructed by filling in either BamHI or the BclI restriction sites with plasmid-borne mat2P and mat3P DNAs (see materials and methods). Importantly, the filling-in process generates a de novo ClaI restriction site at both the BamHI site and the BclI site (Figure 2B). Two sets, consisting of two mutant donor cassettes, [mat2PBamHI*, mat3PBclI*] and [mat2PBclI*, mat3PBamHI*], were sequentially integrated into the genome of strain pG598 using homologous recombination to obtain the two experimental strains studied here, one carrying mat2PBclI* mat3PBamHI* (strain JZ149) and the other mat2PBamHI* mat3PBclI* mutant donor loci (strain TY07; see materials and methods). In addition, two strains were constructed that only contained the PBamHI* or the PBclI* alleles at the mat2 locus, thus carrying mat2PBamHI*mat3M donor loci (strain JZ36) and mat2PBclI*mat3M donor loci (strain JZ67). The latter strains were used to analyze the phenotype of the two mutant P alleles. The P cassette contains two transcriptional units that encode the mating-type-specific transcription factors Pi (sometimes referred to as Pm) and Pc. Both the Pi and Pc activities are required in the zygote for expression of the mei3 gene that encodes an inducer of meiosis and sporulation, while the Pc activity is also required for expression of genes involved in mating/conjugation (Willer et al. 1995). The BamHI* and BclI* mutations utilized here introduce frameshift mutations in the Pc and Pi open-reading frames, respectively. Characterization of the mat2PBamHI*mat3M and mat2PBclI*mat3M strains verifies that these mutations lead to the abolishment of the Pi and Pc gene functions; mat2PBamHI*mat3M cells are unable to mate and sporulate, while mat2PBclI*mat3M cells can mate but not sporulate (Figure 3). Therefore, neither of the two mutant alleles carries enough genetic information for cells to go through the meiosis and sporulation program. However, for these experimental strains SDSA-mediated heteroduplex DNA formation and repair at mat1 will be detectable both biochemically by Southern analysis of restricted DNA, and, in case the heteroduplex DNA repair leads to restoration of the wild-type mat1P-cassette DNA, also genetically by the experimental strains' ability to mate and sporulate with an M-tester strain. First, we employed the genetic approach for detection of wild-type mat1P information (Figure 4). Our experimental mat2PBclI* mat3PBamHI* (JZ149), mat2PBamHI* mat3PBclI* (TY07) strains and a reference mat2P mat3P strain (JZ44), were mixed with a nonswitching M-tester strain on media containing a limiting nitrogen source to induce mating and sporulation. The tester strain (JZ108) carries a donor-region deletion (the Δmat2,3 deletion; Klar and Miglio 1986) in addition to a mat1 cis-acting deletion that abolishes imprinting (the smt-0 deletion; Styrkarsdottir et al. 1993). Importantly, efficient sporulation is observed for both experimental strains, demonstrating that functional mat1P-cassette information is present in a large fraction of cells; the mat2PBclI* mat3PBamHI* and the mat2PBamHI* mat3PBclI* strains display 7 and 20% of the sporulation level observed for the mat2P mat3P reference strain, respectively (Figure 4, A and B, I).
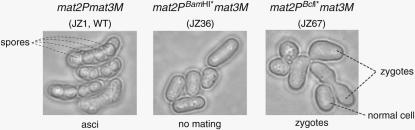
Figure 3.—
The mating- and sporulation-deficient phenotypes of the two mutant mat2PBamHI* mat3M (JZ36) and the mat2PBclI* mat3M (JZ67) strains. Wild-type h90 zygote formation and sporulation (strain JZ1) are shown for reference. Of the two P-cassette genes, Pc is required for conjugation and sporulation, and Pi for sporulation only (Kelly et al. 1988). Spores, zygotes, and cells are identified by dashed lines.
Figure 4.—
Genetic assays detect the wild-type mat1P allele. (A) Graphic outline of the strains is presented. Only strains with the mat2PBamHI* mat3PBclI* allele are shown. smt-0 is a cis-acting mat1-distal deletion that abolishes imprinting and switching. Δmat2,3∷LEU2 is a deletion of the entire mat2 mat3 donor region. Pictographs display the zygotic and azygotic asci quantified. I. “Zygotic” sporulation obtained by mixing constructed strains with an M-tester strain. II. Diploid strains constructed for “azygotic-” sporulation assays. (B) The strain names, sporulation percentages, and the standard deviation (see materials and methods) observed for strains described in A. The sporulation percentage (%) relative to that observed for the reference strain (first column) is given for each strain. For comparison a wild-type h90/h90 diploid strain sporulates >90%. (C) Iodine staining of sporulating colonies for strains. The genotype and the strain names are shown below each image. The bottom image displays a cell that has undergone haploid meiosis creating two spores. However, asci containing four defective spores are also observed. The table indicates the sporulation frequencies observed for the three strains.
The observed difference between the mat2PBclI* mat3PBamHI* and the mat2PBamHI* mat3PBclI* strains could be due to the fact that the mat3 allele is preferentially utilized as the donor in P cells and that in the two experimental strains the two P alleles are introduced at mat1 with different rates. This donor-locus preference is referred to as the “directionality of switching” and acts in a wild-type homothallic strain to ensure that the mat2P locus is used as donor in M cells and the mat3M locus in P cells (Ruusala 1991; Thon and Klar 1993). Importantly, it has been shown that the Pi and Pc transcripts are not required for establishing the directionality of switching (Ruusala 1991). Therefore, the mat2P-donor preference is maintained in the experimental strains and the mating-type cassette located at the mat3 locus will preferentially be copied into the mat1 locus (also see Figure 5 legend). Since the Pi mutation (PBcll*) allows mating, a larger number of diploid cells will form in the mating mix with the mat2PBamHI*mat3PBclI* than with the mat2PBclI*mat3PBamHI* strain. Such diploid cells can directly undergo meiosis and sporulation if wild-type mat1P information is formed by subsequent switching events. To obtain a more comparable measurement of the levels of wild-type mat1P information in the two experimental setups, mat2PBclI*mat3PBamHI* vs. mat2PBamHI*mat3PBclI*, we repeated the experiment while first constructing diploid strains. For this purpose, we used heteroallelic complementation of the ade6 alleles, ade6-M216 and ade6-M210, carried by the strains (two tester strains were used; strains JZ108 and JZ109 are ade6-M210 and ade6-M216, respectively). The experimental strains and the reference strain were allowed to mate with a tester strain of opposite ade6-allele type on media containing limiting nitrogen, and zygotes formed were then shifted to media without adenine. Ade+-prototrophe diploid cells that have escaped meiosis formed colonies after a few days. When these diploid strains are grown on media with a limiting nitrogen source, then both of the experimental diploids display a sporulation frequency of 10% of that observed for the diploid mat2P mat3P reference strain (Figure 4, A and B, II). This observation suggests that in diploid cells mat1P wild-type cassette DNA is formed with similar frequencies for the two genetic mating-type region alleles, mat2PBclI* mat3PBamHI* and the mat2PBamHI* mat3PBclI*. The high level of sporulation observed is therefore consistent with the prediction that switching mediates a high rate of heteroduplex formation and repair at mat1. However, to conclusively establish that the detected wild-type mat1P information is formed by the continuous switching process and not a few, potentially unrelated, gene conversion events amplified through cell divisions, we employed a genetic approach. If both M- and P-cassette information are expressed in haploid cells, the cells will undergo a defective meiosis (generating less than four and/or defective spores) when they experience nutritional starvation. Since spores of S. pombe stain dark when exposed to iodine vapors due to a starch content, switching and sporulation can directly be assayed by staining of colonies (Moreno et al. 1991). For this purpose, M-cassette DNA was introduced in the two experimental strains and the mat2P mat3P reference strain at an ectopic position in the genome (the ade6 locus; see materials and methods). Importantly, sporulating colonies of these strains (>200 observed) display a uniform “salt-and-pepper” staining phenotype (Figure 4C). The colonies' uniform staining establishes that there is a low rate of meiosis and sporulation throughout the surfaces of the individual colonies. Such a staining phenotype is predicted if wild-type P information is generated by the switching program that is continuously propelled by each cell cycle. In contrast, the presence of a few uniformly dark-staining colonies or a colony with darker sectors would have suggested that the wild-type mat1P information originated from a few recombination events that were amplified through cell divisions. This genetic experiment also excludes the occurrence of gene conversion events leading to the formation of wild-type or double-mutant P-donor-cassette DNA at the donor loci, as formation of such strains would lead to formation of colonies or colony sectors with a high-staining or a nonstaining phenotype, respectively. In conclusion, our genetic analysis using both haploid and diploid strains shows that wild-type P-cassette information is continuously formed at mat1, but not at the donor loci, when cells switch using two different mutant P-donor cassettes.
Switching using mutant P-donor cassettes leads to formation of the wild-type P-cassette DNA at mat1:
A Southern blot analysis was employed to directly determine whether wild-type mat1P-cassette DNA is formed (Figure 5A). Chromosomal DNA was purified and digested with three sets of enzymes: SspI; BamHI, BclI, SspI; or ClaI, SspI. The SspI restriction creates three differently sized fragments each containing one of the three loci: mat1, mat2, and mat3. The presence of mutant or wild-type Pi and Pc genes can in this analysis be detected by the presence of the ClaI sites (mutant Pi or Pc) or the BamHI (wild-type Pc) or BclI (wild-type Pi) sites within these SspI fragments (Figure 5A, line drawing). The results confirm the presence of wild-type mat1P-cassette DNA containing both a BclI and a BamHI site in both haploid and diploid experimental strains (Figure 2A, “functional cassette”). The Southern analysis also detects double-mutant mat1P DNA containing two ClaI sites (Figure 5A, signal a, black dot). This fragment potentially corresponds to heteroduplex DNA being repaired in a manner where nucleotides are introduced opposite the single-stranded bulge DNA (Figure 2C, second product). Interestingly, we observe a reproducible difference between the two experimental strains; while similar amounts of the two products, wild-type and double-mutant P-cassette DNA, are formed in the mat2PBamHI*mat3PBclI* strain, less wild-type mat1-cassette DNA is formed in the mat2PBclI*mat3PBamHI* strain (Figure 5A, signal a). This result reflects the greater sporulation observed for the haploid strains carrying the mat2PBamHI*mat3PBclI* allele (Figure 4, A and B, I). This difference is potentially caused by directionality of switching leading to different formation rates for the different species of heteroduplex DNA in the two strains. In concordance, we observe similar levels of wild-type and double-mutant P-cassette signals when the diploid experimental strains are analyzed (Figure 4, A and B, II; data not shown)—an observation that reflects similar levels of sporulation for these strains (see above). In conclusion, the process of mating-type switching leads to efficient restoration of wild-type P-cassette DNA at mat1 in these experimental strains.
Detection of mat1 heteroduplex DNA:
Since ∼50% of dividing cells that carry an imprint undergo mating-type switching during S phase, we expected that it would be possible to directly detect mat1 heteroduplex molecules in DNA purified from exponentially dividing cultures using a gel-shift assay similar to that developed by Lichten et al. (1990). Initial attempts using the strains described above were unsuccessful (data not shown), suggesting that repair occurs rapidly. It has been shown that the Msh6 protein, in complex with Msh2 (Swi8), is required for recognition and repair of bulge heteroduplex DNA in S. pombe (Mansour et al. 2001; Tornier et al. 2001). However, the repair of the mat1 heteroduplex DNA could in this system also involve the Msh3/Swi4 factor. In higher eukaryotes, Msh2/Swi8 and Msh3/Swi4 mediate heteroduplex DNA repair, while the S. pombe factors have been implicated in resolution of mating-type switching intermediates (Egel et al. 1984; Fleck et al. 1992; Rudolph et al. 1999). We decided to analyze the mat1 DNA intermediates in the absence of the Msh2/Msh6 heteroduplex repair pathway. Mutations in the Msh2/Swi8 and Msh3/Swi4 factors cause switching-dependent rearrangements in the mating-type locus (Fleck et al. 1992, 1994), therefore we introduced the Δmsh6 mutation into our experimental strains and the mat2P mat3P reference strain and isolated and analyzed DNA from exponentially dividing cultures (Tornier et al. 2001). Importantly, repair of heteroduplex DNA is still observed in the msh6 mutant background, suggesting the presence of an alternative pathway (Figure 5B; Marti et al. 2002). However, we do detect low levels of unrepaired mat1 heteroduplex DNA (Figure 5C). Importantly, the observed mat1 heteroduplex DNA displays the same mobility when analyzed on a polyacrylamide gel as heteroduplex DNA generated in vitro and, like the in vitro generated molecules, is resistant to ClaI digestion (Figure 5C). The detection of mat1 heteroduplex DNA provides direct support for the SDSA mechanism proposed for mating-type switching.
DISCUSSION
In this study, we present the genetic and biochemical tests of the proposed SDSA recombination mechanism underlying mating-type switching. The experiments utilize artificially constructed strains that carry two different heteroallelic mutant P cassettes at the donor loci, mat2 and mat3. Although this is an artificial situation, where cells are switching between related cassette sequences, the constructed strains display genetic stability, imprinting levels, and recombination frequency between mat1 and the donor loci, which leads to a high rate of heteroduplex formation, supporting that the overall process of switching is not affected by the genetic changes at the donor loci. This reflects previously published experiments where the information of the two donor cassettes was swapped, such that mat2 carried M information and mat3 P information (Thon and Klar 1993). Thus, the observations and conclusions obtained using these strains are likely to reflect the wild-type mechanism where only the H2 homology domain, and not the cassettes, provides the strand annealing described in Figure 1 (note that the limited size of H2, and the inability to use a genetic assay for detection of heteroduplex DNA, excluded the use of only this domain for heteroduplex formation).
Theoretically, alternative mechanisms to the SDSA mechanism could account for the formation of the wild-type and double-mutant mat1P-DNA observed. For example, one such alternative mechanism would involve double-stranded gap repair where the outgoing old cassette DNA is removed such that 3′ ends both at the H2 and the H1 homology domains can invade and be extended at a donor locus. Either an unknown endonuclease or flap endonucleases could account for this removal of the old cassette DNA. In the latter case, flap endonucleases could act after single-stranded H2 DNA had invaded the donor-locus H2 homology domain to create a branched structure. The two 3′ ends formed could then be extended across the donor locus by polymerases. Subsequently, the recombination intermediates could be resolved by helicases or by the establishment of a Holliday junction(s) (HJ) recognized and resolved by an HJ resolvase. However, several observations exclude that these alternative pathways play a significant role during mating-type switching:
While a DNA break can be detected by Southern analysis at the junction between the H1 homology domain and the mat1-cassette DNA, no DNA break has been observed at the junction between the H2 homology domain and the mat1-cassette DNA (Klar and Miglio 1986; Arcangioli 1998).
The resolution of an HJ would potentially allow a crossover event, which would loop out of the intergenic region between mat1 and the utilized donor locus. While such rearrangements do occur in wild-type homothallic strains, they are very rare; only 1 in 5 × 103 cell divisions leads to the formation of such h+L or h−L genetic rearrangements (Beach and Klar 1984).
While it is possible to detect switching intermediates by PCR reactions where a mat1 cen-distal fragment is attached to the mat2P or mat3M cen-proximal fragments, intermediates where the mat1 cen-proximal fragment attached to mat2P and mat3M cen-distal fragments were undetectable (Arcangioli and De Lahondes 2000).
In the case of the experimental strains analyzed here, the alternative recombination mechanisms are predicted to allow heteroduplex DNA to form between the “outgoing” P-cassette strand and the donor locus containing the alternative mutant P allele. Repair of this heteroduplex DNA would potentially lead to the formation of double-mutant or wild-type P-cassette DNA at the donor loci. However, our genetic assay using strains capable of undergoing haploid meiosis, due to the presence of M information at the ade6 locus, establishes that if gene conversions at the donor loci, predicted by the alternative recombination mechanisms, occur, they are rare.
In summary, the detection of mat1 heteroduplex formation and repair presented here, combined with (i) the previously published PCR analysis of switching intermediates (Arcangioli and De Lahondes 2000), (ii) the observation that in the wild-type homothallic strains both strands of a newly switched cassette are newly synthesized (Arcangioli 2000), and (iii) the characterization of naturally occurring rearrangements in homothallic strains, and their rate of occurrence (Beach and Klar 1984), establish conclusively that mating-type switching predominantly occurs by a mechanism where only one strand of the donor cassette is copied and the newly synthesized strand is transferred to the mat1 locus. This SDSA mechanism (Mcgill et al. 1993; Nassif et al. 1994; Ferguson and Holloman 1996; Weng et al. 1996) is similar to that proposed for the recombination event underlying Saccharomyces cerevisiae mating-type switching (Mcgill et al. 1993; Haber 1998). The similarity is interesting since S. cerevisiae mating-type switching is initiated by the HO-endonuclease catalyzed double-stranded break introduced at the MAT locus, while S. pombe mating-type switching is initiated by a stalled replication fork, thus suggesting that breaks generated by different mechanisms can be repaired by related or identical recombination pathways.
Furthermore, stalled replication forks are thought to be a major contributor to genome instability, leading to mutations, rearrangements, gene amplifications, loss of heterozygosity, and chromosome fragmentation (Branzei and Foiani 2005). Stalling can occur at damaged bases, at bound factors, and due to dysfunctional replication factors, low levels of deoxyribonucleotides and, as shown for S. pombe mating-type switching, the presence of ribonucleotides in the template DNA for the leading-strand polymerase (Vengrova and Dalgaard 2004). Several enzymatic pathways have been described that ensure replication restart to prevent such genetic instability, including, but not limited to, the lesion bypass pathway involving error-prone polymerases, the error-free bypass pathway involving replication-fork regression, and the recombination-mediated replication reinitiation pathway involving D-loop formation (Mcilwraith et al. 2005). The latter pathway is similar to the recombination mechanism described here; however, in the case of mating-type switching, replication stalling efficiently induces recombination with one of the donor loci. A recent study might explain why stalled replication forks are not reinitiated at mat1 (Akamatsu et al. 2003). Here the Swi2, Swi5, and Rhp51 factors were identified as part of a complex responsible for mediating the recombination event underlying mating-type switching. However, the Swi5 and Rhp51 factors also form a complex with the transacting factor Sfr1 that has a general role in recombination. Thus, an interesting possibility is that the Swi2 activity might have evolved to channel the intermediates of a genomewide pathway for recombination into a locus-specific pathway mediating mating-type switching.
Finally, the genetic backgrounds constructed here constitute unique tools for studying heteroduplex DNA formation and repair during the mitotic S phase. Although meiotic heteroduplex DNA has previously been directly detected (Lichten et al. 1990), we are for the first time able to detect mitotic heteroduplex intermediates. Some of our observations contrast with what has been previously shown for heteroduplex repair (Marti et al. 2002; Kunkel and Erie 2005) but might reflect that the repair analyzed here occurs during the mitotic S phase and at a specialized locus. We observed for the mat2PBamHI* mat3PBclI* strain high levels of both wild-type as well as double-mutant mat1P-cassette DNA; thus, there is no evidence that the repair enzymes favor old over newly synthesized strand as template when the heteroduplex DNA is repaired or that cleavage of one DNA strand at one bulge affects which strand is cleaved at the other bulge. In addition, we show that the mat1 heteroduplex repair can occur independently of the Msh6 activity thought to be required for heretoduplex DNA repair in S. pombe. The Msh2/Swi8 and Msh3/Swi4 factors could mediate this alternative repair, as they also are required for resolution of switching intermediates (Fleck et al. 1992; Rudolph et al. 1999). Potentially, Msh2/Swi8 and Msh3/Swi4 could mediate heteroduplex DNA repair at the mating-type locus exclusively, or these factors could have a genomewide role in mediating heteroduplex DNA repair specifically during S phase.
Acknowledgments
We thank S. Vengrova, T. Inagawa, and T. Eydmann for advice on genetic manipulation and technical assistance; Oliver Fleck and Geneviève Thon for providing strains and plasmids; and Rob Cross for help with the manuscript. We thank our colleagues at Marie Curie Research Institute for helpful discussions and suggestions. This work was supported by the Intramural Research Program of the National Cancer Institute of the National Institutes of Health (A.J.S.K.) and Marie Curie Cancer Care (T.Y-I. and J.Z.D.).
References
- Akamatsu, Y., D. Dziadkowiec, M. Ikeguchi, H. Shinagawa and H. Iwasaki, 2003. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl. Acad. Sci. USA 100: 15770–15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli, B., 1998. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 17: 4503–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli, B., 2000. Fate of mat1 DNA strands during mating-type switching in fission yeast. EMBO Rep. 1: 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli, B., and R. de Lahondes, 2000. Fission yeast switches mating type by a replication-recombination coupled process. EMBO J. 19: 1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach, D. H., 1983. Cell type switching by DNA transposition in fission yeast. Nature 305: 682–688. [Google Scholar]
- Beach, D. H., and A. J. Klar, 1984. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J. 3: 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei, D., and M. Foiani, 2005. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17: 568–575. [DOI] [PubMed] [Google Scholar]
- Dalgaard, J. Z., and A. J. Klar, 1999. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400: 181–184. [DOI] [PubMed] [Google Scholar]
- Dalgaard, J. Z., and A. J. Klar, 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102: 745–751. [DOI] [PubMed] [Google Scholar]
- Dalgaard, J. Z., and A. J. Klar, 2001. a A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev. 15: 2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard, J. Z., and A. J. Klar, 2001. b Does S. pombe exploit the intrinsic asymmetry of DNA synthesis to imprint daughter cells for mating-type switching? Trends Genet. 17: 153–157. [DOI] [PubMed] [Google Scholar]
- Dalgaard, J. Z., and S. Vengrova, 2004. Selective gene expression in multigene families from yeast to mammals. Sci. STKE 2004: re17. [DOI] [PubMed] [Google Scholar]
- Egel, R., 1977. Frequency of mating-type switching in homothallic fission yeast. Nature 266: 172–174. [DOI] [PubMed] [Google Scholar]
- Egel, R., 1984. The pedigree pattern of mating-type switching in Schizosaccharomyces pombe. Curr. Genet. 8: 205–210. [DOI] [PubMed] [Google Scholar]
- Egel, R., 1989. Mating-type genes, meiosis and sporulation, pp. 31–73 in Molecular Biology of the Fission Yeast, edited by E. Anwar Nasim, P. Young and B. F. Johnson. Academic Press, New York.
- Egel, R., and B. Eie, 1987. Cell lineage asymmetry in Schizosaccharomyces pombe: unilateral transmission of a high-frequency state for mating-type switching in diploid pedigrees. Curr. Genet. 12: 429–433. [Google Scholar]
- Egel, R., and H. Gutz, 1981. Gene activation by copy transposition in mating-type switching of a homothallic fission yeast. Curr. Genet. 3: 5–12. [DOI] [PubMed] [Google Scholar]
- Egel, R., D. H. Beach and A. J. Klar, 1984. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc. Natl. Acad. Sci. USA 81: 3481–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, D. O., and W. K. Holloman, 1996. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc. Natl. Acad. Sci. USA 93: 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck, O., H. Michael and L. Heim, 1992. The swi4+ gene of Schizosaccharomyces pombe encodes a homologue of mismatch repair enzymes. Nucleic Acids Res. 20: 2271–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck, O., C. Rudolph, A. Albrecht, A. Lorentz, P. Schar et al., 1994. The mutator gene swi8 effects specific mutations in the mating-type region of Schizosaccharomyces pombe. Genetics 138: 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J. E., 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32: 561–599. [DOI] [PubMed] [Google Scholar]
- Holmes, A. M., A. Kaykov and B. Arcangioli, 2005. Molecular and cellular dissection of mating-type switching steps in Schizosaccharomyces pombe. Mol. Cell. Biol. 25: 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaykov, A., and B. Arcangioli, 2004. A programmed strand-specific and modified nick in S. pombe constitutes a novel type of chromosomal imprint. Curr. Biol. 14: 1924–1928. [DOI] [PubMed] [Google Scholar]
- Kaykov, A., A. M. Holmes and B. Arcangioli, 2004. Formation, maintenance and consequences of the imprint at the mating-type locus in fission yeast. EMBO J. 23: 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, M., J. Burke, M. Smith, A. Klar and D. Beach, 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 7: 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar, A. J., 1987. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature 326: 466–470. [DOI] [PubMed] [Google Scholar]
- Klar, A. J., 1990. a Regulation of fission yeast mating-type interconversion by chromosome imprinting. Dev. Suppl., 3–8. [PubMed]
- Klar, A. J., 1990. b The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands. EMBO J. 9: 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar, A. J., and M. J. Bonaduce, 1993. The mechanism of fission yeast mating-type interconversion: evidence for two types of epigenetically inherited chromosomal imprinted events. Cold Spring Harbor Symp. Quant. Biol. 58: 457–465. [DOI] [PubMed] [Google Scholar]
- Klar, A. J., and L. M. Miglio, 1986. Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell 46: 725–731. [DOI] [PubMed] [Google Scholar]
- Klar, A. J., M. J. Bonaduce and R. Cafferkey, 1991. The mechanism of fission yeast mating type interconversion: seal/replicate/cleave model of replication across the double-stranded break site at mat1. Genetics 127: 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, T. A., and D. A. Erie, 2005. DNA mismatch repair. Annu. Rev. Biochem. 74: 681–710. [DOI] [PubMed] [Google Scholar]
- Leupold, U., 1950. Die Vererbung von Homothallie und Heterothallie bei Schizosaccharomyces pombe. Compt. Rend. Trav. Lab. Carlsberg, Sér. Physiol. 24: 381–480. [Google Scholar]
- Lichten, M., C. Goyon, N. P. Schultes, D. Treco, J. W. Szostak et al., 1990. Detection of heteroduplex DNA molecules among the products of Saccharomyces cerevisiae meiosis. Proc. Natl. Acad. Sci. USA 87: 7653–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour, A. A., C. Tornier, E. Lehmann, M. Darmon and O. Fleck, 2001. Control of GT repeat stability in Schizosaccharomyces pombe by mismatch repair factors. Genetics 158: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, T. M., C. Kunz and O. Fleck, 2002. DNA mismatch repair and mutation avoidance pathways. J. Cell. Physiol. 191: 28–41. [DOI] [PubMed] [Google Scholar]
- McGill, C. B., B. K. Shafer, L. K. Derr and J. N. Strathern, 1993. Recombination initiated by double-strand breaks. Curr. Genet. 23: 305–314. [DOI] [PubMed] [Google Scholar]
- McIlwraith, M. J., A. Vaisman, Y. Liu, E. Fanning, R. Woodgate et al., 2005. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell 20: 783–792. [DOI] [PubMed] [Google Scholar]
- Miyata, H., and M. Miyata, 1981. Mode of conjugation in homothallic cells of Schizosaccharomyces pombe. J. Gen. Appl. Microbiol. 27: 365–371. [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Nagamine, C. M., K. Chan and Y.-F. C. Lau, 1989. A PCR Artifact: generation of heteroduplexes. Am. J. Hum. Genet. 45: 337–339. [PMC free article] [PubMed] [Google Scholar]
- Nassif, N., J. Penney, S. Pal, W. R. Engels and G. B. Gloor, 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, C., C. Kunz, S. Parisi, E. Lehmann, E. Hartsuiker et al., 1999. The msh2 gene of Schizosaccharomyces pombe is involved in mismatch repair, mating-type switching, and meiotic chromosome organization. Mol. Cell. Biol. 19: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruusala, T., 1991. The mating type in fission yeast is switched independently of its expression. Curr. Genet. 20: 379–383. [DOI] [PubMed] [Google Scholar]
- Schmidt, H., P. Kapitza-Fecke, E. R. Stephen and H. Gutz, 1989. Some of the swi genes of Schizosaccharomyces pombe also have a function in the repair of radiation damage. Curr. Genet. 16: 89–94. [DOI] [PubMed] [Google Scholar]
- Singh, J., and A. J. Klar, 1993. DNA polymerase-alpha is essential for mating-type switching in fission yeast. Nature 361: 271–273. [DOI] [PubMed] [Google Scholar]
- Styrkarsdottir, U., R. Egel and O. Nielsen, 1993. The smt-0 mutation which abolishes mating-type switching in fission yeast is a deletion. Curr. Genet. 23: 184–186. [DOI] [PubMed] [Google Scholar]
- Thon, G., and A. Klar, 1992. The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics 131: 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon, G., and A. J. Klar, 1993. Directionality of fission yeast mating-type interconversion is controlled by the location of the donor loci. Genetics 134: 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornier, C., S. Bessone, I. Varlet, C. Rudolph, M. Darmon et al., 2001. Requirement for Msh6, but not for Swi4 (Msh3), in Msh2-dependent repair of base-base mismatches and mononucleotide loops in Schizosaccharomyces pombe. Genetics 158: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova, S., and J. Z. Dalgaard, 2004. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 18: 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova, S., and J. Z. Dalgaard, 2006. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 7: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, Y. S., J. Whelden, L. Gunn and J. A. Nickoloff, 1996. Double-strand break-induced mitotic gene conversion: examination of tract polarity and products of multiple recombinational repair events. Curr. Genet. 29: 335–343. [DOI] [PubMed] [Google Scholar]
- Willer, M., L. Hoffmann, U. Styrkarsdottir, R. Egel, J. Davey et al., 1995. Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe. Mol. Cell. Biol. 15: 4964–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]