Abstract
The extent of conservation of synteny and gene order in the Lepidoptera has been investigated previously only by comparing a small subset of linkage groups between the moth Bombyx mori and the butterfly Heliconius melpomene. Here we report the mapping of 64 additional conserved genes in H. melpomene, which contributed 47 markers to a comparative framework of 72 orthologous loci spanning all 21 H. melpomene chromosomes and 27 of the 28 B. mori chromosomes. Comparison of the maps revealed conserved synteny across all chromosomes for the 72 loci, as well as evidence for six cases of chromosome fusion in the Heliconius lineage that contributed to the derived 21-chromosome karyotype. Comparisons of gene order on these fused chromosomes revealed two instances of colinearity between H. melpomene and B. mori, but also one instance of likely chromosomal rearrangement. B. mori is the first lepidopteran species to have its genome sequenced, and the finding that there is conserved synteny and gene order among Lepidoptera indicates that the genomic tools developed in B. mori will be broadly useful in other species.
THE domesticated silkmoth Bombyx mori (Bombycidae: Bombycinae) was the first lepidopteran species to become a focus of genomic studies, due to its tractability as a study organism and importance to the silk industry. In combination with a draft genome sequence completed in 2004 (Xia et al. 2004), recent high-density linkage maps provide a comprehensive picture of chromosomal linkage in B. mori (Yasukochi 1998; Miao et al. 2005; Yoshido et al. 2005; Yasukochi et al. 2006). Increasingly, however, genetic studies of other Lepidoptera are also reaching genomic scale as similar tools are applied in nonmodel species (Papanicolaou et al. 2005; Beldade et al. 2006; Turner et al. 2006). In particular, these studies have focused on groups of specific ecological and evolutionary interest. Heliconius butterflies (Nymphalidae: Heliconiinae: Heliconiini) are one such group, notable for their highly accurate Müllerian mimicry in which unpalatable species converge in aposematic coloration.
Genome mapping studies in diverse taxa, such as in the Heliconiini, will allow the first comparisons of linkage across the Lepidoptera. To draw conclusions from comparative mapping between B. mori and Heliconius, it is necessary to consider the phylogenetic relationships and evolution of karyotype in the Lepidoptera as a whole. Rhopalocera, the lineage that contains heliconiines and the rest of the true butterflies (Papilionoidea) is younger than the Bombycoidea, the lineage that contains the silkworm moths (Grimaldi and Engel 2005). Derived heliconiines have a chromosome number of 21, contrasting with the 31 chromosomes of the basal genera in the Heliconiini and basal Lepidoptera (Suomalainen 1979). Thus, both B. mori (28 chromosomes) and derived Heliconiini appear to have undergone independent karyotype reductions from more basal taxa.
Studies using traditional genetic methods located several chromosomal regions responsible for color-pattern changes in the comimicking species Heliconius melpomene and H. erato (Sheppard et al. 1985; Mallet 1989; Jiggins and McMillan 1997; Gilbert 2003; Naisbit et al. 2003). However, it is only very recently that genomic tools, including high-density linkage maps and expressed sequence tag (EST) libraries have begun to be developed for these butterflies as a means for identifying the particular genes underlying changes in color patterns (Jiggins et al. 2005; Tobler et al. 2005; Joron et al. 2006b; Kapan et al. 2006; Kronforst et al. 2006). An earlier linkage map of H. melpomene localized amplified fragment length polymorphisms (AFLPs), microsatellites, and single-copy nuclear loci (SCNLs) to 21 distinct linkage groups (LGs) (Jiggins et al. 2005), which correspond to the 21 H. melpomene chromosomes. Importantly, that study identified and mapped 19 SCNLs that are homologous to annotated genes from other organisms and represent anchor loci for comparisons with B. mori (Yasukochi et al. 2006), H. erato, and Heliconius numata (Joron et al. 2006b). In addition, recently developed EST libraries for H. melpomene and H. erato have allowed the identification of additional anchor loci that are conserved in hexapods or in eukaryotes generally (Papanicolaou et al. 2005). Yasukochi et al. (2006) reported finding conserved synteny (chromosomal linkage of genes) and colinearity among 13 orthologous genes in four LGs in a comparison between B. mori and H. melpomene. However, the linkage map for H. melpomene that was available at that time (Jiggins et al. 2005) lacked additional orthologs that would have allowed a more comprehensive assessment of comparative synteny between the two species.
Comparative mapping can facilitate not only the investigation of specific evolutionary questions, such as the mechanisms of color-pattern changes in the Heliconius clade (Joron et al. 2006a,b), but also the study of chromosome evolution more generally. For example, the examination of synteny at genomic scales can elucidate chromosome homology and provide a framework for predicting the locations of genes in other species (Brown et al. 2001). In addition, knowledge of gene order on homologous chromosomes allows investigation of the types and prevalence of chromosome rearrangements. In particular, although the precise mechanisms of chromosomal rearrangements may be difficult to determine using linkage mapping alone (Eichler and Sankoff 2003), sufficient conservation of synteny should allow at least the detection of chromosomal fusions or fissions.
Here we report the mapping of 64 additional cDNA-derived markers in H. melpomene, contributing 47 markers to a total of 72 markers now mapped in this species (see also Jiggins et al. 2005; Papanicolaou et al. 2005; Joron et al. 2006b) that are orthologous to those recently mapped in B. mori (Yasukochi et al. 2006). We examine the extent of synteny conservation between these species and then propose putative chromosomal fusions that led to the derived chromosome number of heliconiine butterflies.
MATERIALS AND METHODS
Time of divergence:
DNA sequences for Elongation factor 1α (Ef1α) and Wingless (Wg) were downloaded from GenBank for H. melpomene (AY747994, AY745485) and B. mori (D13338, D14169), for two species from the lepidopteran subfamily Nymphalinae, Vanessa annabella (AY788823, AY788583) and Vanessa virginiensis (AY248808, AY248827), and for an outgroup species, Drosophila melanogaster (Diptera) (NM_165850, NM_164746). A tree was built on the basis of taxonomic assignments (Grimaldi and Engel 2005; Figure 1). The Ef1α and Wg sequences were aligned in MacClade 4.06, and branch lengths were calculated via maximum likelihood with the model of evolution GTR + Γ + I in PAUP 4.0b10 without enforcing a molecular clock (Swofford 2002). Node-divergence times were then estimated using penalized likelihood via the truncated Newton algorithm in r8s 1.71, with a smoothing parameter of 3.2 (Sanderson 2002). The time of divergence for V. annabella and V. virginiensis was recently estimated at 27.9–29.3 million years (MY) using a Bayesian relaxed-clock method in a parsimony topology of the subfamily Nymphalinae calibrated with five fossils (Wahlberg 2006). The minimum estimate of 27.9 MY was used as a minimum age for the Vanessa node of the tree. The maximum age of the divergence of Diptera and Lepidoptera was constrained to 190 MY, which is a minimal estimate corresponding to the oldest known lepidopteran fossil (Grimaldi and Engel 2005). Age standard deviations were calculated using nonparametric bootstrapping.
Figure 1.—
Phylogeny used to estimate time of divergence between B. mori and H. melpomene. The dashed line indicates the approximate age of divergence between these lineages (∼103 MY). Arrows indicate constrained nodes (see materials and methods). Numbers in circles indicate the haploid number of chromosomes for B. mori, H. melpomene, and basal taxa.
Collection and crosses:
Butterfly collection, crosses, and DNA extraction were as described previously (Jiggins et al. 2005). Briefly, parental crosses were made between H. melpomene cythera from Ecuador and H. melpomene melpomene from French Guiana; subsequent crossing between F1 offspring produced F2 progeny that segregated for color-pattern genes and also showed considerable variation at sequence-based markers, facilitating linkage mapping (Jiggins et al. 2005). The brood of a single F1 female, brood 33, was used for all of the positional mapping presented here, except in the case of Ribosomal Protein S16, which did not exhibit polymorphic variation in brood 33. Here, RpS16 was mapped to LG13 in the brood of a separate F1 female (brood 44) by comparison with microsatellite marker Hm20 (Jiggins et al. 2005).
Gene identification and primer design:
Methodology for generating ESTs from H. melpomene and H. erato was described previously (Papanicolaou et al. 2005). EST traces were clustered using PartiGene software (http://www.nematodes.org) and are available at ButterflyBase (http://www.heliconius.org) and GenBank (http://www.ncbi.nlm.nih.gov). We used the Basic Local Alignment Search Tool via nucleotide (blastn) and translated (tblastx) comparisons of the coding sequences of the 347 B. mori genes that were identified with GenBank accession numbers by Yasukochi et al. (2006) against the Heliconius ESTs. A total of 127 B. mori coding sequences bore similarity to Heliconius ESTs from the database; of these, 106 ESTs had alignment bit scores >80 and were considered to be candidate orthologous loci for comparison between species. Genes predicted to be orthologs of single-copy loci in the B. mori genome (85 of 106 candidates) were then selected for genetic mapping in H. melpomene. In particular, we concentrated on ribosomal proteins, which are mostly single copy and widely distributed across the genome; although other genes were also included, we avoided members of gene families in Bombyx where possible. Primers for some of these loci had been designed previously on the basis of alignments between B. mori and other species (Galleria mellonella, Hyphantria cunea, Manduca sexta, Spodoptera frugiperda) (Papanicolaou et al. 2005). Additional primers were designed from the Heliconius alignments with B. mori sequences for better amplification of some previous markers as well as for amplification of the new markers. ESTs from H. melpomene or H. erato were aligned with B. mori coding sequences in MacVector 7.2.3. Blastn of B. mori coding sequences against the B. mori whole-genome sequence revealed the location of introns in B. mori, and primers were designed away from intron/exon boundaries to avoid potentially conserved locations of introns. We also attempted to include one or more introns from B. mori between each primer pair in H. melpomene to increase the probability of indel polymorphisms. New primers were designed for 53 candidate genes within the Heliconius spp. coding sequences.
Linkage analysis:
Linkage analysis was performed by taking advantage of the lack of crossing over in female Lepidoptera and the method of forbidden recombinants (Shi et al. 1995). The inheritance pattern of each gene was matched to a known “chromosome print” from brood 33 that had previously been developed using AFLPs, microsatellites, and genes for each of the 21 LGs in H. melpomene (Jiggins et al. 2005). Markers were first amplified in eight test members of the F2 brood to check for visible length polymorphisms that would suggest indels that could be used for mapping. If present, the indel polymorphism was scored for each individual of the brood. If no length polymorphism was observed, the marker was then amplified in the F1 mother and father of the brood and sequenced. PCR reactions for subsequent sequencing contained 5–50 ng of DNA, 0.5 units of BIOTAQ DNA polymerase, 61.5 mm 10× NH4 reaction buffer, 2 mm MgCl2, 0.1 mm dNTPs, and 0.5 μm of each primer in a 20-μl reaction mixture. PCR amplification followed a standard protocol as follows: 95° for 2 min; 35 cycles of 94° for 20 sec, 50°–64° for 30 sec, 72° for 1 min, and a final elongation step of 72° for 10 min. An annealing temperature between 50° and 64° was determined for each primer pair using a gradient PCR with the same conditions. Purified PCR product (1 μl) was used as a template in a 10-μl sequencing reaction containing 1 μl Big Dye, 0.5 μm primer, buffer, and distilled and deionized water. Cycling conditions followed Applied Biosystems (Foster City, CA) protocols. Samples were run on an ABI 3730 DNA analyzer by the School of Biological Sciences Sequencing Service, University of Edinburgh. Single nucleotide polymorphisms (SNPs) were then identified in the mother's sequence and used to create a map of polymorphic restriction sites. An enzyme was chosen that cut one allele and not the other, thereby producing a unique pattern for each of the mother's alleles. For each cDNA-derived marker, 24–48 offspring were genotyped for the maternally derived allele. Restriction products were separated on a 1.5–3% agarose electrophoretic gel of Tris–borate–EDTA (TBE) buffer containing ethidium bromide (1 μg/ml). There was visible segregating variation in the maternal allele for 64 H. melpomene cDNA-derived markers.
Analysis of synteny, possible chromosomal fusions, and gene order:
Following the assignment of markers to specific H. melpomene LGs, the LGs were compared between H. melpomene and B. mori to assess conserved synteny. Chromosomes were considered homologous and syntenically conserved if all linked cDNA-derived markers fell in one H. melpomene LG and one B. mori LG. If markers on one H. melpomene LG were found in two B. mori LGs (which was the case for H. melpomene LGs 7, 10, 12, 13, and 18), this was considered evidence of possible chromosome fusion, and markers were assessed for male polymorphism in H. melpomene for positional recombination mapping. Unless male-informative length polymorphisms due to indels were immediately detected, restriction enzymes were chosen based on SNPs identified in the male sequences (see above) to score genotypes. In total, 73 individuals were scored for each male-informative SNP, although genotyping failures meant that the number of individuals analyzed was generally somewhat less than this (between 30 and 73 with a median of 62 per locus). Some markers that were assigned to LGs on the basis of female-informative polymorphism did not have visible male-informative polymorphism and therefore could not be positionally mapped. Thus, in most cases, there were fewer markers mapped positionally than total markers assigned to each LG (Table 2 and Figure 2). After manual adjustment of linked markers to the same linkage phase, the cDNA-derived markers were added to the previously published linkage maps (Jiggins et al. 2005).
TABLE 2.
Homologous linkage group summary and color-pattern markers
| H. melpomene linkage group | B. mori linkage group | No. of genes common to both species | H. melpomene color pattern markera |
|---|---|---|---|
| 1 | 4 | 4 | K |
| 2 | 16 | 2 | — |
| 3 | 6 | 2 | — |
| 4 | 21 | 3 | — |
| 5 | 3 | 2 | — |
| 6 | 9 | 2 | — |
| 7 | 2, 11 | 7 | — |
| 8 | 25 | 1 | — |
| 9 | 7 | 3 | — |
| 10 | 5, P | 6 | Ac |
| 11 | 15 | 7 | — |
| 12 | 8, 20 | 4 | — |
| 13 | 14, 22 | 5 | — |
| 14 | 19 | 1 | — |
| 15 | 17 | 3 | Yb/Sb/N |
| 16 | 18 | 1 | — |
| 17 | 13, 24 | 7 | — |
| 18 | 23, U | 5 | B/D |
| 19 | 12 | 3 | — |
| 20 | 10 | 2 | — |
| 21 | 1 | 2 | — |
| NA | 26 | 0 | — |
| Total: 21 chromosomes | Total: 28 chromosomes | Total: 72 |
Previously appeared in Joron et al. (2006b).
Figure 2.—
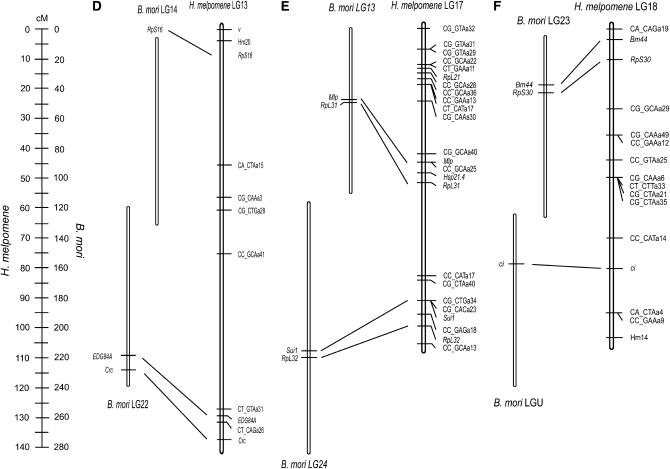
Linkage maps of putatively fused chromosomes in H. melpomene with comparison to maps of conserved markers in B. mori (A–F, corresponding to the six different putatively fused chromosomes in H. melpomene). Note the difference in scale between the maps. The lack of position bars for RpL13, ptc, and Ef1α in B. mori LG5 indicates that these markers were mapped using BAC–FISH instead of recombination linkage mapping (see Yasukochi et al. 2006). The lack of position bars for RpS16 in B. mori LG14 and H. melpomene LG13 indicates the lack of recombination mapping in B. mori and recombination mapping in a different brood (brood 44 as opposed to brood 33; see materials and methods) in H. melpomene.
Each linkage group was analyzed using MapMaker Macintosh 2.0. Linkage between markers was confirmed using the “Group” function with LOD ≥ 3.00 and Θ ≤ 0.40. Initial linkage orders were determined for six loci using “Compare,” requiring a log likelihood ≥ 0.5 between the first suggested order and the next most likely alternative. Smaller log-likelihood differences between the two most likely orders for these initial six loci could usually be attributed to the presence of two very closely linked loci, in which case one of these was replaced with another marker thought to be less closely linked before running the data through “Compare” again. Additional loci were then added one by one using “Try” to check for the likelihood of each new marker at every place in the order, and the final order was tested against the likelihood of permutations of adjacent triplets using “Ripple.” Finally, LGs were mapped using the “Map” function. For more details, see Jiggins et al. (2005). It should be noted that for the previous H. melpomene map, recombination distances were calculated with the MapMaker “error detection” function turned on. We have subsequently observed that genuine recombinant individuals can be excluded from the analysis by this method, leading to artificially reduced recombination distances. Therefore, we do not use the error detection function here, with a corresponding increase in map distance. The final relative positions of the mapped markers in the two species were subsequently compared to assess colinearity.
RESULTS
Time of divergence:
To estimate the time of divergence of lepidopteran lineages, we calculated a molecular clock using a tree based on well-established taxonomic assignments and the DNA sequences of two highly conserved proteins (Figure 1). The estimated time of divergence between the B. mori and H. melpomene lineages was 103 ± 8.6 MY, based on an estimated age of 190 MY for the Lepidoptera–Diptera divergence.
Linkage analysis:
In total, 64 cDNA-derived markers were assigned to H. melpomene linkage groups in this study. Of these markers, 8 were assigned to LGs on the basis of visible length polymorphisms from indels, and the other 56 markers were assigned to LGs on the basis of genotype scoring from restriction digestion at SNPs (Table 1 and supplemental Table 1 at http://www.genetics.org/supplemental/). High sequence similarity of amplified products to EST consensus sequences and B. mori coding sequences supported our hypothesis of orthology.
TABLE 1.
Conserved loci mapped in H. melpomene and orthologs in B. mori
| Marker name | Abbreviation | H. melpomene linkage group | GenBank accession no. | B. mori linkage groupa | GenBank accession no. |
|---|---|---|---|---|---|
| Alanyl-tRNA synthetase | Aats-ala | 1 | EF207962 | 4 | M55993 |
| Dopa decarboxylaseb | DDC | 1 | AY437802 | 4 | AF372836 |
| Ribosomal protein L3b | RpL3 | 1 | EE743523 | 4 | AB024901 |
| Winglessc | Wg | 1 | AY745485 | 4 | D14169 |
| Ribosomal protein L6 | RpL6 | 2 | EF207960 | 16 | AY769273 |
| Ribosomal protein P2 | RpP2 | 2 | EF207959 | 16 | AY769269 |
| Glutathione S-transferase | GST | 3 | EF207961 | 6 | AJ006502 |
| Ribosomal protein L15d | RpL15 | 3 | DN172764 | 6 | AY769285 |
| Mannose–phosphate isomerasec | MPI | 3 | AY332460 | — | — |
| Ribosomal protein S6 | RpS6 | 4 | EF207950 | 21 | AY769320 |
| Ribosomal protein S15 | RpS15 | 4 | EF207951 | 21 | AY706957 |
| Ribosomal protein S17 | RpS17 | 4 | EF207952 | 21 | AY769333 |
| Ribosomal protein L11b | RpL11 | 5 | CO729501 | 3 | AY769280 |
| Ribosomal protein L13A | RpL13A | 5 | EF207949 | 3 | AY769283 |
| Ecdysteroid-inducible angiotensin-converting enzyme-related gene product | Ance | 6 | EF207953 | 9 | AB026110 |
| Ribosomal protein S14d | RpS14 | 6 | CX700812 | 9 | AY706956 |
| Engrailed | eng | 7 | AY745328 | 2 | M64335 |
| Invectedc | Inv | 7 | DQ445457 | 2 | M64336 |
| Ribosomal protein S21 | RpS21 | 7 | DN172654, CX700448 | 2 | AY578154 |
| Ribosomal protein S28 | RpS28 | 7 | EF452418 | 2 | AY583363 |
| Ribosomal protein L14 | RpL14 | 7 | EF207954 | 11 | AY769284 |
| Ribosomal protein L18 | RpL18 | 7 | EF207955, EF211970 | 11 | AY769287 |
| Ribosomal Sop2 | Sop2 | 7 | DT663968, EF211973 | 11 | AY763110 |
| Distal lessc | Dll | 7 | DQ445415 | — | — |
| Mitotic checkpoint control protein (bub3) gene | Bub3 | 7 | CX700513 | — | — |
| Polycomb protein Su(z)12 | Su(z)12 | 7 | DT662097 | — | — |
| Ribosomal protein S25 | RpS25 | 8 | EF207956 | 25 | AY769340 |
| chiB (chitinase precursor) | Cht | 9 | CX700556, EF211966 | 7 | AF052914 |
| Ribonuclease L inhibitor homolog | RLI | 9 | EF207958 | 7 | AB164193 |
| Ribosomal protein S27 | RpS27 | 9 | EF207957 | 7 | AY769342 |
| Cyclin-dependent kinases regulatory subunit | Cks | 9 | CX700558 | — | — |
| Elongation factor 1αb | Ef1a | 10 | AY747994 | 5 | D13338 |
| Elongation factor 1δ | Ef1d | 10 | CX700886 | 5 | AB046366 |
| Patchedb | Ptc | 10 | AY745373 | 5 | AADK01000387 |
| Ribosomal protein L13d | RpL13 | 10 | CO729603 | 5 | AY769282 |
| Ribosomal protein L19b | RpL19 | 10 | CX700796 | 5 | AY769289 |
| Ribosomal protein S11d | RpS11 | 10 | CX700450 | P | AY706955 |
| Opsin1b | OPS1 | 11 | AF126751 | 15 | AB047924 |
| Ribosomal protein L5b | RpL5 | 11 | CO729889 | 15 | AY769272 |
| Ribosomal protein L7A | RpL7A | 11 | EF207963 | 15 | AY769275 |
| Ribosomal protein L10Ab | RpL10A | 11 | CO729740 | 15 | AY769279 |
| Ribosomal protein P0b | RpP0 | 11 | CO729821 | 15 | AJ457827 |
| Ribosomal protein S5b | RpS5 | 11 | CO729660 | 15 | AY769319 |
| Ribosomal protein S8b | RpS8 | 11 | CX700851 | 15 | AY769322 |
| Ribosomal protein L8 | RpL8 | 11 | EF207977, EF211969 | — | — |
| Ribosomal protein L30d | RpL30 | 11 | CO729949 | — | — |
| Glycine-rich protein | GRP | 12 | EF207964, EF211967 | 8 | AB197877 |
| Beta-tubulin | Btub | 12 | EF207965, EF211964 | 20 | AB003287 |
| Ribosomal protein S7 | RpS7 | 12 | EF207966 | 20 | AY769321 |
| Ribosomal protein S20 | RpS20 | 12 | CX700684 | 20 | AY769336 |
| Enolase | Eno | 12 | EF207979 | — | — |
| Ribosomal protein L7d | RpL7 | 12 | CX700625 | — | — |
| Ribosomal protein L27 | RpL27 | 12 | EF207978 | — | — |
| Ribosomal protein S12d | RpS12 | 12 | CX700631 | — | — |
| Ribosomal protein S16 | RpS16 | 13 | EF207967 | 14 | AY769332 |
| Calreticulin | Crc | 13 | EF207968 | 22 | AB090887 |
| Cuticle protein (EDG84A homolog) | EDG84A | 13 | CO729743 | 22 | AB017550 |
| PCNA | PCNA | 13 | CV526328 | 22 | AB002264, AB002265 |
| Ribosomal protein L37 | RpL37 | 13 | DN172717 | 22 | AY769308 |
| Ribosomal protein S4 | RpS4 | 13 | CO729938 | — | — |
| Vermillionb | v | 13 | AY691422 | — | — |
| Ribosomal protein L12 | RpL12 | 14 | EF207969 | 19 | AY769281 |
| Eukaryotic translation elongation factor 2 | eEF2 | 14 | CX700527 | — | — |
| Ribosomal protein S9c | RpS9 | 14 | CX700565 | — | — |
| Ribosomal protein L22c | RpL22 | 15 | CX700470 | 17 | AY769291 |
| Ribosomal protein P40c | RpP40 | 15 | CX700776 | 17 | AB062685 |
| Ribosomal protein S24 | RpS24 | 15 | EF207970, EF211972 | 17 | AY578155 |
| Eukaryotic initiation factor 3B | eiF3B | 15 | EF207980 | — | — |
| Forkhead box J1c | Fox | 15 | CR974474 | — | — |
| Rab geranygeranyl transferasec | GerTra | 15 | CR974474 | — | — |
| Elongation factor 1γ | Ef1g | 16 | EF207971 | 18 | AB046361 |
| Heat shock protein hsp21.4 | Hsp21.4 | 17 | EF207972 | 13 | AB195972 |
| Lim protein | Mlp | 17 | DT663321 | 13 | AY461436 |
| Ribosomal protein L21 | RpL21 | 17 | CO729978 | 13 | AY769290 |
| Ribosomal protein L31c | RpL31 | 17 | CX700740 | 13 | AY769301 |
| ADP/ATP translocase | ANT | 17 | EF207974, EF211962 | 24 | AY227000 |
| Ribosomal protein L32 | RpL32 | 17 | EF207973 | 24 | AB048205 |
| Sui1 | Sui1 | 17 | CO729706, EF211974 | 24 | AY426343 |
| Ribosomal protein L27a | RpL27a | 17 | EF207981 | — | — |
| Ribosomal protein S10 | RpS10 | 17 | EF207982 | — | — |
| Scallopedc | Sd | 17 | DQ674429 | — | — |
| Bm44 | Bm44 | 18 | DT664299 | 23 | AB158647 |
| Inhibitor of Apoptosis protein | IAP | 18 | CV526245, EF211968 | 23 | AF281073 |
| Ribosomal protein S30 | RpS30 | 18 | CX700724 | 23 | AY769346 |
| Cubitus interruptusb | ci | 18 | AY429297 | U | AF529422 |
| 90-kDa heat-shock protein | 90hsp | 18 | CO729719, EF211960 | U | AB060275 |
| α-Tubulin | atub | 18 | EF207983, EF211963 | — | — |
| O-Glycosyltransferased | Ogt | 18 | CV526007 | — | — |
| Decapentaplegicb | Dpp | 19 | AY747899 | 12 | BAAB01102755 |
| J-domain-containing protein | JDP | 19 | DT662955 | 12 | AF176014 |
| Ribosomal protein L9 | RpL9 | 19 | EF207975 | 12 | AY769277 |
| Muscular protein 20 | Mp20 | 19 | CO729543 | — | — |
| Prophenol oxidase-activating enzyme precursor | PPAE | 19 | CO729777 | — | — |
| Ribosomal protein L44c | RpL44 | 19 | CX700847 | — | — |
| Caspase-1 | caspase | 20 | EF207976, EF211965 | 10 | AF448494 |
| Cytosolic juvenile hormone binding protein | Jhbp | 20 | DT661817 | 10 | AF098303 |
| Actin 1 | Act | 20 | EF207985, EF211961 | — | — |
| Calcium ATPase | Ca-P | 20 | CO729824 | — | — |
| Ribosomal protein L23A | RpL23A | 20 | EF207984, EF211971 | — | — |
| Apterousb | apt | 21 (Z) | AY747887 | 1(Z) | AB024903 |
| Triose–phosphate isomeraseb | TPI | 21 (Z) | AY548151 | 1(Z) | AY734490 |
Previously appeared in Yasukochi et al. (2006).
Previously appeared in Jiggins et al. (2005).
Previously appeared in Joron et al. (2006b).
Previously appeared in Papanicolaou et al. (2005).
Synteny analysis:
Of the newly mapped cDNA-derived markers, 47 were orthologous to those mapped in B. mori, which, along with 25 orthologous markers mapped previously (Jiggins et al. 2005; Papanicolaou et al. 2005; Joron et al. 2006b), resulted in 72 orthologous markers mapped in both species. These markers span all 21 chromosomes of H. melpomene and 27 of the 28 chromosomes of B. mori (Tables 1 and 2). These markers showed completely conserved syntenic relationships between the two species and allowed identification of homologous chromosomes defined by conserved groups of anchor loci. Thirty-eight markers fell into 15 H. melpomene LGs, each of which corresponded to a single homologous chromosome in B. mori; each of these LGs contained between one and seven orthologous markers (Table 2).
Chromosomal fusions:
Consistent with the difference in chromosome number between B. mori (28) and H. melpomene (21), we found evidence for 6 of the 10 predicted chromosomal fusions in the derived Heliconiini (Figure 2). Each case suggested that two chromosomes from basal taxa, as still represented by two chromosomes in B. mori, had fused to form one H. melpomene chromosome. The additional predicted fusions could have gone undetected if they had involved either a pair of chromosomes that had also fused in the B. mori lineage or the homolog of B. mori chromosome 26, the only B. mori chromosome for which we had no shared markers in H. melpomene (Table 2), fusing either to a chromosome already identified as fused (i.e., a three-way fusion) or to another chromosome that currently appears to be in 1:1 homology with a B. mori chromosome.
Conservation of gene order:
The six putatively fused chromosomes for which we positionally mapped new markers (Figure 2) had a combined map length of 627 cM, in comparison with an estimated 424 cM for these same six chromosomes using the markers available previously (Jiggins et al. 2005). This is largely due to the error detection function in MapMaker, leading to an artificially reduced estimate of recombination distance in the previous map (see materials and methods). Thus, for example, LG7 had a map length of 51.5 cM in the previous study, which increases to 61.4 cM with the current data set using error detection. Without error detection, however, the length of the same chromosome increases to 98 cM (Figure 2A). It therefore seems likely that the overall recombination length of the H. melpomene genome is significantly larger than that previously reported. Mapping also revealed the probable orientation of most of the chromosomal fusions (Figure 2), assuming conservation of gene order on chromosomes with only two markers positionally mapped. Conservation of gene order was evident in the comparisons of H. melpomene chromosome 7 to B. mori chromosome 2 and of H. melpomene chromosome 10 to B. mori chromosome 5 (Figure 2, A and B). The latter conclusion relies on comparison with markers mapped by BAC–FISH in B. mori (Yasukochi et al. 2006; Figure 2B). An apparent reversal in gene order has occurred between Patched (ptc) and Ef1α (Figure 2B), suggesting a chromosomal inversion in one lineage or the other. Although these loci are tightly linked, reversing their order causes a significant reduction in overall likelihood (log likelihood = 2.38, P < 0.004, of the reversed order compared to that shown in Figure 2B); mapping additional markers to this region would provide a test of this conclusion.
DISCUSSION
These findings represent the most comprehensive comparative assessment of synteny in lepidopteran insects so far and indicate a high degree of conserved synteny between the B. mori and H. melpomene lineages. According to our molecular clock analyses, these lineages diverged ∼103 million years ago (MYA) (but see Vane-Wright 2004 for a discussion of different estimates for the age of butterflies), so the conserved synteny among 72 genes, after accounting for putative chromosomal fusions, indicates that Lepidoptera may have unusually high levels of linkage conservation through evolutionary time. In addition to patterns of synteny, gene order was also largely conserved on the two LGs in which comparisons were possible in this study (H. melpomene LG7, LG10) in addition to a third LG found to have conserved gene order in Yasukochi et al. (2006) (H. melpomene LG11). As in Yasukochi et al. (2006), we found evidence for a reversal of gene order between ptc and Ef1α on H. melpomene LG10 and B. mori LG5. If synteny and gene order are indeed strongly conserved across the Lepidoptera, utilization of genomic resources developed in B. mori, such as a fully assembled genome, will become a powerful resource for predictive gene finding, chromosome identification, and comparative linkage mapping in other Lepidoptera.
Other insect comparisons show a gradual decrease in shared synteny and conserved gene order over time, as might be expected. For example, a comparison of Rhagoletis pomonella with D. melanogaster (divergence time of 50–55 MYA) revealed broad conservation of synteny between several D. melanogaster chromosomal arms and individual R. pomonella chromosomes (Roethele et al. 2001). At increasing evolutionary distance—for example, comparing D. melanogaster and Anopheles gambiae (Zdobnov et al. 2002), which diverged an estimated 250 MYA—it remains possible to identify chromosomal homology but rearrangements are frequent. Thus, on the most similar chromosomal arms, Dm2L and Ag3R, 76% of orthologs in Dm2L map to Ag3R. The percentages of shared orthologs are far lower on other chromosome arms (Zdobnov et al. 2002). Finally, between D. melanogaster and the honeybee, Apis mellifera, which diverged ∼300 MYA, only a few correspondences can be established between major chromosomal elements (Weinstock et al. 2006). In contrast, mammals show much greater conservation of synteny, with considerable gene order shared between humans and chickens, which diverged ∼310 MYA (Hillier et al. 2004). Thus, the high degree of conserved synteny found between H. melpomene and B. mori is perhaps surprising when compared to the pattern seen in the Diptera and indicates that interchromosomal rearrangements have been limited throughout a large section of the Lepidoptera. Among the 72 genes studied, 15 homologous chromosomes were conserved between the two species on the basis of a comparison of 38 markers. The only cases in which genes on one H. melpomene LG were found on more than one B. mori LG appeared to be evidence of chromosomal fusions, as expected given the reductions in chromosome number for each species compared to the 31-chromosome karyotype present in the basal members of both lineages (Figure 1). Thus, the Lepidoptera may be quite unusual among groups of insects, particularly if higher-resolution linkage maps continue to support our finding of conservation of gene order (Coghlan and Wolfe 2002; Sharakhov et al. 2002).
It has been hypothesized that chromosomal fusions among holocentric chromosomes, such as those of Lepidoptera and nematodes, may be more likely than among chromosomes with localized spindles, as there is a greater possibility that fragments from chromosome fissions may attach to other chromosomes' mitotic and meiotic kinetochore microtubules during cell division (Kandul et al. 2004). However, our data do not support this suggestion and the high degree of conserved synteny between H. melpomene and B. mori suggests that chromosome fissions have not been common. Fusions may have occurred simply when two full chromosomes attached to the same set of microtubules with the concomitant loss of some genetic material. It bears noting that we would not have detected rearrangements that occurred twice (Sankoff 1999, cited in Coghlan and Wolfe 2002) or chromosome fissions followed by fusions of similar segments, but, because of the high degree of conserved synteny that we have found, it is most parsimonious to accept the hypothesis that very limited rearrangement of large segments has occurred in the Leipidoptera.
In contrast to broad-scale comparisons of synteny, also known as macrosynteny, whole-genome comparisons at finer scales can show a very different pattern. Thus, in a comparison of 12 insect genomes, very little conservation of gene order was found among species in different genera, much less across families or orders (Zdobnov and Bork 2007). Similarly, recent genome comparison between the nematodes Caenorhabditis elegans and C. briggsae, which diverged ∼100 MYA, revealed significant chromosomal rearrangements that had gone undetected with broad-scale mapping (Blaxter 2003; Gupta and Sternberg 2003; Stein et al. 2003). Between C. elegans and C. briggsae, ∼95% of inversions had taken place in segments <100 kb long (Coghlan and Wolfe 2002). In H. melpomene, there are ∼180 kb/cM (Jiggins et al. 2005); thus, if extensive reorganization between H. melpomene and B. mori were detectable only at scales similar to those in Caenorhabditis, it would be difficult to detect the majority of inversions unless markers were within 1 cM of each other. Thus, our results do not rule out extensive reorganization at a microsynteny scale, and indeed it would be surprising if such reorganization were not found between Bombyx and Heliconius. Ongoing BAC sequencing projects in Heliconius will allow direct comparison of microsynteny with Bombyx and will therefore begin to address this question (Joron et al. 2006b).
Lepidoptera chromosome numbers range from n = 7 to n = 220; however, n = 29–31 is most common. There are far more species with <29 chromosomes than there are with >31, suggesting that chromosomal fusions have been more frequent and evolutionarily successful than fissions through time (White 1973). However, even within Heliconius there is variation in chromosome number; for example, H. sapho has a large number of small chromosomes (n = 56–57; Brown et al. 1992). Thus, the anchor loci identified here would likely be of limited use in identifying conserved chromosomal linkage groups in H. sapho, and it remains an open question as to why certain species have undergone very rapid chromosomal evolution against a background of considerable stability.
Extensive comparative mapping between species and use of comparable molecular markers may contribute substantially to our understanding of speciation, insecticide resistance, and the evolutionary history of insect chromosomes (Heckel 1993; Behura 2006). Specifically, these additional anchor loci mapped in H. melpomene should assist in continuing studies of homology of color-pattern genes (Joron et al. 2006a,b). In particular, Joron et al. (2006b) mapped a locus encoding a H. melpomene wing-color phenotype to a genomic region near the genes Fox (CR974474) and GerTra (CR974474) on LG15. It will be interesting to map these genes in B. mori, as two loci with wing mutant phenotypes have already been mapped to the homologous chromosome 17: Wild wing spot (Ws) at position 14.7 and Black moth (Bm) at position 0.0 (Goldsmith 1995). More generally, it will now be possible to use the set of anchor loci identified here to determine chromosomal homology among other macrolepidopterans and as a tool to facilitate positional cloning of genes of interest controlling other traits, such as insecticide resistance and ecological adaptations.
Acknowledgments
John Pringle, Santiago Ramírez, and an anonymous reviewer provided useful comments on the manuscript. We thank Santiago Ramírez for advice concerning the molecular clock analysis and Mark Blaxter and Claire Conlon for help in generating ESTs and developing markers. We also thank Autoridad Nacional del Ambiente in Panama and the Ministerio del Medio Ambiente in Ecuador for collecting permits, the Biotechnology and Biological Sciences Research Council for financial support, and the Royal Society for a University Research Fellowship to C.D.J.
References
- Behura, S. K., 2006. Molecular marker systems in insects: current trends and future avenues. Mol. Ecol. 15: 3087–3113. [DOI] [PubMed] [Google Scholar]
- Beldade, P., S. Rudd, J. D. Gruber and A. D. Long, 2006. A wing expressed sequence tag resource for Bicyclus anynana butterflies, an evo-devo model. BMC Genomics 7: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter, M., 2003. Comparative genomics: two worms are better than one. Nature 426: 395–396. [DOI] [PubMed] [Google Scholar]
- Brown, G. R., E. E. Kadel, D. L. Bassoni, K. L. Kiehne, B. Temesgen et al., 2001. Anchored reference loci in loblolly pine (Pinus taeda L.) for integrating pine genomics. Genetics 159: 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, K. S., T. C. Emmel, P. J. Eliazar and E. Suomalainen, 1992. Evolutionary patterns in chromosome-numbers in neotropical Lepidoptera.1. Chromosomes of the Heliconiini (family Nymphalidae, subfamily Nymphalinae). Hereditas 117: 109–125. [DOI] [PubMed] [Google Scholar]
- Coghlan, A., and K. H. Wolfe, 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 12: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler, E. E., and D. Sankoff, 2003. Structural dynamics of eukaryotic chromosome evolution. Science 301: 793–797. [DOI] [PubMed] [Google Scholar]
- Gilbert, L. E., 2003. Adaptive novelty through introgression in Heliconius wing patterns: evidence for a shared genetic “toolbox” from synthetic hybrid zones and a theory of diversification, pp. 281–318 in Butterflies: Ecology and Evolution Taking Flight, edited by C. L. Boggs, W. B. Watt and P. R. Ehrlich. University of Chicago Press, Chicago.
- Goldsmith, M. R., 1995. Genetics of the silkworm: revisiting an ancient model system, pp. 21–76 in Molecular Model Systems in the Lepidoptera, edited by M. R. Goldsmith and A. S. Wilkins. Cambridge University Press, Cambridge, UK.
- Grimaldi, D., and M. S. Engel, 2005. Evolution of the Insects. Cambridge University Press, New York.
- Gupta, B. P., and P. W. Sternberg, 2003. The draft genome sequence of the nematode Caenorhabditis briggsae, a companion to C. elegans. Genome Biol. 4: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel, D. G., 1993. Comparative genetic linkage mapping in insects. Annu. Rev. Entomol. 38: 381–408. [Google Scholar]
- Hillier, L. W., W. Miller, E. Birney, W. Warren, R. C. Hardison et al., 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. [DOI] [PubMed] [Google Scholar]
- Jiggins, C. D., and W. O. McMillan, 1997. The genetic basis of an adaptive radiation: warning colour in two Heliconius species. Proc. R. Soc. Lond. Ser. B Biol. Sci. 264: 1167–1175. [Google Scholar]
- Jiggins, C. D., J. Mavarez, M. Beltran, W. O. McMillan, J. S. Johnston et al., 2005. A genetic linkage map of the mimetic butterfly Heliconius melpomene. Genetics 171: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron, M., C. D. Jiggins, A. Papanicolaou and W. O. McMillan, 2006. a Heliconius wing patterns: an evo-devo model for understanding phenotypic diversity. Heredity 97: 157–167. [DOI] [PubMed] [Google Scholar]
- Joron, M., R. Papa, M. Beltran, N. Chamberlain, J. Mavarez et al., 2006. b A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. Plos Biol. 4: 1831–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandul, N. P., V. A. Lukhtanov, A. V. Dantchenko, J. W. S. Coleman, C. H. Sekercioglu et al., 2004. Phylogeny of Agrodiaetus Hübner 1822 (Lepidoptera: Lycaenidae) inferred from mtDNA sequences of COI and COII and nuclear sequences of EF1-α: karyotype diversification and species radiation. Syst. Biol. 53: 278–298. [DOI] [PubMed] [Google Scholar]
- Kapan, D. D., N. S. Flanagan, A. Tobler, R. Papa, R. D. Reed et al., 2006. Localization of Müllerian mimicry genes on a dense linkage map of Heliconius erato. Genetics 173: 735–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst, M. R., D. D. Kapan and L. E. Gilbert, 2006. Parallel genetic architecture of parallel adaptive radiations in mimetic Heliconius butterflies. Genetics 174: 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet, J., 1989. The genetics of warning color in Peruvian hybrid zones of Heliconius erato and Heliconius melpomene. Proc. R. Soc. Lond. Ser. B Biol. Sci. 236: 163–185. [Google Scholar]
- Miao, X. X., S. J. Xu, M. H. Li, M. W. Li, J. H. Huang et al., 2005. Simple sequence repeat-based consensus linkage map of Bombyx mori. Proc. Natl. Acad. Sci. USA 102: 16303–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbit, R. E., C. D. Jiggins and J. Mallet, 2003. Mimicry: developmental genes that contribute to speciation. Evol. Dev. 5: 269–280. [DOI] [PubMed] [Google Scholar]
- Papanicolaou, A., M. Joron, W. O. McMillan, M. L. Blaxter and C. D. Jiggins, 2005. Genomic tools and cDNA derived markers for butterflies. Mol. Ecol. 14: 2883–2897. [DOI] [PubMed] [Google Scholar]
- Roethele, J. B., J. Romero-Severson and J. L. Feder, 2001. Evidence for broad-scale conservation of linkage map relationships between Rhagoletis pomonella (Diptera: Tephritidae) and Drosophila melanogaster (Diptera: Drosophilidae). Ann. Entomol. Soc. Am. 94: 936–947. [Google Scholar]
- Sanderson, M. J., 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19: 101–109. [DOI] [PubMed] [Google Scholar]
- Sankoff, D., 1999. Comparative mapping and genome rearrangement, pp. 124–134 in From Jay Lush to Genomics: Visions for Animal Breeding and Genetics, edited by J. C. M. Dekkers, S. J. Lamont and M. F. Rothschild. Iowa State University, Ames, IA.
- Sharakhov, I. V., A. C. Serazin, O. G. Grushko, A. Dana, N. Lobo et al., 2002. Inversions and gene order shuffling in Anopheles gambiae and A. funestus. Science 298: 182–185. [DOI] [PubMed] [Google Scholar]
- Sheppard, P. M., J. R. G. Turner, K. S. Brown, W. W. Benson and M. C. Singer, 1985. Genetics and the evolution of Muellerian mimicry in Heliconius butterflies. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 308: 433–613. [Google Scholar]
- Shi, J. R., D. G. Heckel and M. R. Goldsmith, 1995. A genetic linkage map for the domesticated silkworm, Bombyx mori, based on restriction fragment length polymorphisms. Genet. Res. 66: 109–126. [Google Scholar]
- Stein, L. D., Z. R. Bao, D. Blasiar, T. Blumenthal, M. R. Brent et al., 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. Plos Biol. 1: 166–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen, E., 1979. Chromosomal evolution of Lepidoptera. Hereditas 91: 316. [Google Scholar]
- Swofford, D. L., 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10. Sinauer Associates, Sunderland, MA.
- Tobler, A., D. Kapan, N. S. Flanagan, C. Gonzalez, E. Peterson et al., 2005. First-generation linkage map of the warningly colored butterfly Heliconius erato. Heredity 94: 408–417. [DOI] [PubMed] [Google Scholar]
- Turner, C. T., M. W. Davy, R. M. Macdiarmid, K. M. Plummer, N. P. Birch et al., 2006. RNA interference in the light brown apple moth, Epiphyas postvittana (Walker), induced by double-stranded RNA feeding. Insect Mol. Biol. 15: 383–391. [DOI] [PubMed] [Google Scholar]
- Vane-Wright, D., 2004. Entomology: butterflies at that awkward age. Nature 428: 477–480. [DOI] [PubMed] [Google Scholar]
- Wahlberg, N., 2006. That awkward age for butterflies: insights from the age of the butterfly subfamily Nymphalinae (Lepidoptera: Nymphalidae). Syst. Biol. 55: 703–714. [DOI] [PubMed] [Google Scholar]
- Weinstock, G. M., G. E. Robinson, R. A. Gibbs, K. C. Worley, J. D. Evans et al., 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443: 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, M. J. D., 1973. Animal Cytology and Evolution. Cambridge University Press, Cambridge, UK.
- Xia, Q. Y., Z. Y. Zhou, C. Lu, D. J. Cheng, F. Y. Dai et al., 2004. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306: 1937–1940. [DOI] [PubMed] [Google Scholar]
- Yasukochi, Y., 1998. A dense genetic map of the silkworm, Bombyx mori, covering all chromosomes based on 1018 molecular markers. Genetics 150: 1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukochi, Y., L. A. Ashakumary, K. Baba, A. Yoshido and K. Sahara, 2006. A second-generation integrated map of the silkworm reveals synteny and conserved gene order between lepidopteran insects. Genetics 173: 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshido, A., H. Bando, Y. Yasukochi and K. Sahara, 2005. The Bombyx mori karyotype and the assignment of linkage groups. Genetics 170: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov, E. M., and P. Bork, 2007. Quantification of insect genome divergence. Trends Genet. 23: 16–20. [DOI] [PubMed] [Google Scholar]
- Zdobnov, E. M., C. Von Mering, I. Letunic, D. Torrents, M. Suyama et al., 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149–159. [DOI] [PubMed] [Google Scholar]