Abstract
The tissue polarity pathway is required for the establishment of epithelial polarity in a variety of vertebrate and invertebrate organs. Core tissue polarity proteins act in a dynamically regulated complex to direct the polarization of the Drosophila eye. We report the identification and characterization of bedraggled (bdg), a novel gene that regulates one output of the tissue polarity pathway—the establishment of the R3/R4 photoreceptor fates. bdg encodes a novel, putative transporter protein and interacts genetically with all of the core polarity genes to influence the specification of the R3 and R4 cell fates. Finally, bdg is required for both viability and the initial stages of imaginal disc development.
THE polarized orientation of cells within an epithelium, known as tissue or planar cell polarity, is essential to the development of functional organs. The core tissue polarity complex, composed of a conserved group of proteins, is required for patterning the polarized structures of both vertebrate and invertebrate epithelia. In mammals, for example, the uniform orientation of stereocilia (Dabdoub et al. 2003) and the polarized movements of cells in convergent extension (Myers et al. 2002) require the activity of this complex. The core tissue polarity proteins are also essential for patterning the polarized epithelia in Drosophila, including micro- and macrochaete, legs, and ommatidia. Tissue and/or cell function-specific modulators of the tissue polarity pathway control the differentiation of diverse epithelial organs.
The Drosophila eye is a planar epithelium consisting of ∼800 unit eyes, called ommatidia. Eight of the 20 cells that compose each ommatidium are photoreceptors. The rhabdomeres, or light-sensitive organelles of the photoreceptors, are arranged in characteristic trapezoids that come in two chiral forms that show mirror-image symmetry across a midline, the equator (Figure 1A).
Figure 1.—
The overexpression of bdg suppresses sev-stbm and generates an ommatidial polarity phenotype. Tangential sections through adult eyes (left) and corresponding schematics (right) are shown. In a wild-type eye (A), chiral ommatidia are arranged with respect to the dorsal/ventral midline of mirror symmetry known as the equator (red line), such that those in the dorsal (blue trapezoids) and ventral (red trapezoids) hemispheres orient toward the dorsal and ventral poles, respectively. (B–D) bdgGMREP suppresses the mild sev-stbm phenotype from one in which ∼15% of ommatidia have polarity errors (B and C) to one in which only ∼3% have defects (B and D). Error bars represent standard deviation (SD) and triple asterisks indicate P < 10−9. Many of the results that follow are displayed in both histogram and table format. The histograms indicate SD between individuals, whereas the P-values in the tables provide a more stringent evaluation of the data, as the R × C test of independence accounts for each type of polarity error as an independent event (see materials and methods). It is therefore the P-value that indicates the robustness of the interaction. (E) Flies with two copies of bdgGMREP have an ommatidial phenotype, and (F) sev>bdg transgenic animals have a similar, but less penetrant, phenotype. Green trapezoid: AP/DV inversion. Yellow shapes denote symmetrical ommatidia (rectangles, R3/R3; circles, R4/R4) and asterisks indicate missing photoreceptors.
The Drosophila eye is precisely patterned during development. While thousands of genes cooperate to build an eye, a relatively small subset is required to polarize the epithelium (reviewed in Mlodzik 2005). Polarization of the eye is a multitiered process involving the cooperation of a long-range signal, the activity of the core tissue polarity complex, and Notch signaling. Long-range patterning systems initiate polarization in the eye with the establishment of an organizing center at the dorsoventral (D/V) boundary. Through the activity of a number of signaling molecules and pathways, dorsal and ventral fates are specified. The long-range polarity signal is transmitted by probably two parallel, nonredundant systems. The first of these systems is a set of gradients of at least three genes, four-jointed [a type II transmembrane protein (Zeidler et al. 1999; Strutt et al. 2004)], fat, and dachsous (atypical cadherins), which act via an unknown mechanism (Rawls et al. 2002; Yang et al. 2002). The second of these is the nonautonomous activity of frizzled (fz) and strabismus (stbm; also known as Van Gogh), two of the core tissue polarity genes. A complex consisting of the proteins encoded by stbm (Taylor et al. 1998; Wolff and Rubin 1998), fz (Vinson and Adler 1987; Vinson et al. 1989; Zheng et al. 1995), flamingo (fmi, also known as starry night) (Chae et al. 1999; Usui et al. 1999; Rawls and Wolff 2003), dishevelled (dsh) (Klingensmith et al. 1994; Theisen et al. 1994), diego (dgo) (Feiguin et al. 2001; Das et al. 2004), and prickle (pk) (Gubb et al. 1999; Tree et al. 2002), or the core tissue polarity complex, is a dynamically regulated signaling center that receives the global polarizing signal. Proper interpretation and execution of downstream events that establish the polarized epithelium requires that these proteins form asymmetric complexes in the photoreceptor (R) precursor cells, R3 and R4. The ultimate readout of the D/V signal is specification of the R3 and R4 cell fates via Notch signaling.
Two key events establish tissue polarity in the eye: specification of the R3 and R4 fates and the appropriate direction (and degree) of ommatidial rotation. In wild-type eyes, the polar, or more lateral, cell of the R3/R4 precursor pair adopts the R4 cell fate and the equatorial cell, which is located closer to the midline, adopts the R3 cell fate. It is believed that fate specification precedes the second event, ommatidial rotation, in which precursors rotate 90° counterclockwise in the dorsal half of the eye and 90° clockwise in the ventral half. Furthermore, it is also thought that the R3 and R4 cells instruct the ommatidial precursor to rotate in the appropriate direction of rotation with respect to its dorsal or ventral location in the eye. In the discussion that follows, all definitions are based on the model that the direction of ommatidial rotation occurs with respect to the assigned R3 and R4 fates.
In the tissue polarity mutants, one or both of these two key events can be misprogrammed, leading to a distinct set of subclasses of mutant ommatidia, including inversions on the anterior/posterior (A/P) axis, the dorsal/ventral (D/V) axis, or both axes (AP/DV) (Wolff and Rubin 1998). In AP/DV inversions, the R3 and R4 fates are correctly specified, yet ommatidia rotate in the wrong direction with respect to those fates. In D/V inversions, the R3 and R4 fates are reversed, yet rotation still occurs in the correct direction with respect to the misspecified cells. A/P inversions arise when the R3 and R4 fates are reversed and the direction of rotation is inconsistent with respect to those fates [refer to Figure 1 for detailed description of subclasses (Wolff and Rubin 1998; Wolff et al. 2007)]. A fourth class of defects, known as “symmetric ommatidia,” includes ommatidia with two R3 cells and no R4 cells (R3/R3) or two R4 cells and no R3 cells (R4/R4). The identities of cells comprising symmetric ommatidia were characterized in a landmark study by Cooper and Bray (1999), in which they correlated the molecular identity of R3 and R4 with the placement of rhabdomeres in an ommatidium. This study showed that symmetric R3/R3-type ommatidia are rectangular in shape whereas symmetric R4/R4-type ommatidia are square in shape.
The ommatidial defects described above, in combination with the fact that the mutant ommatidia often fail to rotate precisely 90°, cause a disruption of the normally smooth ommatidial lattice, giving the eye a “rough” texture. The ability to rapidly detect this phenotype enabled the identification of several of the core tissue polarity genes in large-scale loss-of-function screens. However, genes that contribute to the establishment of polarity, yet have either no or a very weak polarity phenotype, go undetected using this strategy. To circumvent this limitation in a search for new regulators of ommatidial polarity, we conducted a genetic modifier screen in a sensitized stbm background. Such modifier screens also provide an opportunity to identify genes that act redundantly with, downstream of, or in parallel to the core polarity complex to direct its output in a process-appropriate manner.
This screen identified bdg, a novel gene that is predicted to encode a transporter protein. bdg modifies the core tissue polarity genes to influence the R3 and R4 cell fates. An extensive genetic analysis demonstrates that bdg interacts with all of the core tissue polarity genes, but perhaps not with Notch, to influence the R3/R4 fate decision. While overexpression of bdg generates a moderate ommatidial polarity phenotype, polarity defects in bdg loss-of-function mutants are rare, suggesting functional redundancy with the core polarity complex. In addition, Bdg is required for viability and early imaginal disc development. Finally, bdg mutant escapers display several locomotor phenotypes typically seen in neurotransmission-deficient flies.
MATERIALS AND METHODS
Genetics and P-element screen:
The Glass Multimer Reporter Enhancer-Promoter (GMREP) collection (generous gift from B. Hay) was used, which included bdgGMREP, CG8291KG07083, bdg36, bdg90, bdg164, sev-stbm14-1, sev-stbm3-2, sev-stbm3-4, sev-stbm9-4, stbm6cn, stbm153, sev-phyl, sev-fz, sev-dsh, sev-N, sev-Gal4, UAS-bdgS1, UAS-fmi, UAS-dgo, GMR-phyl, GMR-Gal4, L(2)Pin/Kr-GFP, CyO, pksple, dsh1, fz19 (fzN21), fz20(fzJ22), dgo380, fmifrz3, dIAP2GMREP, P{ry+t7.2=PZ}l(2)0524805248 cn1/CyO; ry506, SpCyO; Δ2-3 Sb/TM6, w1118, and Canton-S.
For the dominant modifier screen, transgenic flies carrying two copies of the sev-stbm construct were crossed to ∼1800 GMREP lines. The F1 progeny were scored under the dissecting microscope for dominant modification of the sev-stbm rough eye phenotype; eyes from the 69 GMREP lines that showed an interaction were subsequently sectioned (as described by Wolff 2000a,b) and the phenotypes quantitated. Thirty-five enhancers and one suppressor of sev-stbm were confirmed as dominant modifiers of the sev-stbm phenotype (see Wolff et al. 2007 for details).
Phenotypic, statistic, and mosaic analyses:
Adult eyes were fixed, embedded, and sectioned according to Wolff (2000). The number of ommatidia, N, and number of eyes (in parentheses) scored per genotype are included in detailed tables reporting phenotypic analyses. Because bdg90 is lethal, and since bdg90, bdg36, and bdg164 show consistent loss-of-function phenotypes and genetic interactions in the seven cases tested (supplemental Table 1 at http://www.genetics.org/supplemental/), bdg164 was used for all double-mutant analyses.
A G-test of independence was performed to determine if the sev-stbm/P-element lines displayed statistically significant differences in the classes of ommatidial defects compared to sev-stbm. The G-test is similar to the commonly used Pearson's chi-square test, but produces more accurate results for small sample sizes and makes possible a distinction between the component parts that comprise the overall change in phenotype. In other words, while two genotypes may have the same overall ommatidial phenotype, there can be dramatic differences in the subclasses of ommatidial phenotypes; the G-test of independence extracts these differences. Therefore, even though a standard deviation (SD) for a given interaction may be quite large, if the more diagnostic P-value is very small, the interaction is robust. For this test, between 411 and 2135 ommatidia from a minimum of five eyes of each modifier and background line were placed into the following categories: A/P inversions, D/V inversions, AP/DV inversions, R3/R3, R4/R4, fail to rotate (or missing photoreceptors), and normal. Two MATLAB scripts were written to calculate the G statistic (corrected by William's factor) as described in (Sokal 1995). These scripts can be downloaded from (http://www.genetics.wustl.edu/rmlab/gtest/).
bdgGMREP overexpression clones were generated using standard FLP/FRT methods and mosaic R3/R4 pairs were scored for expression of the transgene. bdg90 clones were also generated using standard FLP/FRT methods using ey-FLP or using the FLP/FRT strategy in a Minute background.
In situ hybridization and Northern blot analysis:
For in situ hybridization studies of candidate genes, third instar eye discs were dissected and processed as described (Wolff 2000a,b). Antisense and sense DIG-labeled RNA probes of the following candidate genes were generated from EST clones (Research Genetics, Birmingham, AL): CG8291 (bdg) (clone SD06837), CG8297 (clone SD23639), and MLF (clone SD02769), according to manufacturer's protocol (Roche Molecular Biochemicals). In situ hybridization was carried out according to established protocol, using 1 μg of DIG-labeled RNA probe (Wolff 2000a,b). For Northern blot analysis of bdg, 30 third instar larvae were homogenized in Trizol reagent, and total RNA was extracted according to the manufacturer's protocol (GIBCO, Grand Island, NY). Twenty-five nanograms of RNA were analyzed according to standard protocol (Sambrook et al. 1989) using 32P-labeled probe (∼2 × 107 CPM) generated from cDNA clone SD06837.
Immunohistology:
Third larval instar eye discs were dissected and processed (Wolff 2000a,b). Primary antibody incubations were conducted at 4° overnight, at the following concentrations: α-dIAP2 1:50 (Huh et al. 2007), α-Stbm 1:500, α-Fmi 1:10 (Usui et al. 1999; generous gift from T. Uemura), α-Arm 1:10 (Developmental Studies Hybridoma Bank, University of Iowa), and α-GABA 1:100 (gift of R. Wong). Secondary antibodies conjugated to Alexafluor fluorescent dyes were used at 1:200 according to the manufacturer's protocol (Molecular Probes, Eugene, OR).
Phylogenetic analysis of Bdg:
Nine proteins most similar to the Bdg amino acid sequence were determined using NCBI-BlastP (Altschul and Lipman 1990). The Drosophila serotonin transporter (Ser T) was also included. A multiple sequence alignment was generated using Clustal X (Thompson et al. 1997). The neighbor-joining tree was generated from 1000 iterations of the unweighted pair group method with arithmetic means (UPGMA).
P-element excision screen for bdg deletion alleles:
bdgGMREP females were crossed to Sp/CyO; Δ2-3 Sb/TM6 males and 20,000 F2 genomes were scored for loss of the w+ transgene. Individuals with potential excisions were subjected to a PCR-based analysis of the genomic region. Three bdg alleles, bdg36, bdg90, and bdg164, were identified using this approach, and PCR was used to map minimal deletions in these alleles (see supplemental methods at http://www.genetics.org/supplemental/). The bdg locus encodes three transcripts, each of which produces an identical 1331-amino-acid protein (FlyBase, Indiana University; Figure 2A) due to the use of a shared translational start in exon 2. The deletions in bdg36 and bdg164 alleles are contained within the large intron, leaving exons 1 and 2 intact. Exon 2 is deleted in bdg90, the predicted null allele.
Figure 2.—
Annotated gene CG8291 is bdg and encodes a novel transporter protein. (A) The CG8291 locus at chromosomal position 52D2 generates three mRNA transcripts (RA, RB, and RC), each of which encodes the same protein due to use of a common translation start site (yellow boxes). The bdgGMREP and KG07083 P-element insertions map to the large intron (orange triangles). The regions deleted in each of the bdg loss-of-function alleles are illustrated as blue lines for each allele. (B) In situ hybridization of CG8291-RB probe in third instar eye discs suggests that bdg is CG8291. (C) Phylogenetic analysis of bdg reveals that it is a novel member of the sodium-dependent neurotransmitter transporter family, most closely related to the GABA transporters.
Generation of UAS-bdg transgenic flies:
The CG8291-RB (bdg) transcript was amplified by PCR from the full-length cDNA clone, SD06851 (Research Genetics), using 5′ GACAAATCAGCTGCGACATC and 3′ AAGCAAGCGGATATGTGGAT primers. The 5′ and 3′ primer included BglII and NotI sites, respectively, and the PCR product was directionally cloned into pUASt. Automated sequencing (Big Dye V3 chemistry; ABI Prism 3100 sequencer, Applied Biosystems, Foster City, CA) confirmed that the construct represented the full-length wild-type bdg cDNA. pUASt-bdg DNA was injected into nondechorionated embryos within 1 hr after egg laying at 200 ng/λ with 50 ng/λ of s129A helper DNA (Beall et al. 2002), according to established protocol (Rubin and Spradling 1982). Six hundred embryos were injected. Ninety surviving founders were backcrossed to w1118 and F1 progeny were screened for the w+ transgene. Six transformants were isolated, and UAS-bdgS1 was used for overexpression and rescue experiments.
RESULTS
bdgGMREP is a dominant suppressor of sev-stbm:
bedraggled (bdg), a novel gene that encodes a putative transporter protein, was identified in an F1 dominant modifier screen as a suppressor of the tissue polarity gene stbm, under the control of the sevenless (sev) promoter. This screen was carried out in a sensitized genetic background in which the sev promoter was used to drive high levels of stbm expression (sev-stbm) in photoreceptors R3, R4, R7, and the nonneuronal cone cells. Misexpression of stbm in this subset of cells results in a mild ommatidial polarity phenotype: flies carrying one copy of the sev-stbm transgene inserted on the second chromosome (sev-stbm14-1) exhibit polarity defects in 15.4% of ommatidia (Figure 1C) (Rawls and Wolff 2003). Since this degree of disruption, as well as enhancement and suppression of this phenotype, can be readily detected at the dissecting microscope level, sev-stbm14-1 was used as the genetic background for the screen. Briefly, flies carrying two copies of the sev-stbm14-1 insertion were independently crossed to 1800 uncharacterized P-element lines [GMREP collection (Hay et al. 1997); generous gift of B. Hay] and the F1 progeny were examined for an enhanced or suppressed degree of eye roughness. Thirty-five GMREP lines were found to enhance (Wolff et al. 2007) and one was found to suppress the sev-stbm mild rough-eye phenotype (Figure 1, B and D). Notably, it is rare to identify suppressors of tissue polarity genes—bdg represents one of only three suppressors of sev-stbm that have been identified in our lab in ∼4000 lines screened. Characterization of the suppressor identified in this screen, which we have named bedraggled for its appearance after getting stuck in the food due to defects in motor coordination, is described here.
One copy of the bdg P-element line, bdgGMREP, suppresses the sev-stbm14-1/+ phenotype from one in which 15.4% of ommatidia have defects in polarity to one in which only 3.4% are mutant (Figure 1, B and D; Table 1); note that the P-value, not the SD, is the indicator of significance of the interaction, as discussed in materials and methods). This genetic interaction was reproducible in three additional sev-stbm lines tested: the sev-stbm3-2 phenotype is suppressed from 8.8 to 1.4%, sev-stbm3-4 from 9.4 to 1.7%, and sev-stbm9-4 from 21.0 to 0.9%. The genetic interaction between sev-stbm and bdgGMREP is specific to the function of these genes and is not due to a nonspecific effect on the promoters, as bdgGMREP does not dominantly modify the sev-phyllopod (phyl) phenotype nor does sev-stbm modify the GMR-phyl phenotype (data not shown). To confirm the role for bdg in the tissue polarity signaling pathway suggested by these overexpression data, we carried out extensive loss-of-function genetic analyses, as described below.
TABLE 1.
bdg interacts with the core tissue polarity genes in overexpression and loss-of-function analyses
| Genotype | A/P (%) | D/V (%) | AP/DV (%) | R3/R3 (%) | R4/R4 (%) | FTR (%) | Total errors (%) | N |
|---|---|---|---|---|---|---|---|---|
| sev-stbm14-1/+ | 5.5 | 6.3 | 0.4 | 1.2 | 2.2 | 0 | 15.5 | 2135 (20) |
| sev-stbm14-1/bdgGMREP*** | 1.2 | 1.5 | 0.2 | 0.3 | 0.3 | 0 | 3.4 | 1892 (20) |
| sev-stbm14-1/bdg164*** | 7.0 | 11.5 | 3.1 | 0.7 | 2.6 | 0.7 | 25.5 | 1537 (13) |
| stbm6cn/stbm6cn | 10.6 | 21.9 | 3.6 | 4.3 | 1.6 | 2.3 | 44.3 | 1328 (12) |
| stbm6cn, bdgGMREP/stbm6cn*** | 12.6 | 27.7 | 2.3 | 1.0 | 1.3 | 0 | 45.0 | 1240 (16) |
| stbm6cn, bdg164/stbm6cn, bdg164*** | 9.7 | 26.3 | 4.5 | 13.8 | 4.9 | 1.7 | 61.1 | 1488 (15) |
| stbm153/stbm153 | 11.3 | 18.6 | 3.0 | 0.4 | 1.4 | 1.5 | 36.2 | 1049 (11) |
| stbm153, bdg164/stbm153, bdg164*** | 8.8 | 12.9 | 3.0 | 23.2 | 4.4 | 1.8 | 53.9 | 799 (8) |
| sev-dsh/+ | 3.4 | 2.3 | 2.4 | 1.9 | 0.6 | 0 | 10.5 | 1006 (9) |
| sev-dsh/bdgGMREP* | 5.0 | 3.8 | 2.6 | 3.7 | 1.6 | 0 | 16.7 | 1336 (11) |
| sev-dsh/bdg164 | 3.0 | 2.6 | 3.5 | 1.0 | 1.0 | 0 | 11.1 | 800 (7) |
| dsh1/Y | 9.4 | 6.3 | 4.9 | 2.2 | 0.6 | 1.3 | 24.7 | 956 (9) |
| dsh1/Y; bdgGMREP/+*** | 14.3 | 11.8 | 6.3 | 3.6 | 5.2 | 1.1 | 42.4 | 441 (5) |
| dsh1/Y; bdg164/+*** | 8.4 | 7.0 | 4.9 | 4.4 | 6.4 | 0.4 | 31.6 | 699 (6) |
| dsh1/Y; bdg164/bdg164*** | 12.9 | 10.3 | 5.4 | 25.6 | 7.0 | 0.8 | 61.9 | 1268 (8) |
| pksple/pksple | 1.1 | 41.5 | 1.9 | 1.6 | 0.8 | 0 | 46.9 | 744 (7) |
| pksple, bdgGMREP/pksple | 2.1 | 39.2 | 1.2 | 0.5 | 0.9 | 0 | 43.9 | 864 (10) |
| pksple, bdg164/pksple, bdg164*** | 2.7 | 31.1 | 1.1 | 9.2 | 3.4 | 0.1 | 47.6 | 818 (9) |
| sev-fmi/Y | 9.7 | 31.9 | 2.2 | 0.7 | 0.4 | 0.6 | 45.6 | 1253 (10) |
| sev-fmi/Y; bdgGMREP/+* | 13.3 | 27.3 | 2.8 | 0.8 | 1.1 | 0.1 | 45.5 | 714 (6) |
| sev-fmi/Y; bdg164/+* | 7.8 | 24.9 | 2.5 | 1.9 | 2.3 | 0.2 | 39.6 | 606 (6) |
| fmifrz3/fmifrz3 | 9.6 | 5.4 | 2.7 | 3.6 | 1.2 | 0.7 | 23.2 | 1073 (12) |
| fmifrz3, bdgGMREP/fmifrz3*** | 13.5 | 23.7 | 4.2 | 2.0 | 0.6 | 0.9 | 44.9 | 801 (9) |
| fmifrz3, bdg164/fmifrz3, bdg164*** | 6.1 | 5.2 | 3.6 | 12.5 | 8.7 | 0 | 35.9 | 473 (6) |
| sev∷dgo2C/+ | 19.6 | 0.8 | 1.4 | 2.8 | 0.4 | 3.0 | 28.1 | 855 (9) |
| sev∷dgo2C; bdgGMREP/+*** | 31.5 | 2.4 | 0.8 | 6.7 | 4.4 | 1.5 | 47.3 | 723 (8) |
| sev∷dgo2C; bdg164/+*** | 18.8 | 1.0 | 0.4 | 10.4 | 3.0 | 3.7 | 37.3 | 789 (8) |
| dgo380/dgo380 | 3.6 | 1.4 | 0.5 | 13.2 | 14.1 | 0.6 | 33.5 | 937 (9) |
| dgo380, bdgGMREP/dgo380*** | 6.3 | 3.2 | 1.1 | 4.3 | 5.0 | 0.1 | 20.0 | 916 (10) |
| dgo380, bdg164/dgo380*** | 2.2 | 0.9 | 0.9 | 8.6 | 5.4 | 0.2 | 18.1 | 778 (9) |
| dgo380, bdg164/dgo380, bdg164 | Lethal | |||||||
| sev-fz/+ | 11.2 | 6.3 | 6.1 | 29.9 | 13.2 | 0 | 66.7 | 839 (9) |
| bdgGMREP/+; sev-fz/+** | 17.1 | 8.1 | 4.6 | 18.8 | 14.6 | 0 | 63.1 | 919 (7) |
| bdg164/+; sev-fz | 10.0 | 7.1 | 4.4 | 32.7 | 12.4 | 0 | 66.6 | 661 (7) |
| fzN21/fzJ22 | 5.6 | 4.9 | 3.1 | 0.1 | 0.3 | 1.5 | 15.6 | 744 (8) |
| bdgGMREP/+; fzN21/fzJ22* | 6.3 | 6.8 | 5.5 | 0.2 | 0.7 | 0.2 | 19.7 | 867 (9) |
| bdg164/+; fzN21/fzJ22 | 5.2 | 4.0 | 1.6 | 0.4 | 0.5 | 0.2 | 11.9 | 965 (10) |
| bdg164/bdg164; fzN21/fzJ22*** | 7.8 | 7.3 | 3.5 | 5.8 | 1.6 | 0.3 | 26.4 | 1180 (7) |
FTR, failure to rotate; N, number of ommatidia scored (eyes scored). Statistical significance is measured by a G-test of independence. The underlined genotype is the reference in each data set. *P-value <10−3; **P-value <10−6; ***P-value <10−9.
bdgGMREP is an overexpression line of annotated gene CG8291:
The P-element transposon in bdgGMREP contains the glass multimer reporter (GMR) with its endogenous enhancer-promoter element (EP). GMR is an eye-specific driver in cells posterior to the morphogenetic furrow of the eye imaginal disc (Hay et al. 1997). The EP element can exert its effect on genes that lie within 10 kb upstream or downstream of the P element (Hay et al. 1997). Consequently, phenotypes in the GMREP, sev-stbm lines can be the result of (1) disruption of the gene into which the P element inserts or (2) overexpression of a gene that lies within 10 kb of the insertion site.
As a first step in identifying the gene responsible for the interaction with sev-stbm and characterizing the nature of the effect [i.e., loss of function due to the disruption of a gene vs. mis- or overexpression of a nearby gene(s)], we used plasmid rescue to isolate the genomic DNA surrounding the GMREP insertion and subsequently cloned and sequenced this DNA. The P element is inserted at cytological position 52D2 and disrupts the 5′ region of annotated gene CG8291 (Figure 2A). Three additional genes, Drosophila inhibitor of apoptosis 2 (dIAP2), myologenous leukemia factor (MLF), and annotated gene CG8297, lie within 20 kb (+10 to −10 kb) of the insertion site and were therefore also considered candidate interactors. However, genetic interaction data (not shown) and in situ hybridization analysis eliminated these three genes as candidates. In situ hybridization of wild-type and bdgGMREP third larval instar eye imaginal discs revealed that CG8297 and MLF transcripts are expressed at wild-type levels in bdgGMREP discs (data not shown), whereas CG8291 is overexpressed in bdgGMREP discs (Figure 2B). dIAP2 protein is also present at wild-type levels in bdgGMREP discs (data not shown). Finally, Northern blot analysis revealed that the CG8291 transcript is more abundant in total RNA isolated from bdgGMREP larvae than in total RNA isolated from wild-type larvae (supplemental Figure 1 at http://www.genetics.org/supplemental/).
Additional evidence that CG8291 is the gene responsible for the bdgGMREP phenotypes comes from the analysis of a second P-element insertion in CG8291, CG8291KG07083 (BDGP). This experimentally uncharacterized P element also maps to the 5′ region of CG8291 (Figure 2A) and is predicted to disrupt the transcriptional activity of CG8291. CG8291KG07083 dominantly enhances the phenotype: the sev-stbm phenotype is enhanced approximately twofold, from 15.4% ommatidial errors to 30% ommatidial errors (supplemental Figure 2 at http://www.genetics.org/supplemental/). (A further characterization of this interaction was conducted using confirmed loss-of-function alleles of bdg, generated by imprecise excision, as discussed below.)
Finally, the bdg cDNA was used to generate UAS-bdg transgenic animals. Standard rescue experiments were not possible, as overexpression of bdg throughout the body, using either actin- or hs-Gal4, is lethal. As an alternative, we demonstrated that bdg driven by GMR recapitulates the bdgGMREP homozygous phenotype (see below; Table 2). Additionally, UAS-bdg∷sev-Gal4 transgenic flies reproduce, albeit weakly, the bdgGMREP eye phenotype (Figure 1F; Table 2). Together, these molecular and genetic data demonstrate that CG8291 is bdg.
TABLE 2.
All core tissue polarity genes, except fz, dominantly suppress bdgGMREP overexpression phenotype
| Genotype | A/P (%) | D/V (%) | AP/DV (%) | R3/R3 (%) | R4/R4 (%) | Missing R (%) | Total errors (%) | N |
|---|---|---|---|---|---|---|---|---|
| bdgGMREP/bdgGMREP | 1.6 | 0.4 | 0.2 | 6.8 | 3.0 | 8.1 | 20.1 | 894 (10) |
| stbm6cn, bdgGMREP/bdgGMREP*** | 0 | 0.1 | 0.1 | 0.7 | 0.2 | 2.0 | 3.1 | 912 (8) |
| dsh1/+; bdgGMREP/bdgGMREP*** | 0.4 | 0.1 | 0 | 0.1 | 0.3 | 0.3 | 1.2 | 730 (9) |
| pksple, bdgGMREP/bdgGMREP*** | 0.4 | 1.6 | 0 | 2.8 | 1.9 | 1.9 | 8.6 | 573 (5) |
| fmifrz3, bdgGMREP/bdgGMREP*** | 0.4 | 0.6 | 0 | 0.6 | 1.0 | 0.8 | 3.4 | 495 (6) |
| dgo380, bdgGMREP/bdgGMREP*** | 0.2 | 0.7 | 0.2 | 1.1 | 0.7 | 1.8 | 4.6 | 565 (6) |
| bdgGMREP/bdgGMREP; fzJ22/+ | 1.5 | 0.5 | 0 | 2.9 | 4.4 | 7.1 | 16.3 | 411 (5) |
| GMR-Gal4RC/+;UAS-bdg/UAS-bdg | 1.1 | 0.1 | 0.1 | 5.2 | 2.0 | 5.3 | 13.9 | 699 (13) |
| sev-Gal4/sev-Gal4;UAS-bdg/UAS-bdg | 0.1 | 0 | 0.3 | 1.2 | 0 | 0.4 | 2.0 | 1852 (12) |
N, number of ommatidia scored (eyes scored). Statistical significance was measured by a G-test of independence. The underlined genotype is the reference for this data set. ***P-value <10−9.
Bdg is a putative transporter protein:
Bdg is encoded by a novel gene and is annotated as a neurotransmitter:sodium symporter (Berkeley Drosophila Genome Project, BDGP), a protein that uses energy from the cotransport of Na+ and Cl− to transport a neurotransmitter against its concentration gradient. Our phylogenetic analysis indicates that Bdg is most closely related to the sodium-dependent GABA transporter family (Figure 2C). A hydrophobicity analysis of Bdg predicts that it contains 11 or 12 transmembrane domains [derived using TMHMM, a transmembrane helix prediction program (Sonnhammer et al. 1998)], in agreement with the BDGP annotation of CG8291 as an integral membrane protein. This finding is consistent with the idea that Bdg is a transporter with a hydrophobic core that constructs a channel for sodium-dependent transport.
To test the possibility that Bdg is transporting GABA, we (1) assayed GABA expression in the developing eye and (2) studied the effect of inhibition of GABA transport. While we did detect high levels of GABA in the larval brain, GABA was undetectable in the third larval eye imaginal disc (supplemental Figure 3 at http://www.genetics.org/supplemental/). Furthermore, nipecotic acid, a pharmacological inhibitor of GABA transport, did not result in an ommatidial polarity phenotype nor did it modify the sev-stbm phenotype (data not shown; methods according to Leal and Neckameyer 2002). These data suggest that Bdg does not transport GABA in the developing eye.
The only convincing non-drosophilid homolog of Bdg is a novel transporter in the genome of the mosquito, Anopheles gambiae (gene identifier: agCG55776). While there is no functional or structural information available for this homolog, Bdg and agCG55776 may represent a novel insect-specific transporter. Expression studies of CG8291 in the embryo indicate that bdg transcript is abundant in the developing CNS, a pattern consistent with its predicted role as a neurotransmitter transporter (Kearney et al. 2004). The hydrophobicity data, together with our phylogenetic analysis and the published embryonic expression study, make a correlative argument that Bdg functions in the transport of a small molecule (on the order of 100–200 Da); this small molecule could be a neurotransmitter, an ion, or an amino acid. An alternative possibility is that bdg encodes a signaling molecule.
Trapezoid morphing: trapezoid shape can be plane dependent:
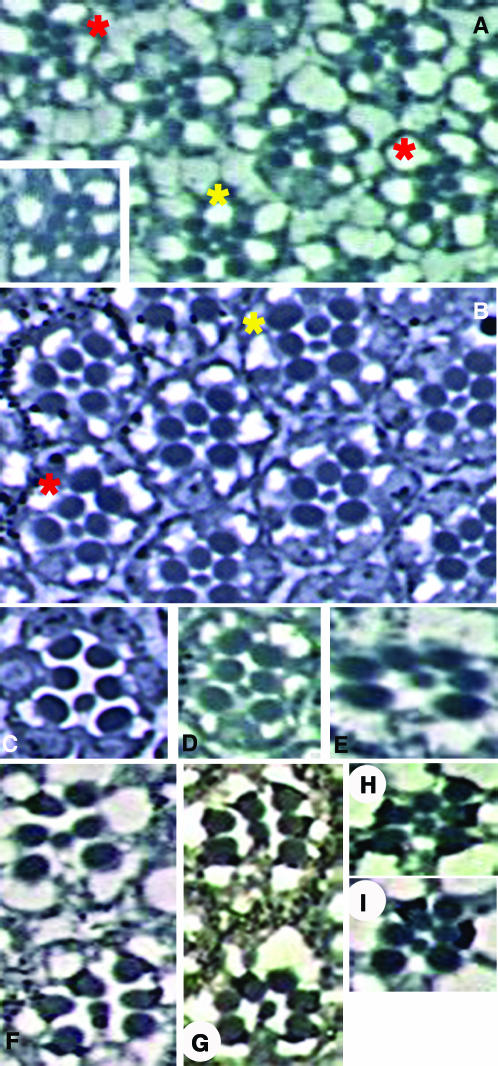
Tomlinson and Struhl (1999) noted that the rhabdomeres of mutant ommatidia can assume different arrangements relative to one another at different depths within the retina. For example, an ommatidium that appears squat in shape and is characteristic of symmetric R4/R4-type ommatidia (Fanto et al. 1998; Cooper and Bray 1999) can look distinctly different at a more apical or basal plane. Due to the nature of the phenotypes explored in our studies, it was necessary to extend the original observation by Tomlinson and Struhl to include wild type, several mutants, and a sufficiently large sample size to establish whether their observation was a rare or a common phenomenon and if this phenomenon occurs in wild type or if it is unique to ommatidia with certain phenotypes.
Ommatidia were analyzed in at least three focal planes at the level of the R7 rhabdomere to exclude differences in rhabdomeral arrangement that occur at the R8 level (Figure 3). Our analysis indicates that “trapezoid morphing” does not occur in wild type (zero incidents in ∼1200 ommatidia scored from 12 eyes), but does occur in assorted mutants and therefore is not unique to bdg. We analyzed eyes from dsh;bdg and bdg;fz double mutants, as well as from a random GMREP line that exhibits polarity defects, and found that a significant percentage of ommatidia undergo trapezoid morphing: 39% of mutant ommatidia morph in dsh;bdg double-mutant eyes (320 ommatidia analyzed in 13 eyes) (Figure 3, see legend for details). Some aberrant ommatidia were likely missed in the quantification of the bdg/tissue polarity genetic interactions due to trapezoid morphing. However, since these mutant ommatidia would be missed with equal probability in all genotypes, the data reported in Table 1 accurately reflect the genetic interactions.
Figure 3.—
Trapezoid morphology of mutant ommatidia can change between focal planes. Thirty-nine percent of mutant ommatidia in dsh1/dsh1;bdgGMREP/bdgGMREP eyes “morph” throughout the apical portion of the eye. A subset of morphed ommatidia is highlighted; each color represents a single ommatidium at three distinct focal planes that bisect the eye at the level of the R7 rhabdomere. For example, the ommatidium circled in orange appears to have two R3 cells in the most apical plane (A), wild-type morphology in an intermediate plane (B), and two R4 cells in the most basal plane (C).
Overexpression of bdg generates an eye phenotype:
Eyes from bdgGMREP flies exhibit a tissue polarity phenotype. Flies carrying one copy of the bdgGMREP transgene are wild type (data not shown), but in the presence of two copies, 12% of ommatidia have defects in polarity. An additional 8% are missing photoreceptors, so the polarity of these ommatidia could not be assayed (Figure 1E; Table 2). Of these polarity defects, 82%—a strikingly large proportion—are due to problems in specification of the R3 and R4 cell fates (Table 2), as the trapezoids are symmetric in shape. Fifty-seven percent of these ommatidia have a characteristic “rectangular trapezoid” shape, a shape indicative of two R3 cells and no R4 cells (symmetric R3/R3-type ommatidia) and 25% of ommatidia are “squat” in shape, likely a consequence of a failure to specify the R3 fate, giving rise to ommatidia that have two R4 cells and no R3 cells (symmetric R4/R4-type ommatidia) (Figure 1E). (An analysis of the molecular identity of these cells was reserved for the loss-of-function tissue polarity studies described below.)
Notably, despite the significant number of symmetric ommatidia in bdgGMREP flies, the external surface of the eye remains smooth. In contrast with tissue polarity mutants, the lattice is perfectly maintained due to wild-type rotation of these mutant ommatidia.
bdgGMREP modifies the ommatidial polarity phenotypes of core tissue polarity genes:
A detailed description of the interactions discussed in this and the following sections is found in Table 1. Note that statistical significance indicated may be due to modification of specific subclasses rather than the overall phenotype, as explained in materials and methods.
bdgGMREP significantly modifies the eye polarity phenotypes of stbm, dsh, fmi, dgo, and fz, but not of pk. While stbm, dsh, fmi, and fz are all enhanced by bdgGMREP, dgo is the only core tissue polarity gene that is suppressed by bdgGMREP. The R3 and R4 fate decisions are predominantly modified in these interactions, with the exception of the bdg, fz interaction. This initial genetic characterization suggests that bdg (i) is important for the specification of a single R3 cell and a single R4 cell, (ii) can uniquely modify members of the polarity complex, and (iii) interacts with the core polarity genes (with the possible exception of fz) to influence the R3 and R4 fates.
R3 and R4 cell identities in the analyses described above were characterized on the basis of ommatidial morphology, as defined by Cooper and Bray (1999). We confirmed these assignments of cell fate using the R4-specific marker mδ-lacZ (Cooper and Bray 1999). Ommatidial precursors from dsh/Y; bdgGMREP/+; mδ-lacZ/+ exhibit either symmetric staining in the R3 and R4 precursors, including clusters with two or zero lacZ-positive cells, or the wild-type asymmetry (Figure 4). These results are consistent with the assignments made in adult eyes and thereby confirm our interpretation that Bdg is important for the R3/R4 cell fate decision. Since the adult phenotype (scored on the basis of morphology) is consistent with the larval phenotype (scored on the basis of molecular markers), all subsequent analyses were conducted in the adult to enable the sampling of a sufficient number of ommatidia required for our rigorous statistical analysis.
Figure 4.—
An R4-specific marker reveals that the R3 and R4 cells are incorrectly specified in dsh, bdgGMREP imaginal discs. (A–C) mδ 0.5 in a wild-type background and (D–F) dsh1/Y; bdgGMREP/+; mδ 0.5/+ third instar eye imaginal discs stained with α-β-gal to identify the R4-specific Notch target, mδ 0.5 (green; A and D), and a-Elav (red; B and E), a pan-neuronal marker used here to label photoreceptor nuclei. Overlays are shown in C and F. In wild-type discs, one cell per cluster is identified as R4 by mδ 0.5 (A and C, arrows), whereas in the mutant tissue, ommatidial precursors exhibit a variety of defects, including: two mδ 0.5-positive cells, indicative of two R4 cells (D and F, arrowheads); zero mδ 0.5-positive cells, indicative of two R3 cells (D and F, arrows); or one mδ 0.5-cell, indicating a wild-type complement of cells (one R3 and one R4; D and F, asterisks). These results are consistent with the assignments of symmetric R3/R3- and R4/R4-type ommatidia in adult sections. Anterior is to the right.
bdg is required broadly for organismal viability, imaginal disc development, and motor coordination:
Multiple loss-of-function alleles of bdg were generated by imprecise excision of the bdgGMREP transgene. Three of these alleles were characterized in detail at the molecular, phenotypic, and genetic levels. Two of these three alleles, bdg36 and bdg164, are likely to be hypomorphic alleles while the third, bdg90, is likely null, based on molecular and genetic analyses. PCR-based analysis indicates that exon 2, which encodes the translation start site (Figure 2A), is deleted in bdg90 mutants. In situ hybridization reveals that larvae homozygous for the bdg36 allele appear to have wild-type to slightly reduced levels of bdg transcript (data not shown) and bdg164 larvae have reduced transcript levels (Figure 2B). The bdg transcript in bdg90 larvae cannot be accurately measured due to their severely reduced eye imaginal discs (see below).
Bdg is required for viability: each of the three alleles described above is homozygous lethal, although occasional bdg36 and bdg164 escapers survive to eclosion. Bdg plays a role in early morphogenesis of the imaginal discs, as these structures are dramatically diminished in size in third instar larvae homozygous for bdg90; notably, overall larval size is normal (see Figure 5 for eye and antennal discs). bdg36 and bdg164 escapers do not exhibit abnormalities in thorax or wing hair polarity (data not shown). However, on the basis of the eye phenotype (described below), we speculate that if there are phenotypes in these tissues, they would be of such low penetrance that it would not be feasible to unambiguously define them as genetic defects (for example, an occasional misoriented hair could be due to mechanical disruption), so we cannot determine if bdg plays a role in these aspects of development.
Figure 5.—
Eye and antennal imaginal discs are significantly reduced in bdg null larvae. Wild-type (A and B) and bdg90 (C and D) eye-antennal imaginal discs are shown. Eye-antennal imaginal discs were removed from the brain (B) to better illustrate the size of wild-type discs. bdg null discs are ill-defined masses of tissue (C and D). A bdg eye disc attached to the brain via the optic stalk can be distinguished in D.
bdg also plays critical roles in later developmental events in the eye, and at least one of these roles is likely to be redundant with other genes (perhaps the core tissue polarity genes). Analysis of bdg164 escaper eyes and bdg90 mutant clones revealed that while the vast majority of bdg mutant ommatidia are wild type, two phenotypes do occur: some ommatidia are missing photoreceptors and a small fraction show defects in R3/R4 fate specification (Figure 6, A–E; both phenotypes are consistent with the overexpression line, bdgGMREP); of >2200 bdg90 ommatidia surveyed, only 10 symmetric ommatidia were identified in bdg null clones. The low frequency of symmetric ommatidia in null clones raises the possibility that bdg acts redundantly with other genes involved in R3/R4 fate specification.
Figure 6.—
bdg mutant phenotype. bdg mutant ommatidia are generally phenotypically wild type, although rare ommatidia are missing photoreceptors (not shown), have one extra photoreceptor opposite the R3 cell (yellow asterisks, A and B), are of the R3/R3-type symmetric ommatidia (red asterisks, A and B; inset in A; C and D), or are of the R4/R4-type symmetric ommatidia (E). These phenotypes are evident in a hypomorphic allele, bdg164 (A) as well as a null allele, bdg90 (B–E). Ommatidia mosaic for the R3/R4 pair are either R3/R3-type (F; R3−/R4+) or R4/R4-type (R3−/R4+) ommatidia (G; note that in F and G, one wild-type ommatidium is included to illustrate anterior and posterior), or these mosaic ommatidia are phenotypically wild type (H and I). Phenotypically wild-type mosaic ommatidia can be either R3+/R4− (H) or R3−/R4+ (I). Anterior is to the right.
Finally, bdg is essential for motor coordination. bdg36 and bdg164 escapers are grossly uncoordinated—upon eclosion, they immediately fall into the food and die (hence the name, bedraggled). If these flies are retrieved before getting stuck in the food, they are unable to fly, they exhibit a severely delayed righting reflex, and although escapers appear to walk normally, they have impaired climbing behavior. The behavioral defects we observe in bdg mutants are similar to those reported for some flies with defective neurotransmission (Arredondo et al. 1998; Leal and Neckameyer 2002; Nichols et al. 2002; Godenschwege et al. 2004), raising the possibility that the bdg phenotype arises as a consequence of a deficiency in neurotransmitter transport.
bdg loss-of-function alleles interact with all core tissue polarity genes:
Loss-of-function bdg also interacts with sev-stbm: bdg164 dominantly enhances the sev-stbm phenotype from one in which 15.4% of ommatidia have polarity errors to one in which 25.5% have defects. This enhancement results primarily from an increase in D/V inversions, which have their basis in an R3/R4 fate reversal (Figure 7, A, B, and G; Table 1). Additionally, bdg164 dominantly suppresses sev>fmi (45.6–39.6%), again due almost entirely to suppression of D/V inversions (Figure 7, C, D, and G; Table 1). sev>dgo is also enhanced by bdg164 (28.1–37.3%), but this enhancement results entirely from an increase of symmetric R3/R3- and R4/R4-type ommatidia (Figure 7, E–G; Table 1). Interestingly, loss of bdg function does not modify the sev-dsh or sev-fz phenotypes (Table 1). bdg164 was used in all loss-of-function analyses; bdg164 and bdg90 show consistent genetic interactions (see materials and methods and supplemental Table 1 at http://www.genetics.org/supplemental/).
Figure 7.—
Reduced bdg function enhances sev-driven tissue polarity phenotypes. Sections of adult eyes (left) and corresponding schematics (right) are shown. bdg164, a hypomorphic allele, enhances the phenotypes of sev-stbm (A and B), sev>fmi (C and D), and sev>dgo (E and F). These genetic interactions are quantified in G (see Table 1 for phenotypic details). Each bar represents the total number of polarity defects; blue represents that portion that are R3/R4 errors and all other classes are depicted as red. Error bars represent SD; single and triple asterisks indicate P < 10−3 and 10−9, respectively. Colored trapezoids: blue, wild type; red, D/V inversions; black, A/P inversions; green, AP/DV inversions; yellow rectangles, R3/R3; yellow circles, R4/R4; +, extra cell; *, missing cell. Anterior is to the right.
None of the bdg alleles tested—bdgGMREP, bdg90, or bdg164—exhibited genetic interactions with sev-Notch (supplemental Figure 4 at http://www.genetics.org/supplemental/, see legend for details). Even though all of the core tissue polarity genes interacted with bdg in at least one of these genetic assays, we cannot rule out the possibility that Notch interacts with bdg in other contexts.
Loss-of-function bdg also displays genetic interactions with loss-of-function alleles of the tissue polarity genes. bdg164 dominantly enhances dsh1, and this interaction can be attributed entirely to a specific enhancement of R3/R3- and R4/R4-type defects (Figure 8, A, B, and E; Table 1). Interestingly, bdg164 dominantly suppresses dgo380. Again, as with previously discussed bdg/dgo interactions, the most significant suppression occurred in the symmetric class of defects (Figure 8, C–E; Table 1).
Figure 8.—
bdg dominantly modifies the dsh and dgo phenotypes. Tangential sections through adult eyes (left) and corresponding schematics (right) are shown. bdg164 dominantly enhances dsh1 (A and B) and dominantly suppresses dgo380 (C and D). These genetic interactions were quantified, as illustrated in E. (See Table 1 for phenotypic details.) Each bar represents the total number of polarity defects; blue represents that portion that are R3/R4 errors and all other classes are depicted as red. Error bars represent SD; triple asterisks indicate P < 10−9. Colored trapezoids: blue, wild type; red, D/V inversions; black, A/P inversions; green, AP/DV inversions; yellow rectangles, R3/R3; yellow circles, R4/R4; +, extra cell; *, missing cell. Anterior is to the right.
bdg acts synergistically with all of the core tissue polarity genes (Figure 9). Double homozygous combinations of bdg164 and mutations in the core tissue polarity genes stbm, dsh, pk, fmi, and fz result in a highly statistically significant (P < 10−9) enhancement of the homozygous tissue polarity phenotype in all cases (Table 2); homozygosity for both bdg164 and dgo380 is lethal. The striking enhancement of the stbm6cn/stbm6cn, dsh1 /Y, fmifrz3/fmifrz3, and fzN21/fzJ22 phenotypes by bdg164 homozygosity is due almost entirely to the specific increase of both symmetric R3/R3- and R4/R4-type defects (Figure 9, A–D, G, J, and K, respectively; Table 1). The highly statistically significant modification of the pksple/ pksple phenotype by bdg164 homozygosity is unique: while the R3/R3- and R4/R4-type defects are significantly enhanced, D/V inversions are strongly suppressed, resulting in only a modest change in the total percentage of ommatidia with polarity errors (Figure 9, E, F, and K; Table 1).
Figure 9.—
bdg acts synergistically with the core tissue polarity pathway. Sections of adult eyes (left) and corresponding schematics (right) are shown. Double-mutant analysis reveals that bdg164 homozygosity enhances the R3/R4 phenotypes of stbm6cn (A and B), dsh1 (C and D), pksple (E and F), fmifrz3 (G and H), and fzN21/fzJ22 (I and J). Notably, the double-mutant combination of bdg164and dgo380 is lethal. Quantification of these genetic interactions is shown in K. (See Table 1 for phenotypic details.) Each bar represents the total number of polarity defects; blue represents that portion that are R3/R4 errors and all other classes are depicted as red. Error bars represent SD; triple asterisks indicate P < 10−9. Colored trapezoids: blue, wild type; red, D/V inversions; black, A/P inversions; green, AP/DV inversions; yellow rectangles, R3/R3; yellow circles, R4/R4; +, extra cell; *, missing cell. Anterior is to the right.
Bdg influences the regulation of the R3/R4 fates through the tissue polarity complex:
The bdgGMREP overexpression phenotype suggests that bdg may influence the R3/R4 cell fates. To determine if bdg is required to specify either the R3 or R4 fate, we performed a mosaic analysis of bdg90 (a null allele) mutant clones. bdg null clones, generated using ey-FLP, were notably smaller than standard ey clones, typically encompassing as few as several cells up to 10–20 ommatidia (one virtually phenotypically wild-type outlier included ∼50 ommatidia). The small size of bdg clones is consistent with the small imaginal discs described for bdg90 larval escapers.
Two mutant phenotypes occur in ommatidia mosaic for bdg90. First, numerous ommatidia on clonal borders are missing photoreceptors (data not shown). These ommatidia are evidently of mosaic descent, as the remaining photoreceptors in these ommatidia can be either genetically mutant or genetically wild type. Although the genotype of the missing receptors cannot be determined, they are likely to be bdg− since ommatidia with missing photoreceptors are characteristic of the bdg phenotype.
Second, symmetric mosaic ommatidia also occur, although they are relatively rare (Figure 6). Each mosaic symmetric ommatidium identified was mosaic for bdg in those cells occupying the R3 and R4 positions, so symmetric ommatidia do not arise unless an ommatidium is mosaic for the R3/R4 pair. Notably, the absence of bdg in the remaining photoreceptors (R1, R2, and R5–R8) did not affect the R3/R4 fate decision. The majority of symmetric mosaic ommatidia identified were of the R4/R4 type (Figure 6G), although R3/R3 ommatidia were also found (Figure 6F). In these ommatidia, the cell on the anterior side (i.e., the R3 side) was always mutant whereas the cell on the posterior was always wild type. These observations (i) suggest that bdg activity is necessary, but not sufficient, to promote the R3 fate and (ii) raise the possibility that the relative levels of bdg activity in R3 and R4 can tip the overall balance in the feedback loop(s) that operate among the tissue polarity proteins in these two cells.
The vast majority of mosaic ommatidia are phenotypically wild type, as is the case with ommatidia composed entirely of photoreceptors that are null for bdg. Analysis of mosaic, phenotypically wild-type ommatidia indicates that they can be of the R3+/R4− (Figure 6H) or R3−/R4+ (Figure 6I) genotypes, although more were of the R3−/R4+genotype.
We also generated bdgGMREP overexpression clones and found that the photoreceptor that overexpressed bdg was modestly biased toward the R4 cell fate: in 65% of R3/R4 pairs mosaic for bdgGMREP, the bdgGMREP cell adopted the R4 fate (data not shown). The consequence of this bias is not clear, as all of these ommatidia are phenotypically wild type. Taken together, the loss-of-function and overexpression mosaic analyses described here do not enable us to make a clear distinction between a potential requirement for bdg in either R3 or R4. Rather, they suggest that bdg is important generally for the R3/R4 fate decision. In addition, the missing photoreceptor phenotype indicates that bdg is necessary but not sufficient for cell viability or recruitment.
bdgGMREP is dominantly rescued by all core tissue polarity genes except fz:
Haploinsufficiency of stbm rescues the bdgGMREP homozygous phenotype from one in which 20.1% of ommatidia have developmental defects to one in which only 3.1% have errors (Table 2). These data suggest that bdg may act upstream of stbm since reduction in the dose of a downstream signaling molecule can rescue an overexpression phenotype. Interactions between bdgGMREP and the tissue polarity genes were quantitated to define the position of bdg in the signaling pathway. Loss-of-function alleles of dsh, pk, fmi, and dgo, but not fz, also dominantly suppress the homozygous bdgGMREP phenotype (Table 2). Although reduction in an upstream gene can sometimes rescue an overexpression phenotype, here we argue that loss of a downstream gene is the most common mechanism for the suppression of an overexpression phenotype. Under this assumption, these data are consistent with a model in which bdg acts upstream of (or in parallel to) stbm, dsh, pk, fmi, and dgo, but downstream or independently of fz.
DISCUSSION
Bdg is a novel regulator of ommatidial polarity that plays a nonessential but integral role in ommatidial polarity by regulating the activity of the tissue polarity complex to influence the R3/R4 cell fates. bdg is a unique component of the tissue polarity signaling pathway, as it is the first gene to show selectivity in its interactions with members of the core tissue polarity complex; it acts upstream of, or in parallel to, and yet redundantly with, the core tissue polarity complex to direct the establishment of ommatidial polarity; and it is the first reported suppressor of a loss-of-function tissue polarity phenotype (the symmetrical defects in dgo, as described). Furthermore, the nature of the molecule—a predicted transporter—provides an unexpected twist to the current model of how tissue polarity is regulated and suggests a need to think more broadly about the mechanisms that drive this process. In addition to its unique role in regulating polarity, bdg plays more global roles as well, as it is also required for viability and the early development of imaginal discs.
Bdg plays a critical and novel role in influencing the R3 and R4 cell fates:
bdg represents a novel regulator of the R3/R4 fate decision, and the evidence for this role is compelling. First, loss of bdg function in tissue polarity mutant backgrounds robustly enhances or suppresses the number of symmetric ommatidia; all other subclasses of polarity defects (for example, AP/DV) are affected to a significantly smaller degree. This specificity suggests that bdg acts during the R3/R4 cell fate decision, perhaps at the level of feedback between the presumptive R3 and R4 cells, to ensure that just one cell of each fate is specified in each ommatidium. Second, symmetric ommatidia can arise when bdg expression either is reduced or exceeds wild-type levels. While this phenotype reveals a role for bdg in the specification of the R3 and R4 cells, it appears to be a redundant role, given the small number of phenotypically mutant ommatidia in bdg null clones. However, this role is clearly not insignificant, as indicated by the critical role revealed for bdg in the sensitized background of the tissue polarity mutants. Third, symmetric mosaic ommatidia occur only when the R3/R4 pair of cells is mosaic for bdg. bdg occupies a novel position in the specification of the R3 and R4 cell fates in that it interacts with the tissue polarity genes but perhaps not with the downstream effector and ultimate determinant of the R3/R4 fate decision, Notch.
Furthermore, Bdg is partially redundant with the tissue polarity complex in specifying a single R3 and R4 cell, as mutations in bdg synergistically enhance the symmetric errors of all the core tissue polarity genes (with the notable exception of dgo). The observation that loss-of-function bdg can strongly influence this fate choice in both fz and stbm mutant backgrounds, gene products for which there is a requirement in R3 and R4, respectively (Zheng et al. 1995; Wolff and Rubin 1998), suggests that Bdg function affects the output of the complex at the level of the R3/R4 fates.
bdg acts uniquely in the context of dgo:
dgo is the most recently identified, and least understood, core tissue polarity gene. As such, the cellular function of Dgo has not been fully elucidated. It has been proposed that Dgo is enriched on the R3 side of the R3/R4 border and acts to anchor the polarity proteins to the membrane, a role that is redundant with Pk and Stbm (Mihaly et al. 2005).The loss-of-function interaction between bdg and dgo is unique in that it is the only tissue polarity gene in which the number of symmetric errors is dominantly suppressed by loss-of-function bdg, as this class of error is enhanced in fz, dsh, stbm, pk, and fmi. Similarly, dgo is the only tissue polarity gene that is suppressed by bdgGMREP.
Dgo's remarkably distinct response to Bdg function reveals that Dgo is somehow functionally distinct from other members of the complex. Indeed, of all the tissue polarity mutants, dgo mutants exhibit, by far, the most penetrant R3/R4 phenotype: ∼30% of ommatidia are symmetric (Das et al. 2004). This phenotype is particularly interesting in light of the unique interaction between dgo and bdg, in which bdg selectively suppresses the R3/R3- and R4/R4-type ommatidia in dgo. The bdg/dgo interaction could also have a basis in the cellular requirements for, or physical associations between, bdg and dgo. For example, bdg may be localized to either the R3 or the R4 cell, but its cargo required in the other cell. In this case, bdg and dgo could physically associate in the R3 cell, but bdg could be required in both the R3 cell (localization) and the R4 cell (cargo). It is also interesting to note that, in contrast to all other tissue polarity, bdg double mutants, dgo, bdg homozygotes are lethal. The significance of this interaction is unclear, but would indicate that dgo acts with bdg in additional developmental contexts.
Neurotransmitter transport may be broadly required during development:
The role of neurotransmitter transport in the establishment and maintenance of neuromuscular junction biology is well established; however, only a handful of studies implicate the role of neurotransmission in developmental events. For example, acetylcholine transport is important for the morphology and establishment of neuronal connections during development (Maruyama et al. 2001). A role for GABA in Drosophila morphogenesis has also been established—pharmaceutical antagonism of, and RNA interference directed against, the GABA (β(1)) subunit results in reduced hatching, lethality, runting, and trachaeal folding (Dzitoyeva et al. 2005). Our identification of a novel role for a putative neurotransmitter transporter in additional developmental processes indicates that roles for neurotransmission may be more diverse than previously recognized.
bdg is predicted to encode a neurotransmitter transporter and belongs to a family of transporters that use the energy generated by the cotransport of Na+ and Cl− to move neurotransmitter molecules against a concentration gradient. This protein family shares a common 12-transmembrane helix motif, as well as a sodium:neurotransmitter symporter family (SNF) domain. The Na+/Cl− neurotransmitter transporters can be further divided into four subfamilies on the basis of sequence homology: transporters for GABA, monoamines, the amino acids proline and glycine, and a group of orphan transporters (Lill and Nelson 1998). Our phylogenetic analysis supports the prediction that bdg encodes a neurotransmitter transporter and also indicates that Bdg is closely related to the GABA transporter family. While the Bdg cargo has not yet been experimentally defined, we have shown that Bdg does not appear to transport GABA during eye development.
While Bdg's cargo may not be a neurotransmitter, at the very least we can conclude that its cargo is small, as neurotransmitters are only ∼100–200 Da. Possible cargoes that fall within these size restrictions include single amino acids (110 Da, on average), ions, and neurotransmitters, but not proteins (for example, Wg is >50 kDa). It is also formally possible that Bdg shares homology with transporters yet functions primarily as a signaling molecule. This is the case with the Drosophila protein Pathetic, which, although annotated as an amino acid transporter, regulates growth via a nutrient-sensing mechanism that functions independently of bulk amino acid transport (Goberdhan et al. 2005).
Bedraggled—transporter of elusive factor X?
A long-standing model for ommatidial polarity invokes the activity of a long-range morphogen that establishes a gradient of polarizing activity across the developing eye (Wehrli and Tomlinson 1998). In spite of extensive efforts dedicated to the identification of this predicted morphogen, termed Factor X, it has not yet been uncovered. It has long been assumed that the morphogen is a protein. However, if the morphogen is instead a small molecule, it would have been missed in genetic screens since its identity would have precluded it from the pool of candidates. The discovery of Bdg, the only transporter protein implicated in ommatidial polarity to date, points to a possible role for Bdg as a transporter of a non-Wnt morphogen, provided Bdg acts independently of fz (our genetic analysis does indicate that bdg acts downstream or independently of fz). Several observations support the validity of this possibility. First, Bdg makes a critical contribution to the R3/R4 fate decision, which is thought to be a functional readout of Factor X, indicating that the transport of some small molecule plays a role in this fate decision. Second, bdg transcript is enriched at the wings of the third instar eye imaginal disc, a pattern that has been predicted for a factor that acts in long-range signaling in the eye (Wehrli and Tomlinson 1998). Third, Bdg has been predicted to symport an unknown neurotransmitter along with sodium, a cation (BDGP annotation). Intriguingly, cations are among a handful of small molecules that have established roles in long-range activity in some developmental contexts, for example, the role of calcium in setting up the D/V axis in embryos (Creton et al. 2000) and the essential role of the H+/K+-ATPase transporter in orienting the left–right body axis in Xenopus (Levin et al. 2002).
While this global transport hypothesis is within the realm of possibility, we favor a model in which Bdg contributes to the establishment of ommatidial polarity by facilitating local (i.e., between R3 and R4), rather than global, transport. More specifically, Bdg may transport a signal between members of the tissue polarity complex to modulate R3/R4 cell fate specification. Such a locally transported signal could have global results if it is perpetuated via cell–cell relay across the eye primordium. This revised hypothesis would indicate that Bdg represents a reasonable candidate for the transporter of “Factor X.” The identification of the Bdg cargo will be necessary to test this potential mechanism.
Acknowledgments
We thank B. Hay for generously providing the Glass Multimer Reporter Enhancer-Promoter (GMREP) collection; J. Glasscock for analyzing the bdg intronic region; J. First for technical assistance; S. Dutcher for assistance with the phylogenetic analysis; B. Hay, D. Strutt, D. Gubb, M. Mlodzik, T. Uemura, and the Bloomington Stock Center for providing fly stocks; B. Hay and T. Uemura for providing antibodies; B. Hay, D. Wassarman, and H. Chang for scientific guidance; and D. Strutt, H. Chang, and A. DiAntonio for critical comments on the manuscript. This work was supported by a Lucille P. Markey Pathway fellowship to A.S.R. and by National Institutes of Health grant R01 EY13136 to T.W.
References
- Altschul, S. F., and D. J. Lipman, 1990. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. USA 87: 5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredondo, L., H. B. Nelson, K. Beckingham and M. Stern, 1998. Increased transmitter release and aberrant synapse morphology in a Drosophila Calmodulin mutant. Genetics 150: 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall, E. L., M. B. Mahoney and D. C. Rio, 2002. Identification and analysis of a hyperactive mutant form of Drosophila P-element transposase. Genetics 162: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, J., M. J. Kim, J. H. Goo, S. Collier, D. Gubb et al., 1999. The Drosophila tissue polarity gene starry night encodes a member of the protocadherin family. Development 126: 5421–5429. [DOI] [PubMed] [Google Scholar]
- Cooper, M. T., and S. J. Bray, 1999. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature 397: 526–530. [DOI] [PubMed] [Google Scholar]
- Creton, R., J. A. Kreiling and L. F. Jaffe, 2000. Presence and roles of calcium gradients along the dorsal-ventral axis in Drosophila embryos. Dev. Biol. 217: 375–385. [DOI] [PubMed] [Google Scholar]
- Dabdoub, A., M. J. Donohue, A. Brennan, V. Wolf, M. Montcouquiol et al., 2003. Wnt signaling mediates reorientation of outer hair cell stereociliary bundles in the mammalian cochlea. Development 130: 2375–2384. [DOI] [PubMed] [Google Scholar]
- Das, G., A. Jenny, T. J. Klein, S. Eaton and M. Mlodzik, 2004. Diego interacts with Prickle and Strabismus/Van Gogh to localize planar cell polarity complexes. Development 131: 4467–4476. [DOI] [PubMed] [Google Scholar]
- Dzitoyeva, S., A. Gutnov, M. Imbesi, N. Dimitrijevic and H. Manev, 2005. Developmental role of GABAB(1) receptors in Drosophila. Brain Res. Dev. Brain Res. 158: 111–114. [DOI] [PubMed] [Google Scholar]
- Fanto, M., C. A. Mayes and M. Mlodzik, 1998. Linking cell-fate specification to planar polarity: determination of the R3/R4 photoreceptors is a prerequisite for the interpretation of the Frizzled mediated polarity signal. Mech. Dev. 74: 51–58. [DOI] [PubMed] [Google Scholar]
- Feiguin, F., M. Hannus, M. Mlodzik and S. Eaton, 2001. The ankyrin repeat protein Diego mediates Frizzled-dependent planar polarization. Dev. Cell 1: 93–101. [DOI] [PubMed] [Google Scholar]
- Goberdhan, D. C., D. Meredith, C. A. Boyd and C. Wilson, 2005. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development 132: 2365–2375. [DOI] [PubMed] [Google Scholar]
- Godenschwege, T. A., D. Reisch, S. Diegelmann, K. Eberle, N. Funk et al., 2004. Flies lacking all synapsins are unexpectedly healthy but are impaired in complex behaviour. Eur. J. Neurosci. 20: 611–622. [DOI] [PubMed] [Google Scholar]
- Gubb, D., C. Green, D. Huen, D. Coulson, G. Johnson et al., 1999. The balance between isoforms of the prickle LIM domain protein is critical for planar polarity in Drosophila imaginal discs. Genes Dev. 13: 2315–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, B. A., R. Maile and G. M. Rubin, 1997. P element insertion-dependent gene activation in the Drosophila eye. Proc. Natl. Acad. Sci. USA 94: 5195–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, J. R., I. Foe, I. Muro, C. H. Chen, J. H. Seol et al., 2007. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J. Biol. Chem. 282: 2056–2068. [DOI] [PubMed] [Google Scholar]
- Kearney, J. B., S. R. Wheeler, P. Estes, B. Parente and S. T. Crews, 2004. Gene expression profiling of the developing Drosophila CNS midline cells. Dev. Biol. 275: 473–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingensmith, J., R. Nusse and N. Perrimon, 1994. The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes Dev. 8: 118–130. [DOI] [PubMed] [Google Scholar]
- Leal, S. M., and W. S. Neckameyer, 2002. Pharmacological evidence for GABAergic regulation of specific behaviors in Drosophila melanogaster. J. Neurobiol. 50: 245–261. [DOI] [PubMed] [Google Scholar]
- Levin, M., T. Thorlin, K. R. Robinson, T. Nogi and M. Mercola, 2002. Asymmetries in H+/K+-ATPase and cell membrane potentials comprise a very early step in left-right patterning. Cell 111: 77–89. [DOI] [PubMed] [Google Scholar]
- Lill, H., and N. Nelson, 1998. Homologies and family relationships among Na+/Cl- neurotransmitter transporters. Methods Enzymol. 296: 425–436. [DOI] [PubMed] [Google Scholar]
- Maruyama, H., T. L. Rakow and I. N. Maruyama, 2001. Synaptic exocytosis and nervous system development impaired in Caenorhabditis elegans unc-13 mutants. Neuroscience 104: 287–297. [DOI] [PubMed] [Google Scholar]
- Mihaly, J., T. Matusek and C. Pataki, 2005. Diego and friends play again. FEBS J. 272: 3241–3252. [DOI] [PubMed] [Google Scholar]
- Mlodzik, V. E. M., 2005. Planar Cell Polarization During Development. Elsevier, New York.
- Myers, D. C., D. S. Sepich and L. Solnica-Krezel, 2002. Bmp activity gradient regulates convergent extension during zebrafish gastrulation. Dev. Biol. 243: 81–98. [DOI] [PubMed] [Google Scholar]
- Nichols, C. D., J. Ronesi, W. Pratt and E. Sanders-Bush, 2002. Hallucinogens and Drosophila: linking serotonin receptor activation to behavior. Neuroscience 115: 979–984. [DOI] [PubMed] [Google Scholar]
- Rawls, A. S., and T. Wolff, 2003. Strabismus requires Flamingo and Prickle function to regulate tissue polarity in the Drosophila eye. Development 130: 1877–1887. [DOI] [PubMed] [Google Scholar]
- Rawls, A. S., J. B. Guinto and T. Wolff, 2002. The cadherins fat and dachsous regulate dorsal/ventral signaling in the Drosophila eye. Curr. Biol. 12: 1021–1026. [DOI] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Sokal, R., and F. Rohlf, 1995. Biometry: The Principles and Practice of Statistics in Biological Research, Ed. 3. W. H. Freeman, San Francisco.
- Sonnhammer, E. L., G. von Heijne and A. Krogh, 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6: 175–182. [PubMed] [Google Scholar]
- Strutt, H., J. Mundy, K. Hofstra and D. Strutt, 2004. Cleavage and secretion is not required for four-jointed function in Drosophila patterning. Development 131: 881–890. [DOI] [PubMed] [Google Scholar]
- Taylor, J., N. Abramova, J. Charlton and P. N. Adler, 1998. Van Gogh: a new Drosophila tissue polarity gene. Genetics 150: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen, H., J. Purcell, M. Bennett, D. Kansagara, A. Syed et al., 1994. dishevelled is required during wingless signaling to establish both cell polarity and cell identity. Development 120: 347–360. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson, A., and G. Struhl, 1999. Decoding vectorial information from a gradient: sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development 126: 5725–5738. [DOI] [PubMed] [Google Scholar]
- Tree, D. R., J. M. Shulman, R. Rousset, M. P. Scott, D. Gubb et al., 2002. Prickle mediates feedback amplification to generate asymmetric planar cell polarity signaling. Cell 109: 371–381. [DOI] [PubMed] [Google Scholar]
- Usui, T., Y. Shima, Y. Shimada, S. Hirano, R. W. Burgess et al., 1999. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98: 585–595. [DOI] [PubMed] [Google Scholar]
- Vinson, C. R., and P. N. Adler, 1987. Directional non-cell autonomy and the transmission of polarity information by the frizzled gene of Drosophila. Nature 329: 549–551. [DOI] [PubMed] [Google Scholar]
- Vinson, C. R., S. Conover and P. N. Adler, 1989. A Drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains. Nature 338: 263–264. [DOI] [PubMed] [Google Scholar]
- Wehrli, M., and A. Tomlinson, 1998. Independent regulation of anterior/posterior and equatorial/polar polarity in the Drosophila eye; evidence for the involvement of Wnt signaling in the equatorial/polar axis. Development 125: 1421–1432. [DOI] [PubMed] [Google Scholar]
- Wolff, T., 2000. a Histological techniques for the Drosophila eye. Part I: larva and pupa, pp. 200–227 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Wolff, T., 2000. b Histological techniques for the Drosophila eye. Part II: adult, pp. 228–243 in Drosophila Protocols, edited by W. Sullivan, M. Ashburner and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Wolff, T., and G. M. Rubin, 1998. Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development 125: 1149–1159. [DOI] [PubMed] [Google Scholar]
- Wolff, T., J. B. Guinto and A. S. Rawls, 2007. Screen for genetic modifiers of stbm reveals that photoreceptor fate and rotation can be genetically uncoupled in the Drosophila eye. PLoS ONE 2: e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. H., J. D. Axelrod and M. A. Simon, 2002. Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell 108: 675–688. [DOI] [PubMed] [Google Scholar]
- Zeidler, M. P., N. Perrimon and D. I. Strutt, 1999. The four-jointed gene is required in the Drosophila eye for ommatidial polarity specification. Curr. Biol. 9: 1363–1372. [DOI] [PubMed] [Google Scholar]
- Zheng, L., J. Zhang and R. W. Carthew, 1995. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development 121: 3045–3055. [DOI] [PubMed] [Google Scholar]