Abstract
A previous study of histone H3 in Saccharomyces cerevisiae identified a mutant with a single amino acid change, leucine 61 to tryptophan, that confers several transcriptional defects. We now present several lines of evidence that this H3 mutant, H3-L61W, is impaired at the level of transcription elongation, likely by altered interactions with the conserved factor Spt16, a subunit of the transcription elongation complex yFACT. First, a selection for suppressors of the H3-L61W cold-sensitive phenotype has identified novel mutations in the gene encoding Spt16. These genetic interactions are allele specific, suggesting a direct interaction between H3 and Spt16. Second, similar to several other elongation and chromatin mutants, including spt16 mutants, an H3-L61W mutant allows transcription from a cryptic promoter within the FLO8 coding region. Finally, chromatin-immunoprecipitation experiments show that in an H3-L61W mutant there is a dramatically altered profile of Spt16 association over transcribed regions, with reduced levels over 5′-coding regions and elevated levels over the 3′ regions. Taken together, these and other results provide strong evidence that the integrity of histone H3 is crucial for ensuring proper distribution of Spt16 across transcribed genes and suggest a model for the mechanism by which Spt16 normally dissociates from DNA following transcription.
THE basic unit of chromatin is the nucleosome, a structure consisting of 146 nucleotides of DNA wrapped around a protein octamer composed of pairs of each of the four core histone proteins (Luger et al. 1997). In addition to directing the condensation of DNA, histone proteins also play crucial and active roles in the regulation of cellular processes that use chromatin as their substrate (Luger 2006). Studies from many laboratories employing a variety of experimental approaches have converged into a general model for chromatin function in which dynamic alterations in chromatin structure are of central importance to processes such as gene transcription and DNA replication.
The presence of nucleosomes over transcription units can pose a structural barrier that is inhibitory to transcription initiation and elongation (Sims et al. 2004; Workman 2006). Several mechanisms have been described that can overcome the repressive nature of nucleosomes in initiation, including covalent chemical modifications of specific histone residues, ATP-dependent remodeling of nucleosomes, and the removal of histones (Berger 2002; Narlikar et al. 2002; Boeger et al. 2003; Reinke and Horz 2003). Furthermore, recent studies have demonstrated that regulatory regions of genes are inherently low in nucleosome density and that histone loss at promoter regions, a process known to be mediated at some genes by the Asf1 histone chaperone, is associated with active transcription (Boeger et al. 2003; Reinke and Horz 2003; Adkins et al. 2004; Bernstein et al. 2004; Ercan and Simpson 2004; Lee et al. 2004; Sekinger et al. 2005; Yuan et al. 2005; Korber et al. 2006; Segal et al. 2006). These and other studies have revealed the dynamic nature of chromatin over gene promoters as it relates to the transcription state and have provided insights into how the factors that mediate these chromatin alterations relate to each other.
The role of chromatin structure in transcription elongation has been the focus of intense research in the last several years. This work has shown that as transcription proceeds across a gene and eventually terminates, the composition of the RNA polymerase II (Pol II) elongation complex changes, with different factors arriving at and departing from the complex in a highly coordinated fashion. These changes are dependent, at least in part, on the phosphorylation status of the C-terminal domain (CTD) of the largest subunit of Pol II (Hartzog 2003; Bentley 2005; Buratowski 2005; Eissenberg and Shilatifard 2006; Rosonina et al. 2006). As the elongation complex traverses a transcription unit, the underlying chromatin undergoes a set of histone modifications, including changes in methylation, acetylation, and ubiquitylation (Workman 2006). In addition to covalent modifications, histones are also believed to be physically displaced in front of an elongating Pol II complex and reassembled in the wake of Pol II passage by a number of factors, including the FACT complex, the elongation factors Spt4, Spt5, and Spt6, and the histone chaperone Asf1 (Hartzog et al. 1998; Formosa et al. 2002; Belotserkovskaya et al. 2003; Kaplan et al. 2003; Kristjuhan and Svejstrup 2004; Lee et al. 2004; Schwabish and Struhl 2004, 2006).
The FACT complex is among the better-characterized transcription elongation complexes studied thus far. The components of FACT were first discovered through a number of genetic and biochemical experiments in Saccharomyces cerevisiae (Kolodrubetz and Burgum 1990; Malone et al. 1991; Rowley et al. 1991; Wittmeyer and Formosa 1997) and have since been found in several larger eukaryotes. The human FACT complex, originally identified as a biochemical activity from HeLa cells that facilitates the passage of Pol II across nucleosomal templates in vitro, is composed of two factors, hSpt16 and SSRP1, which possess distinct but overlapping biochemical activities (Orphanides et al. 1998, 1999). Specifically, hSpt16, but not SSRP1, directly interacts with H2A–H2B dimers and mononucleosomes, whereas SSRP1, but not hSpt16, binds to H3–H4 tetramers (Belotserkovskaya et al. 2003). However, both subunits are needed for the in vitro deposition of the four core histones onto DNA templates (Belotserkovskaya et al. 2003). Yeast FACT (yFACT) is composed of Spt16, Pob3 (the yeast homolog of SSRP1), and Nhp6, which supplies yFACT with an HMG1 DNA-binding motif found in SSRP1 but absent from Pob3 (Brewster et al. 1998, 2001; Formosa et al. 2001). Biochemical studies have shown that yFACT also associates directly with nucleosomes and reorganizes these structures in such a way as to alter the patterns of DNAse I-sensitive sites of DNA molecules within nucleosomes (Formosa et al. 2001; Rhoades et al. 2004). Recent work has provided evidence for a functional relationship between histone H2B monoubiquitination and FACT activity during transcription elongation, thereby establishing a mechanistic link between histone modifications and FACT function (Pavri et al. 2006).
A role for FACT in transcription elongation in vivo is supported by several studies in S. cerevisiae, Drosophila, and mammalian systems, including: (i) immunolocalization studies showing association of the Drosophila melanogaster FACT complex across actively transcribed regions on polytene chromosomes (Saunders et al. 2003), (ii) chromatin immunoprecipitation (ChIP) experiments in yeast showing association of FACT subunits over transcribed regions of active genes (Mason and Struhl 2003; Kim et al. 2004), (iii) experiments showing that spt16 mutations result in aberrant transcription from cryptic promoter sites within coding regions of certain genes (Kaplan et al. 2003; Mason and Struhl 2003), and (iv) genetic studies implicating yFACT in the reconstitution of proper chromatin structure in the wake of Pol II passage (Formosa et al. 2002). These and other findings have converged on a model that proposes that FACT first alters the structure of chromatin to facilitate the movement of Pol II across transcribed genes and then reestablishes proper nucleosomal structure following the passage of Pol II (Reinberg and Sims 2006). The level of reliance on FACT function for efficient transcription appears to be determined by the level of chromatin organization of specific genes, as shown by experiments indicating that genes with positioned nucleosomes located at the 5′ region of the transcribed unit are more dependent on FACT activity than genes with less stable nucleosomes (Jimeno-González et al. 2006). In addition to transcription elongation, the FACT complex is involved in other cellular functions including regulation of transcription initiation, DNA replication, response to DNA damage, and the prevention of heterochromatin spreading in flies (Reinberg and Sims 2006; Nakayama et al. 2007).
In spite of the extensive characterization of FACT and its roles in transcription elongation, there is little understanding of what might regulate its association with transcribed chromatin and whether it interacts directly with nucleosomes in vivo. In this report, we describe studies of histone H3 that provide new information regarding its role in transcription elongation in vivo, in particular as it relates to its functional relationship with Spt16. We present evidence that a single amino acid substitution within the globular domain of histone H3 significantly alters the distribution of Spt16 across transcribed genomic regions, resulting in a dramatic accumulation of Spt16 in the 3′-untranslated regions (3′-UTRs) of transcribed genes. These and other results indicate that the integrity of histone H3 is crucial for proper Spt16 localization across genes and provide initial insights into the molecular mechanisms that regulate the dynamic association of Spt16 with chromatin in vivo.
MATERIALS AND METHODS
Yeast strains, genetic methods, and media:
All S. cerevisiae strains used in this study (Table 1) are GAL2+ derivatives of the S288C strain background (Winston et al. 1995) . The experimental procedures for the integration of the hht2-11 allele into the genome and the replacement of the HHT1-HHF1 and HHT2-HHF2 loci with the different markers have been previously described (Duina and Winston 2004). Construction of the snf2Δ∷LEU2 allele has been described elsewhere (Cairns et al. 1996). The spt16-197, spt6-1004, and spt4Δ2∷HIS3 mutations have been described in previous studies (Malone et al. 1991; Basrai et al. 1996; Kaplan et al. 2003). The dst1Δ∷NATMX allele was created by a one-step PCR-transformation method that resulted in the replacement of the DST1 open reading frame with the NATMX4 cassette (Goldstein and Mccusker 1999). The Ty912Δ35-lacZ∷his4 reporter gene is a derivative of the Ty912Δ44-lacZ∷his4 allele described previously (Dudley et al. 1999). The techniques used for mating, transformation, sporulation, and tetrad analysis have been described previously (Rose et al. 1990). Detailed descriptions of rich yeast extract-peptone-dextrose (YPD), synthetic dextrose (SD), synthetic complete (SC), omission (SC−), 5-fluoroorotic acid (5-FOA), galactose, and sporulation media are presented elsewhere (Rose et al. 1990). Selection for cells containing the NATMX cassette was done on YPD media containing 100 μg/ml clonNAT (Werner BioAgents, Jena, Germany), whereas selection for kanamycin-resistant cells was performed using media containing 200 μg/ml of active G418 (Sigma, St. Louis). Media containing canavanine (SC + Can) contained 50 μg/ml canavanine.
TABLE 1.
Saccharomyces cerevisiae strains
| Strain | Genotype |
|---|---|
| yAAD108 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ∷LEU2 (hht2-hhf2)Δ∷HIS3 Ty912Δ35-lacZ∷his4 〈pAAD11〉 |
| yAAD476 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ∷LEU2 (hht2-hhf2)Δ∷HIS3 Ty912Δ35-lacZ∷his4 〈pAAD11〉 |
| yAAD482 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ∷LEU2 (hht2-hhf2)Δ∷HIS3 Ty912Δ35-lacZ∷his4 SPT16-857 〈pAAD11〉 |
| yAAD563 | MATα leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ∷LEU2 (hht2-hhf2)Δ∷HIS3 Ty912Δ35-lacZ∷his4 〈pAAD11〉 |
| yAAD587 | MATahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ∷LEU2 (hht2-hhf2)Δ∷HIS3 Ty912Δ35-lacZ∷his4 SPT16-790 〈pAAD11〉 |
| yAAD994 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ∷HIS3 (hht2-hhf2)Δ∷HIS3 snf2Δ∷LEU2 〈pDM9〉 〈pAAD11〉 |
| yAAD1046 | MATα his3Δaleu2Δbura3ctrp1Δ63 lys2-128δ met15Δ0 (hht1-hhf1)Δ∷LEU2 |
| tkl2Δ∷URA3 can1Δ∷MFA1pr-HIS3 SPT16-857 | |
| yAAD1048 | MATα his3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 |
| yAAD1049 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 |
| yAAD1052 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 |
| yAAD1053 | MATα his3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 |
| yAAD1060 | MATα his3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 SPT16-857 |
| yAAD1061 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 SPT16-857 |
| yAAD1063 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 SPT16-790 |
| yAAD1118 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht2-hhf2)Δ∷HIS3 SPT16-857 |
| yAAD1119 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht2-hhf2)Δ∷HIS3 |
| yAAD1121 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 SPT16-857 |
| yAAD1127 | MATα his3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 SPT16-790 |
| yAAD1128 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 SPT16-790 |
| yAAD1134 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 SPT16-790 |
| yAAD1135 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht2-hhf2)Δ∷HIS3 SPT16-790 |
| yAAD1159 | MATα his3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht2-hhf2)Δ∷KANMX4 |
| yAAD1200d | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 spt16-197 〈pCC58〉 |
| yAAD2000d | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 spt16-197 〈pCC58〉 |
| yAAD2005 | MATahis3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 spt6-1004 〈pDM9〉 |
| yAAD2006 | MATα his3Δ200 leu2Δ1 ura3-52 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 〈pDM9〉 |
| yAAD2013 | MATα his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 spt4Δ2∷HIS3 〈pDM9〉 |
| yAAD2015 | MATα his3Δ200 leu2Δbura3clys2-128δ (hht1-hhf1)Δ∷LEU2 hht2-11 dst1Δ∷NATMX 〈pDM9〉 |
The allele at this locus is either his3Δ200 or his3Δ1.
The allele at this locus is either leu2Δ1 or leu2Δ0.
The allele at this locus is either ura3-52 or ura3Δ0.
For these strains, the allele at the HIS4 locus is either HIS4 or his4-912δ.
Plasmids:
pDM9 is a centromeric, URA3-marked plasmid harboring the HHT1-HHF1 region. pDM18 is a centromeric, TRP1-marked plasmid carrying the HHT2-HHF2 region. pAAD11 is a centromeric, TRP1-marked plasmid carrying the hht2-11-HHF2 region. The details on the construction of these three plasmids have been presented previously (Duina and Winston 2004). The generation of pCC58, a URA3-marked centromeric plasmid containing a 4.6-kb restriction fragment that includes the SPT16 locus, has been described elsewhere (Malone et al. 1991).
Identification and analysis of the SPT16-790 and SPT16-857 mutations:
Mutations that suppress the H3-L61W Cs− phenotype were isolated by replica plating patches of either strain yAAD108 or strain yAAD476 grown on YPD plates at 30° to fresh YPD plates and incubated at 14° for several weeks. Papillae originating from these patches were isolated and further analyzed. The two intragenic mutations and the SPT16-790 mutation were derived from strain yAAD108, whereas the tco89 and SPT16-857 mutations were derived from strain yAAD476. Thus, the SPT16-790 and SPT16-857 alleles (strains yAAD587 and yAAD482) are of independent origin. Initial evidence that SPT16-790 and SPT16-857 represent mutations in a single gene was obtained by the observation that the Cs+ phenotype segregated 2:2 in tetrads from crosses between strains yAAD587 and yAAD482 with strain yAAD563. The strains used to determine the dominant Spt− phenotypes conferred by SPT16-790 and SPT16-857 shown in Table 2 were obtained by mating the following strains: SPT16/SPT16 = yAAD1119XyAAD1159, SPT16/SPT16-790 = yAAD1135XyAAD1159, and SPT16/SPT16-857 = yAAD1118XyAAD1159. The strains used to determine the dominant suppression of the H3-L61W Cs− phenotype by the SPT16-790 and SPT16-857 alleles shown in Table 2 were obtained by mating the following strains: SPT16/SPT16 = yAAD1049XyAAD1048, SPT16/SPT16-790 = yAAD1049XyAAD1062 (yAAD1062 has the same relevant genotype as strain yAAD1127 in Table 1), and SPT16/SPT16-857 = yAAD1061XyAAD1048.
TABLE 2.
Dominance analysis of SPT16-790 and SPT16-857
| Relevant genotypea | Spt phenotypeb | Cs phenotypec |
|---|---|---|
| SPT16/SPT16 | + | −/+ |
| SPT16/SPT16-790 | − | + |
| SPT16/SPT16-857 | − | + |
Shown are the SPT16 alleles for each test. For testing the Spt phenotype, the diploid strains were also homozygous for (hht2-hhf2)Δ and for lys2-128δ. The SPT16-790 and SPT16-857 mutations confer an Spt− phenotype only in the context of reduced levels of histones H3 and H4. For testing the Cs phenotype, the diploid strains were also homozygous for (hht1-hhf1)Δ and hht2-11. See materials and methods for additional details on the strains used in these tests.
The Spt phenotype was determined with respect to suppression of the insertion mutation lys2-128δ (Simchen et al. 1984). An Spt+ phenotype indicates lack of suppression (Lys−) and an Spt− phenotype indicates suppression (Lys+).
The Cs phenotype was determined by incubation of plates at 14°.
As the hht2-11 suppressors are dominant, we first determined their genetic map position using a modified systematic genetic analysis (SGA) approach with the ordered array of yeast deletion mutants described previously (Tong et al. 2001). For this analysis, we took advantage of the observation that both mutations also confer a strong Spt− phenotype (growth on SC −Lys media in a lys2-128δ background) in strains that contain a deletion of the HHT1-HHF1 locus. We used this phenotype to test linkage between one of the suppressor mutations (now referred to as SPT16-857) and each strain in the deletion set.
For these experiments we made lawns of yeast cells on YPD plates using strain yAAD1046 and mated these cells to the yeast deletion set strains (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 orfΔ∷KANMX) by replica plating. The diploids resulting from these crosses were selected on diploid-selective media (SC −Trp −Ura). The resulting diploids were stimulated to undergo meiosis by replica plating to sporulation media. Sporulated cultures were replica plated to SC −His −Arg +canavanine to select for recombinant MATa spores as the can1Δ∷MFA1pr-HIS3 reporter gene present in yAAD1046 results in expression of HIS3 exclusively in haploid MATa cells. Recombinant MATa spores were then replica plated to SC −His −Arg −Leu −Ura +canavanine +G418 to select for those spores containing the (hht1-hhf1)Δ∷LEU2, lys2-128δ (which is linked to the tkl2Δ∷URA3 allele), and orfΔ∷KANMX alleles. Finally, the selected cells were replica plated to SC −His −Arg −Leu −Ura −Lys +Can +G418 to determine their Spt− phenotype, which would in turn allow us to monitor for the presence of the suppressor mutation. This analysis resulted in the identification of a region on chromosome VII that showed genetic linkage to the suppressor mutation. The identification of the exact location and nature of the two suppressor mutations was carried out as described in the results section.
Reconstitution of the SPT16-857 allele:
To create a strain containing a newly generated SPT16-857 allele, a 746-bp PCR product (from position +2083 to +2829 within the SPT16 open reading frame) containing the base-pair substitution G2569C amplified from an SPT16-857 strain was cloned into the integrative yeast plasmid pRS406 (Brachmann et al. 1998). The resulting plasmid was analyzed by DNA sequencing to ensure that the SPT16 fragment cloned into it contained only the desired mutation. The plasmid was then linearized using the HpaI restriction enzyme and transformed into a homozygote SPT16 wild-type diploid strain. The previously described integration and excision gene-replacement method (Rothstein 1991) was then used to generate a strain heterozygous for the SPT16-857 allele. In subsequent genetic analyses, the newly generated SPT16-857 allele showed suppression of H3-L61W phenotypes.
RNA analysis:
Northern blot analyses of FLO8 transcripts were performed as described previously (Kaplan et al. 2003). Briefly, total RNA from logarithmically growing cells was collected using the hot- phenol method (Ausubel et al. 1991) on a 1% agarose gel and transferred to a nylon membrane. Hybridizations were performed using a radioactively labeled FLO8 probe specific to a region toward the 3′ end of the transcript (from position +1595 to +2349).
Chromatin immunoprecipitation experiments:
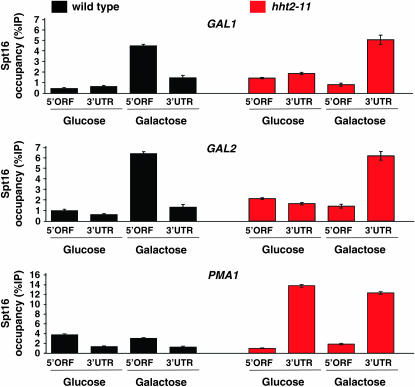
Most chromatin immunoprecipitation (ChIP) experiments were performed as previously described (Martens and Winston 2002). Spt16 ChIP assays were conducted using a rabbit polyclonal antibody specific for Spt16 (a gift from Tim Formosa), the Rpb3 ChIP data were obtained using a Rpb3-specific mouse monoclonal antibody (Neoclone, no. W0012), and the histone H3 ChIP experiments were performed using a rabbit polyclonal antibody specific for histone H3 (AB1791, Abcam). The dependence of the ChIP signals on the specific antibody used was confirmed by performing mock ChIP experiments in the absence of antibody. As a control, an untranscribed region of the genome was assayed (Komarnitsky et al. 2000). In the ChIP experiments presented in Figure 5, cultures were grown in either glucose or galactose media overnight as indicated and the DNAs were quantified by qPCR using the Light Cycler 480 and the qPCR kit (Light Cycler 480 Cyber Green I Master; Roche, Indianapolis). For each DNA, the reaction was done in duplicate. Prior to their use in the qPCR the primers were analyzed for linearity, range, and efficiency. The primer pairs used for the experiments presented in Figure 5 are as follows: GAL1 5′ORF and 3′-UTR, OAD306–OAD307 and OAD345–OAD346, respectively; GAL2 5′ORF and 3′-UTR, AO369–AO370 and OAD347–OAD348, respectively; and PMA1 5′ORF and 3′-UTR, F01810–FO1811 and FO1820–FO1821, respectively. The sequences for these and for all other primers used in this work are listed in supplemental online material at http://www.genetics.org/supplemental/.
Figure 5.—
The accumulation of Spt16 at the 3′-UTRs of the inducible genes GAL1 and GAL2 is dependent upon transcription. Spt16 ChIP experiments were carried out on H3 WT (yAAD 1053) and H3-L61W (yAAD1048) strains grown in the presence of either glucose (repressive condition) or galactose (activating condition). Two primer sets per gene were used to assess Spt16 binding to the 5′ ORF and to the 3′-UTR regions of the GAL1, GAL2, and PMA1 genes. The analysis and quantitation of the signals were performed using real-time PCR technology (see materials and methods). The %IP values are relative to the corresponding input material and represent the average with standard error from three independent clones. Note that accumulation of Spt16 in the 3′-UTR of the constitutively expressed gene PMA1 (bottom) is independent of the carbon source. As a reference point, in these experiments the average levels of Spt16 binding to the control untranscribed region were as follows: H3 WT (glucose), 0.49% ± 0.09; H3 WT (galactose), 0.50% ± 0.09; H3-L61W (glucose), 1.21% ± 0.13; and H3-L61W (galactose) 1.15% ± 0.11.
RESULTS
A functional interaction between histone H3 and Spt16 revealed through the isolation of extragenic suppressors:
Our studies began with the isolation of suppressors of hht2-11, which encodes the histone H3-L61W mutant. This mutation was previously shown to cause several phenotypes, including a defect in the ability of the Swi/Snf chromatin-remodeling complex to interact normally with gene promoters (Duina and Winston 2004). The hht2-11 mutant also has several other phenotypes, including an inability to grow at 14° (Cs− phenotype), suppression of transcription defects resulting from insertion of Ty elements and Ty long-terminal repeats into certain genes (Spt− phenotype), sensitivity to formamide (Fors phenotype), and the inability to grow in the absence of inositol (Ino− phenotype) (Figure 1A and Duina and Winston 2004).
Figure 1.—
Analysis of new SPT16 mutants. (A) Suppression of the hht2-11 phenotypes by the SPT16 mutations. The strains used were as follows: wild type (yAAD1052), hht2-11 (yAAD1048), hht2-11 SPT16-790 (yAAD1063), and hht2-11 SPT16-857 (yAAD1061). Note that each of the strains also harbors a deletion of the HHT1-HHF1 locus such that the HHT2 allele provides the sole source of histone H3 (see Table 1). Approximately 105 cells were spotted on YPD-based media and ∼104 cells on SD-based media. The plates were incubated at either 30° or 14° (where indicated) for the following times: YPD 30°, 2 days; YPD 14°, 14 days; YPD + formamide (form), 4 days; SD, 2 days; and SD with no inositol (SD-ino), 3 days. (B) Nature and location of the amino acid changes present in the mutant Spt16 proteins. Shown is an alignment of several amino acids within a region spanning residues 785–862 of the S. cerevisiae Spt16 protein with those present within the corresponding region of Spt16 homologs from several other organisms. Each of the Spt16 mutants described in this study harbors a single amino acid substitution (indicated in boldface type). In both cases, the change is from a highly conserved acidic residue to a nonacidic amino acid (see the boxed residues). Note that the two mutations are relatively near each other, possibly defining a domain involved in functional interactions with histone H3.
To further characterize the Cs− phenotype, we selected for spontaneous suppressors that allowed hht2-11 cells to grow at 14° (see materials and methods). Of five isolates chosen for further characterization, two were found to harbor intragenic suppressor mutations, both resulting in a change of the H3-W61 residue encoded by the hht2-11allele to a cysteine (H3-W61C). The remaining three suppressors were found to be unlinked to the hht2-11 gene. One of these isolates was found to harbor a recessive mutation in TCO89, a gene encoding a subunit of the TOR complex 1 (Reinke et al. 2004) and was not characterized further. The other two suppressor mutations were analyzed extensively and are the focus of the studies presented here. These two mutations were shown to suppress a subset of hht2-11 phenotypes (Figure 1A). Whereas neither suppressor mutation displays any detectable mutant phenotype in an otherwise wild-type background, both mutations confer a strong Spt− phenotype in the context of reduced intracellular levels of histones H3 and H4 (data not shown). Both mutations were shown to segregate 2:2, indicating that each is caused by a single mutation, and each one was shown to be dominant (Table 2). Furthermore, these two suppressor mutations were shown to be tightly linked to each other (data not shown), suggesting that they are in the same gene.
The gene identified by these dominant suppressor mutations was determined in a series of steps beginning with linkage analysis. To do this, we crossed one of the suppressor mutants to a yeast strain set containing deletions in all of the nonessential genes, testing for linkage between each individual deletion and the suppressor mutation (see materials and methods). These crosses defined a genomic region on chromosome VII. The initial crosses were followed by additional crosses to refine the position of the mutations, and then by DNA sequence analysis of several genes in this region. These experiments led to the discovery that each of the two suppressor mutations consists of a distinct single-base-pair change in the SPT16 gene, each predicted to cause a different single amino acid change in the carboxyl-terminal region of the protein, E790K and E857Q (Figure 1B). Evidence that these alleles are responsible for the suppression of H3-L61W phenotypes came from the demonstration that a reconstituted SPT16-857 allele (see materials and methods) confers suppression of hht2-11 phenotypes (data not shown) and that a perfect correlation between the suppression phenotypes and the presence of the corresponding SPT16 allele was observed in all the strains tested. As shown in Figure 1B, both suppressor mutations are predicted to affect highly conserved acidic residues. The fact that the two affected residues are relatively close to each other suggests that they define an Spt16 domain important for functional interactions with histone H3. Interestingly, both of these residues are located in a region of Spt16 previously shown to be a distinct structural domain of the protein (O'Donnell et al. 2004; Vandemark et al. 2006), and a different amino acid change at position 857 (E857K) has been previously shown to affect Spt16 function in vivo (O'Donnell et al. 2004).
The SPT16 mutations isolated in this study are likely to encode gain-of-function versions of Spt16 as both are dominant for suppression of hht2-11 and for the Spt− phenotype in the context of reduced levels of intracellular histones H3 and H4 (Table 2). In sharp contrast to the suppression effects shown by these novel spt16 mutations, a partial loss-of-function mutation in SPT16 is synthetically lethal in combination with the hht2-11 mutation (see below). The fact that the genes encoding histone H3 and Spt16 display allele-specific interactions supports the notion that the functions of these proteins are intimately related to each other.
Additional evidence that hht2-11 affects transcription elongation:
The genetic interaction between hht2-11 and the SPT16 suppressors suggests that one of the defects conferred by the histone mutant is at the level of transcription elongation. To obtain additional evidence that hht2-11 affects transcription elongation, we took advantage of a recently developed assay that relies on the finding that mutations that impair many elongation factors, including Spt6 and Spt16, result in transcription initiation from cryptic promoters within the coding region of genes (Kaplan et al. 2003; Mason and Struhl 2003). These findings are consistent with the generally accepted notion that one role for elongation factors is to remove the chromatin barrier in front of Pol II to facilitate transcription elongation, and a second role is to reestablish proper chromatin structure in the wake of Pol II passage. Defects in this process would result in the inability to re-form proper nucleosomal structures during transcription elongation, thereby allowing use of cryptic promoters. To test for this defect in hht2-11 mutants, we examined transcription of FLO8, a gene characterized as having a cryptic promoter (Kaplan et al. 2003). Our results show that hht2-11 mutants indeed produce a FLO8 cryptic transcript (Figure 2A, lane 4). In contrast, the SPT16-857 and SPT16-790 mutations do not cause cryptic initiation at FLO8 in a histone H3 wild-type background (Figure 2A, lanes 2 and 3). At the permissive temperature of 30°, the SPT16-857 mutation, but not the SPT16-790 mutation, causes partial suppression of the FLO8 cryptic initiation phenotype seen in hht2-11 cells (Figure 2A, lanes 5 and 6). However, partial suppression of this hht2-11 phenotype by both SPT16-857 and SPT16-790 was observed after a shift to the nonpermissive temperature of 14° for 24 hr (data not shown). These results support the notion that the change in histone H3, H3-L61W, causes transcriptional elongation defects and that these defects can be suppressed by the SPT16 alleles we have isolated.
Figure 2.—
Molecular and genetic evidence that H3-L61W affects transcription elongation. (A) Northern analysis of the FLO8 RNA transcripts in H3 WT SPT16 (yAAD1052), H3 WT SPT16-857 (yAAD1121), H3 WT SPT16-790 (yAAD1134), H3-L61W SPT16 (yAAD1049), H3-L61W SPT16-857 (yAAD1061), and H3-L61W SPT16-790 (yAAD1128) strains. In the top section, the top arrow indicates the location of the full-length transcript and the bottom arrow indicates the location of the short transcript. The intensities of the rRNA bands (bottom section) reflect the amount of total RNA loaded per sample. The data shown are representative of two independent experiments for all of the strains tested except for the H3-L61W SPT16 and H3-L61W SPT16-857 strains, which have been tested in three independent experiments, and the H3 WT SPT16-790 strain, which has been tested in a single experiment. The samples shown were analyzed on the same gel; the order of the lanes has been rearranged to clarify the presentation. (B) H3-L61W cells (strain yAAD1049) were spotted in a 10-fold dilution series from 2.8 × 108 cells/ml to 2.8 × 105 cells/ml (4 μl per spot) onto either synthetic complete (SC) medium (top) or SC medium containing 20 μg/ml MPA (bottom) and incubated at 14° for 28 days. (C) Approximately 1.8 × 106 cells with the indicated genotypes were spotted on either SC media lacking uracil (SC −Ura) or 5-FOA media (SC +FOA) and incubated for 3 days at 30° (the hht2-11 allele encodes the H3-L61W mutant). Strains whose genotypes include pHHT1 contain a URA3-marked plasmid carrying a wild-type copy of the HHT1 gene to cover for the hht2-11 mutation, whereas the strain whose genotype includes pSPT16 (second row) contain a URA3-marked plasmid expressing wild-type Spt16 to cover for the spt16-197 mutation. Before spotting, the cells were grown on nonselective media (YPD) to allow strains containing nonlethal genomic mutations to lose the plasmids and propagate. Inability to grow on 5-FOA media indicates synthetic lethal mutant combinations. Refer to Table 1 for the full genotypes of the strains used in these experiments (yAAD2006, yAAD2000, yAAD2005, yAAD2013, yAAD2015, and yAAD994).
A series of genetic experiments also suggests that hht2-11 affects transcription elongation. In one set of experiments we tested the effects of the drugs mycophenolic acid (MPA) and 6-azarucil (6-AU), both of which are thought to affect transcription elongation by reducing the intracellular pools of nucleotide precursors (Hampsey 1997). We found that the Cs− phenotype of H3-L61W cells is partially suppressed by the inclusion of either MPA (Figure 2B) or 6-AU (data not shown) in the growth media. This phenotype is similar to that previously reported for spt5-242 mutants, whose Cs− phenotype was also suppressed by 6-AU (Hartzog et al. 1998). As previously proposed for spt5-242 (Hartzog et al. 1998), a plausible interpretation of our observations is that slowing down transcription elongation through the addition of MPA or 6-AU to deplete intracellular pools of nucleotide precursors might provide cells with sufficient time to overcome the elongation defects conferred by the H3-L61W mutant. Regardless of the exact mechanism, the finding that MPA and 6-AU suppress the H3-L61W Cs− phenotype is consistent with the hypothesis that H3-L61W affects transcription elongation.
In addition, we tested for genetic interactions between hht2-11 and mutations in genes encoding known elongation factors by construction and analysis of double mutants. Our results show that the hht2-11 mutation causes inviability when combined with partial loss-of-function mutations in either SPT16 or SPT6, or a null mutation in SPT4 (Figure 2C). Since synthetic lethal interactions between two factors are usually indicative of a functional relationship between them (Prelich 1999), these data support the idea that H3-L61W causes defects in transcription elongation. Interestingly, deletion of DST1, the gene encoding the general elongation factor TFIIS, only mildly impairs growth when combined with hht2-11 (Figure 2C), showing that hht2-11 does not indiscriminately interact with mutations in all elongation-factor genes. As previously shown, a mutation that removes the Snf2 component of the transcription-initiation complex Swi/Snf does not result in significant synthetic growth defects in the context of H3-L61W (Figure 2C and Duina and Winston 2004). These results provide genetic evidence in support of the notion that hht2-11 confers transcription elongation defects.
H3-L61W affects localization of Spt16 across transcribed genes:
The allele-specific interactions uncovered between hht2-11 and SPT16 mutations suggest that the functions of Spt16 and those of histone H3 are closely related. One interpretation of our data is that H3-L61W-containing nucleosomes are defective for functional interactions with Spt16 and that certain mutations in Spt16 can alleviate these defects. To learn more about possible effects of H3-L61W on Spt16 functions, we used ChIP assays to examine whether Spt16 association along transcribed genes is affected by the presence of the H3-L61W mutant.
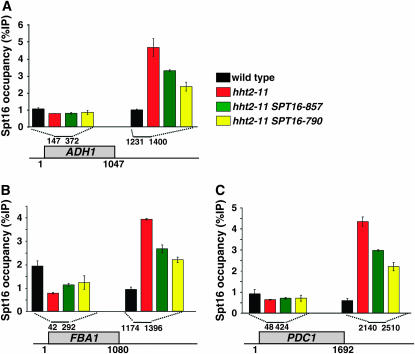
In the first set of experiments, the distribution of Spt16 over the highly transcribed gene PMA1 was compared between an H3 wild-type strain and an hht2-11 mutant. Our results demonstrate that H3-L61W dramatically alters the distribution of Spt16 over the PMA1 gene. Whereas Spt16 is localized predominantly over the PMA1 coding region in a wild-type strain, (Figure 3A, black bars, and see Mason and Struhl 2003; Kim et al. 2004), Spt16 accumulates toward the 3′-UTR in the hht2-11 mutant (Figure 3A, red bars). In contrast, hht2-11 does not cause an altered distribution of RNA Pol II along PMA1 (Figure 3B), suggesting that H3-L61W does not generally affect the distribution of transcription factors across transcribed regions. These latter observations are consistent with our data that hht2-11 causes only minor effects on PMA1 mRNA levels (Duina and Winston 2004 and data not shown). To test if the allele-specific interactions between hht2-11 and the SPT16 suppressor mutations were reflected by this assay, we also did ChIP experiments on the Spt16 mutant proteins. Interestingly, the defect in Spt16 distribution seen in the hht2-11 mutant is significantly suppressed when cells express either one of the two Spt16 mutants described above (Figure 3A, green and yellow bars). To determine if the defect conferred by H3-L61W on Spt16 localization is general, we tested three additional highly transcribed genes. All three showed very similar patterns to what was observed for PMA1 (Figure 4, A–C). Thus, the effects of H3-L61W on the localization of Spt16 across genes and the suppressive effects of the Spt16 mutants are not confined to the PMA1 locus but appear to be widespread across the genome.
Figure 3.—
Analyses of the association of Spt16 and RNA Pol II across the PMA1 gene in the context of wild-type or mutant versions of histone H3 and Spt16. (A) H3-L61W causes Spt16 to accumulate at the 3′-UTR of PMA1, and the two Spt16 mutants isolated in this study alleviate this defect. ChIP experiments were performed on H3 WT (yAAD1052 and yAAD1053), H3-L61W (yAAD1048 and yAAD1049), H3-L61W Spt16-E857Q (yAAD1060 and yAAD1061), and H3-L61W Spt16-E790K (yAAD1127 and yAAD1128) using an Spt16-specific polyclonal antibody (see materials and methods). Nine primer sets were used to assess Spt16 binding across the PMA1 ORF (shaded area) as well as within the 5′- and 3′-UTRs in the different genetic backgrounds. The percentage of immunoprecipitation values (%IP) indicated in the bar graphs for each of the strains at each region across the PMA1 locus are relative to the corresponding input material. Each %IP value represents the average with standard error from three independent experiments. As a reference point, the average levels of Spt16 binding to an untranscribed region (see materials and methods) were as follows: H3 WT, 0.22% ± 0.006; H3-L61W, 0.45% ± 0.009; H3-L61W Spt16-E857Q, 0.47% ± 0.007; and H3-L61W Spt16-E790K, 0.38% ± 0.008. (B) H3-L61W does not appear to greatly affect the distribution of RNA Pol II across the PMA1 locus. ChIP experiments were performed on H3 WT (yAAD1052) and H3-L61W (yAAD1049) strains using a monoclonal antibody specific for the Rpb3 subunit of RNA Pol II (see materials and methods). The two regions assayed are indicated above the PMA1 locus diagram. The %IP values are relative to the input material and in each case represent the average of two independent experiments. As a reference point, the average levels of Rpb3 binding to the untranscribed region were as follows: H3 WT, 0.09% ± 0.01; and H3-L61W, 0.10% ± 0.01. For both sets of experiments, the analysis and quantitation of the signals were carried out using the method described by Martens and Winston (2002) (see materials and methods).
Figure 4.—
H3-L61W affects Spt16 binding across three other transcribed genes and the Spt16 mutants partially suppress these defects. ChIP experiments were performed as described in Figure 3A, except that the genomic regions tested correspond to the 5′ ORF regions and 3′-UTRs across the (A) ADH1 locus, (B) FBA1 locus, and (C) PDC1 locus. Each %IP value represents the average with standard error from three independent experiments.
To assess whether the accumulation of Spt16 at 3′-UTRs in hht2-11 cells is dependent upon transcription, we performed ChIP experiments to determine the levels of Spt16 occupancy at the GAL1 and GAL2 genes under conditions in which these genes are either repressed (glucose medium) or expressed (galactose medium). As shown in the top two sections of Figure 5, abnormally high levels of Spt16 are detected in the 3′-UTRs of both of these genes in hht2-11 cells only when cells were grown under inducing conditions. In contrast, the Spt16-3′-UTR accumulation phenotype in hht2-11 cells is observed at the PMA1 gene regardless of the carbon source present in the growth media (Figure 5, bottom section). These results strongly suggest that the abnormal accumulation of Spt16 at 3′-UTRs is dependent upon active transcription.
H3-L61W does not greatly affect the distribution of nucleosomes across transcribed genes:
Our studies have shown that H3-L61W causes Spt16 to accumulate over the 3′-UTRs of transcribed genes. In an effort to investigate the molecular mechanisms underlying this effect, we developed two testable models that would be consistent with these observations. For the first model, we envision Spt16 as able to normally traverse the length of transcribed genes in the context of H3-L61W, but unable to dissociate from the 3′-UTRs. In this model, Spt16 normally receives a signal that causes departure from the 3′ end of genes once the transcription process is terminated; the H3-L61W mutant might somehow interfere with this signal, resulting in the accumulation of Spt16 at the 3′ ends of genes. For the second model, we envision Spt16 as unable to properly remove and redeposit histones in the context of H3-L61W during the elongation process. This defect might in turn result in the “sliding” and accumulation of nucleosomes toward the 3′ ends of genes as transcription proceeds. If this were to take place, Spt16 might also be expected to accumulate at the 3′ ends of genes since its substrate, the nucleosomes, has been pushed toward the 3′-UTRs.
To differentiate between these two models, we performed ChIP experiments to determine the distribution of nucleosomes across the PMA1 gene in wild-type and hht2-11 strains. For these studies, we used a histone-H3-specific antibody. We note that this antibody was raised against a peptide encompassing residues 124–135 of human histone H3 (Rao et al. 2005) and therefore should still recognize the H3-L61W mutant protein. Importantly, the levels of histone H3 protein in wild type and the hht2-11 mutant are similar as measured by Westerns using this antibody (data not shown). The results from the histone-H3-ChIP experiments show that H3-L61W has only a minor effect on the nucleosomal distribution across the PMA1 gene, resulting in a slight increase in the concentration of nucleosomes in the 3′-UTR when compared to wild-type cells (Figure 6). This minor effect is not expected to be the cause of Spt16-3′-UTR accumulation in H3-L61W cells, since in these cells we see at least a 15-fold increase in Spt16-3′-UTR occupancy compared to wild-type cells. These data are therefore consistent with the first model, raising the possibility that the H3-L61W mutation interferes with a signal that normally causes Spt16 to dissociate from the 3′-UTR of transcribed genes.
Figure 6.—
H3-L61W does not appear to greatly affect the distribution of nucleosomes across the PMA1 gene. ChIP experiments were performed as described in Figure 3A, except that the immunoprecipitations were performed with a histone-H3-specific antibody (see materials and methods). Each %IP value represents the average with standard error from three independent experiments. As a reference point, the average levels of histone H3 binding to the control untranscribed region were as follows: H3 WT, 2.4% ± 0.32; H3-L61W, 4.10% ± 0.57. As anticipated, the %IP of histone H3 at the untranscribed region was relatively high since nucleosomes are present throughout the genome.
DISCUSSION
In this work we provide evidence indicating that the integrity of histone H3 is crucial for the normal association of Spt16 along transcribed regions. We have found that a single amino acid substitution, L61W, within the globular domain of H3, results in a dramatic transcription-dependent accumulation of Spt16 at the 3′-UTRs of transcribed genes. H3-L61W appears to affect Spt16 localization in a general fashion, as all the transcribed genes analyzed thus far are affected in a similar way by this mutant. Furthermore, two specific amino acid changes in Spt16, E790K, and E857Q, partially compensate for the H3-L61W defect. Taken together, these results suggest that a direct Spt16–H3 interaction is required for normal localization of Spt16 on chromatin.
Our results, combined with those from a previous study (Duina and Winston 2004), suggest a working model for the role of the Spt16–H3 interaction. Analysis of the predicted structure of a nucleosome containing H3-L61W suggests that the L61W substitution may lead to increased H3–H4 tetramer stability, potentially leading to stabilization of the H3 N-terminal helix (Duina and Winston 2004). This stabilization, in turn, might prevent the propagation of a structural change within nucleosomes, such as histone modifications or nucleosome remodeling, that might normally serve as a signal for the departure of Spt16 from 3′-UTRs after transcription has terminated. Thus, according to this model, enrichment of Spt16 at 3′-UTRs in H3-L61W cells would be a result of the inability of a “3′-UTR-dissociation signal” to be properly transmitted from the nucleosome to Spt16. In the context of this model, the amino acid changes in Spt16 identified here might mediate their suppressive effects by rendering Spt16 more sensitive to the weakened 3′-UTR dissociation signal in H3-L61W cells, thereby allowing for more efficient departure of Spt16 from 3′-UTRs in the H3-L61W background.
Our studies provide insights into a general process that has not been extensively addressed to date: the mechanisms that regulate the dissociation of transcription-elongation factors during or following gene transcription. As Pol II travels across the polyadenylation [poly(A)] sites of transcribed genes, exchange of certain elongation factors (e.g., the PAF and TREX complexes) for poly(A) factors occurs, while other elongation factors, including the FACT complex, appear to depart from genes along with Pol II downstream of the poly(A) sites (Kim et al. 2004). Whereas it has become clear that processing of nascent RNA transcripts is coupled to transcription termination and the eventual departure of Pol II from transcribed units, the exact molecular mechanisms responsible for the coupling of these processes have yet to be completely understood. Similarly, the processes that control the dynamic exchange of elongation and termination factors on the transcription machinery as it travels across genes are not well- characterized. The finding that H3-L61W appears to prevent normal dissociation of Spt16 from transcribed genes, while apparently not affecting Pol II, raises the possibility that the signals required for dissociation of Pol II and Spt16 (and perhaps other factors) from 3′-UTRs are at least to some degree separable.
Future studies will take advantage of the experimental system described in this work to learn more about the dynamic interactions that occur between chromatin and Spt16, as well as chromatin and other elongation factors, in vivo. Experiments to determine the breadth of the effects of H3-L61W on chromatin association of other factors will provide important information about the specificity of the defects associated with this histone mutant. In addition, isolation of other mutations, both in histone and in nonhistone proteins, that prevent proper dissociation of Spt16 from 3′-UTRs will provide insights into the nature of the putative signaling pathway that regulates Spt16 dissociation from chromatin following transcription.
Acknowledgments
We are grateful to Mark Hickman and Reine Protacio for helpful comments on the manuscript and Kacey Swindle for technical assistance. We thank Tim Formosa for providing the anti-Spt16 antibodies. A.A.D. is grateful to Patricia Wight and members of her laboratory in the Department of Physiology and Biophysics at the University of Arkansas for Medical Sciences for providing assistance in the performance of part of this work. This material is based upon work supported by the National Science Foundation under grant 0543412, the Charles A. King Trust Fellowship from The Medical Foundation, National Institutes of Health (NIH) grants P20RR16460-03 and P20RR16460-03, ARIA 0711-03 from the IDeA Networks of Biomedical Research Excellence (INBRE), Program of the National Center for Research Resources, and start-up funds and a faculty project grant from Hendrix College to A.A.D. This work was also supported by NIH grant GM32967 to F.W. and a Canadian Institutes of Health Research grant to A.N. A.N. holds a Tier 2 Canadian Research Chair.
References
- Adkins, M. W., S. R. Howar and J. K. Tyler, 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14: 657–666. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al. (Editors), 1991. Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York.
- Basrai, M. A., J. Kingsbury, D. Koshland, F. Spencer and P. Hieter, 1996. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2838–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky et al., 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093. [DOI] [PubMed] [Google Scholar]
- Bentley, D. L., 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17: 251–256. [DOI] [PubMed] [Google Scholar]
- Berger, S. L., 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12: 142–148. [DOI] [PubMed] [Google Scholar]
- Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein and S. L. Schreiber, 2004. Global nucleosome occupancy in yeast. Genome Biol. 5: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger, H., J. Griesenbeck, J. S. Strattan and R. D. Kornberg, 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11: 1587–1598. [DOI] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Brewster, N. K., G. C. Johnston and R. A. Singer, 1998. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem. 273: 21972–21979. [DOI] [PubMed] [Google Scholar]
- Brewster, N. K., G. C. Johnston and R. A. Singer, 2001. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21: 3491–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski, S., 2005. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 17: 257–261. [DOI] [PubMed] [Google Scholar]
- Cairns, B. R., R. S. Levinson, K. R. Yamamoto and R. D. Kornberg, 1996. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 10: 2131–2144. [DOI] [PubMed] [Google Scholar]
- Dudley, A. M., L. J. Gansheroff and F. Winston, 1999. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4–912delta promoter in Saccharomyces cerevisiae. Genetics 151: 1365–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina, A. A., and F. Winston, 2004. Analysis of a mutant histone H3 that perturbs the association of Swi/Snf with chromatin. Mol. Cell. Biol. 24: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg, J. C., and A. Shilatifard, 2006. Leaving a mark: the many footprints of the elongating RNA polymerase II. Curr. Opin. Genet. Dev. 16: 184–190. [DOI] [PubMed] [Google Scholar]
- Ercan, S., and R. T. Simpson, 2004. Global chromatin structure of 45,000 base pairs of chromosome III in a- and alpha-cell yeast and during mating-type switching. Mol. Cell. Biol. 24: 10026–10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa, T., P. Eriksson, J. Wittmeyer, J. Ginn, Y. Yu et al., 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20: 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa, T., S. Ruone, M. D. Adams, A. E. Olsen, P. Eriksson et al., 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162: 1557–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Hampsey, M., 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13: 1099–1133. [DOI] [PubMed] [Google Scholar]
- Hartzog, G. A., 2003. Transcription elongation by RNA polymerase II. Curr. Opin. Genet. Dev. 13: 119–126. [DOI] [PubMed] [Google Scholar]
- Hartzog, G. A., T. Wada, H. Handa and F. Winston, 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12: 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno-González, S., F. Gómez-Herreros, P. M. Alepuz and S. Chávez, 2006. A gene-specific requirement for FACT during transcription is related to the chromatin organization of the transcribed region. Mol. Cell. Biol. 26: 8710–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, C. D., L. Laprade and F. Winston, 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099. [DOI] [PubMed] [Google Scholar]
- Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt and S. Buratowski, 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23: 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodrubetz, D., and A. Burgum, 1990. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J. Biol. Chem. 265: 3234–3239. [PubMed] [Google Scholar]
- Komarnitsky, P., E. J. Cho and S. Buratowski, 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14: 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber, P., S. Barbaric, T. Luckenbach, A. Schmid, U. J. Schermer et al., 2006. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 281: 5539–5545. [DOI] [PubMed] [Google Scholar]
- Kristjuhan, A., and J. Q. Svejstrup, 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23: 4243–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl and J. D. Lieb, 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36: 900–905. [DOI] [PubMed] [Google Scholar]
- Luger, K., 2006. Dynamic nucleosomes. Chromosome Res. 14: 5–16. [DOI] [PubMed] [Google Scholar]
- Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent and T. J. Richmond, 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Malone, E. A., C. D. Clark, A. Chiang and F. Winston, 1991. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 5710–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, J. A., and F. Winston, 2002. Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes Dev. 16: 2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, P. B., and K. Struhl, 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23: 8323–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, T., K. Nishioka, Y. Dong, T. Shimojima and S. Hirose, 2007. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 21: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar, G. J., H. Y. Fan and R. E. Kingston, 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487. [DOI] [PubMed] [Google Scholar]
- O'Donnell, A. F., N. K. Brewster, J. Kurniawan, L. V. Minard, G. C. Johnston et al., 2004. Domain organization of the yeast histone chaperone FACT: the conserved N terminal domain of FACT subunit Spt16 mediates recovery from replication stress. Nucleic Acids Res. 32: 5894–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides, G., G. LeRoy, C. H. Chang, D. S. Luse and D. Reinberg, 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116. [DOI] [PubMed] [Google Scholar]
- Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey and D. Reinberg, 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400: 284–288. [DOI] [PubMed] [Google Scholar]
- Pavri, R., B. Zhu, G. Li, P. Trojer, S. Mandal et al., 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717. [DOI] [PubMed] [Google Scholar]
- Prelich, G., 1999. Suppression mechanisms: themes from variations. Trends Genet. 15: 261–266. [DOI] [PubMed] [Google Scholar]
- Rao, B., Y. Shibata, B. D. Strahl and J. D. Lieb, 2005. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 25: 9447–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg, D., and R. J. Sims, 3rd, 2006. de FACTo nucleosome dynamics. J. Biol. Chem. 281: 23297–23301. [DOI] [PubMed] [Google Scholar]
- Reinke, A., S. Anderson, J. M. McCaffery, J. Yates, III, S. Aronova et al., 2004. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279: 14752–14762. [DOI] [PubMed] [Google Scholar]
- Reinke, H., and W. Horz, 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11: 1599–1607. [DOI] [PubMed] [Google Scholar]
- Rhoades, A. R., S. Ruone and T. Formosa, 2004. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 24: 3907–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., F. Winston and P. Hieter, 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Rosonina, E., S. Kaneko and J. L. Manley, 2006. Terminating the transcript: breaking up is hard to do. Genes Dev. 20: 1050–1056. [DOI] [PubMed] [Google Scholar]
- Rothstein, R., 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194: 281–301. [DOI] [PubMed] [Google Scholar]
- Rowley, A., R. A. Singer and G. C. Johnston, 1991. CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol. Cell. Biol. 11: 5718–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, A., J. Werner, E. D. Andrulis, T. Nakayama, S. Hirose et al., 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301: 1094–1096. [DOI] [PubMed] [Google Scholar]
- Schwabish, M. A., and K. Struhl, 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24: 10111–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish, M. A., and K. Struhl, 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22: 415–422. [DOI] [PubMed] [Google Scholar]
- Segal, E., Y. Fondufe-Mittendorf, L. Chen, A. Thastrom, Y. Field et al., 2006. A genomic code for nucleosome positioning. Nature 442: 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger, E. A., Z. Moqtaderi and K. Struhl, 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18: 735–748. [DOI] [PubMed] [Google Scholar]
- Simchen, G., F. Winston, C. A. Styles and G. R. Fink, 1984. Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc. Natl. Acad. Sci. USA 81: 2431–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, III, R. J., R. Belotserkovskaya and D. Reinberg, 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18: 2437–2468. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Vandemark, A. P., M. Blanksma, E. Ferris, A. Heroux, C. P. Hill et al., 2006. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol. Cell 22: 363–374. [DOI] [PubMed] [Google Scholar]
- Winston, F., C. Dollard and S. L. Ricupero-Hovasse, 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55. [DOI] [PubMed] [Google Scholar]
- Wittmeyer, J., and T. Formosa, 1997. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 17: 4178–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman, J. L., 2006. Nucleosome displacement in transcription. Genes Dev. 20: 2009–2017. [DOI] [PubMed] [Google Scholar]
- Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu et al., 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630. [DOI] [PubMed] [Google Scholar]