Abstract
OBJECTIVE
To identify early diabetes-related alterations in gene expression in bladder and erectile tissue that would provide novel diagnostic and therapeutic treatment targets to prevent, delay or ameliorate the ensuing bladder and erectile dysfunction.
MATERIALS AND METHODS
The RG-U34A rat GeneChip® (Affymetrix Inc., Sunnyvale, CA, USA) oligonucleotide microarray (containing ≈8799 genes) was used to evaluate gene expression in corporal and male bladder tissue excised from rats 1 week after confirmation of a diabetic state, but before demonstrable changes in organ function in vivo. A conservative analytical approach was used to detect alterations in gene expression, and gene ontology (GO) classifications were used to identify biological themes/pathways involved in the aetiology of the organ dysfunction.
RESULTS
In all, 320 and 313 genes were differentially expressed in bladder and corporal tissue, respectively. GO analysis in bladder tissue showed prominent increases in biological pathways involved in cell proliferation, metabolism, actin cytoskeleton and myosin, as well as decreases in cell motility, and regulation of muscle contraction. GO analysis in corpora showed increases in pathways related to ion channel transport and ion channel activity, while there were decreases in collagen I and actin genes.
CONCLUSIONS
The changes in gene expression in these initial experiments are consistent with the pathophysiological characteristics of the bladder and erectile dysfunction seen later in the diabetic disease process. Thus, the observed changes in gene expression might be harbingers or biomarkers of impending organ dysfunction, and could provide useful diagnostic and therapeutic targets for a variety of progressive urological diseases/conditions (i.e. lower urinary tract symptoms related to benign prostatic hyperplasia, erectile dysfunction, etc.).
Keywords: diabetes mellitus, streptozotocin, erectile dysfunction, bladder dysfunction, microarray analysis, gene chips, genomics
INTRODUCTION
Diabetes mellitus (DM) is a chronic multifactorial disease reaching epidemic proportions in the USA, where ≈ 18 million individuals are affected; worldwide, current estimates are that ≈ 150 million people are affected [1-5]. DM results in neural, metabolic, endocrine and vascular changes culminating in many well-documented end-organ complications. The best known of these are retinopathy, neuropathy, cardiovascular and neuropathic effects (http://www.jdrf.org/ and http://diabetes.niddk.nih.gov/complications/index.htm). However, it is increasingly clear that many male patients with DM eventually develop bladder and/or erectile dysfunction (ED) [6-15].
The streptozotocin (STZ)-DM rat is a well-established animal model that recapitulates aspects of the human urological complications, including both bladder dysfunction [15-25] and ED [26-31]. In the STZ-DM rat model and in patients with DM, the ensuing bladder dysfunction and ED are thought to be progressive, and to depend on both the duration and severity of disease. Nonetheless, diabetic bladder dysfunction and ED are complex, multifactorial diseases, and the precise molecular mechanisms associated with progression from hyperglycaemia to organ dysfunction remain elusive. The importance of changes in gene expression to hypertrophic remodelling of the obstructed and diabetic bladder have long been recognized [32], and a recent study confirmed that many gene alterations accompany DM-related ED [30]. However, while various molecular alterations attend urological diabetic complications, we are unaware of any pathway-based analyses of gene expression changes in urogenital smooth muscle from diabetic animals before the onset of physiologically significant diabetic complications. It seems that early molecular analyses of progressive diseases such as DM, whose clinical sequelae depend on both the duration and severity of disease, might provide valuable insights into molecular changes that are precursors or harbingers of organ failure (i.e. diagnostic and preventative), as well as changes that represent mechanisms of organ failure (i.e. therapeutic targets).
To this end, we conducted gene chip microarray studies on smooth muscle excised from the bladder and corpora of male STZ-DM rats 1 week after confirmation of the diabetic state. The 1 week time point is associated with severe hyperglycaemia, but little change in bladder (Christ et al. unpublished observation) or erectile function in vivo [31]. This is in contrast to ≈2 months of STZ-DM, which is associated with significant, reproducible alterations in both bladder [20,27,33,34] and erectile function [26,29-31,35]. The aim of these initial studies was to develop a method for identifying/defining DM-induced, organ-specific alterations in gene expression. This information can then be used to identify novel and improved diagnostic and therapeutic targets that might prevent, delay or ameliorate the debilitating consequences of DM-related bladder dysfunction and ED. Such a strategy also has broad implications for the diagnosis, prevention and treatment of other urological diseases, i.e. LUTS related to BPH.
MATERIALS AND METHODS
DM was induced in male F-344 rats (Taconic Farms, Germantown, NY, USA; 8–10-weeks-old and 200–240 g) by one i.p. injection of STZ (35 mg/kg, dissolved in citrate buffer, 0.6 M citric acid/0.08 M Na2HPO4, pH 4.6). Age-matched control rats were injected with vehicle. DM was confirmed by the presence of blood glucose levels of >250 mg/dL for 3 consecutive days. After confirming the diabetic state, rats were killed 1 week later in a CO2 asphyxiation chamber, and the bladder and corpora were excised. The bladder was quickly denuded of the urothelial and suburothelial layers, and both bladder and corpora were flash-frozen in liquid nitrogen and stored at −70 °C until RNA preparation. All experimental protocols were approved by the Animal Institute Committee of the Albert Einstein College of Medicine.
For the GeneChip® studies, the RG-U34A rat GeneChip (Affymetrix Inc., Sunnyvale, CA, USA) containing ≈8799 genes was used. RNA was prepared from the corpora and bladder of age-matched control and STZ-DM rats using a Mixer Miller (MM3, Qiagen, CA, USA). To determine that the isolated RNA was not degraded, agarose gel-electrophoresis was performed, followed by staining with ethidium bromide. After confirming its quality, the isolated RNA was reverse transcribed to cDNA, labelled with biotin and fragmented according to Affymetrix protocols (http://www.affymetrix.com). Subsequent hybridization to microarray chips was done at the AECOM microarray core facilities using an Affymetrix Fluidics Station. Test chips were used to screen for the quality of the labelled cRNA, before proceeding to the RG-U34A chip. Data were from: three age-matched control bladders; six 1-week STZ-DM bladders; five age-matched control corpora; and two 1-week DM-corpora. The very conservative analytical approach we took in this study offsets the small sample size; the major impact of the small sample size is expected to be that fewer gene changes are detected. Nonetheless, the 5 × 2 comparison for corpora and the 6 × 3 comparison for the bladder still permitted the identification of hundreds of statistically significant differentially expressed genes.
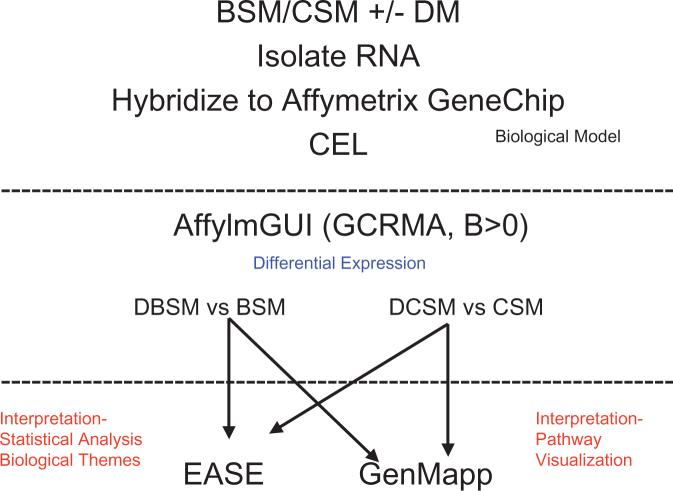
A flow chart of the data analysis is shown in Fig. 1 and is described elsewhere [36]. Briefly, raw cell intensity (CEL) files from the AECOM core facility were analysed with affylmGUI software (Affymetrix LIMMA, Linear Models for Microarray Data, Graphical User Interface) [37,38]. Within affylmGUI, gene expression values were summarized with RMA and GCRMA. RMA adjusts for background noise, performs a quantile normalization, transforms the data into log base 2, and summarizes the multiple probes into one intensity [39-43]. GCRMA is an adaptation of RMA that uses a model-based background correction based on G-C content and perfect match-mismatch [44].
FIG. 1.
Schematic depiction of the algorithm used for identifying differentially expressed genes, as well as for illustrating prominent biological themes and pathways. BSM, bladder smooth muscle; CSM, corporal smooth muscle; DBSM, diabetic-bladder smooth muscle; DCSM, diabetic-corporal smooth muscle.
GCRMA is more accurate than MAS5 without losing much precision. Quantification of relative differences in gene expression among the groups of interest used affylmGUI, the sister package of limmaGUI [37,38], which is a comprehensive software package freely available at: http://bioinf.wehi.edu.au/afflmGUI/. This reads the Affymetrix CEL files directly, summarizes the gene expression values using GCRMA, and uses LIMMA [45] to identify statistically significant differences in gene expression. The package uses LIMMA to fit a linear model for every gene (similar to ANOVA or multiple regression analyses), and adjusts the P values for multiple hypothesis testing [45,46]. Differentially expressed genes were identified with a B statistic >0. This statistic, also known as a likelihood-of-odds score, is a moderated t-statistic with posterior residual SDs [46].
For the pathway/ontological analysis, genes identified as differentially expressed were analysed with the Expression Analysis Systematic Explorer (EASE) and the Gene Map Annotator and Pathway Profiler (GenMapp). Both are free programs to evaluate the importance of detected changes to biologically relevant pathways and processes. The former uses statistical methods to evaluate alterations in terms of groups or sets of genes; the latter is used to visualize those changes. The goal of both programs is a more macroscopic biological interpretation of the patterns of change in gene expression, rather than focusing on individual targets.
The EASE program provides the tools and statistical methods for uncovering enriched biological themes within diverse and disparate gene lists. EASE allows identification of over-represented biological themes or pathways present in a data set based on their gene ontologies (GOs) [46-48]. Development of GOs is a collaborative effort to create a controlled vocabulary that describes gene expression in terms of biological processes, cellular components and molecular functions [49]. Once a list of altered genes is loaded into EASE (in this case, genes with a B statistic >0 from bladder and corporal smooth muscle cells), each gene is matched to all possible GOs. The results are then compared to all possible GOs for all genes on the microarray platform, and an EASE score (P-value) is calculated, based on a conservative variant of the Fisher's exact probability test. We selected those pathways/processes with a P < 0.05.
The GenMapp is designed to visualize gene expression data on maps representing biological pathways and groupings of genes. GenMapp consists of hundreds of pre-made pathway maps [50,51], and we used it to identify pathway level changes amongst multiple disparate data sets. In this program, data sets are assigned a colour corresponding to treatment (e.g. DM or control), or tissue type (bladder or corporal smooth muscle), and direction of change in gene expression (up or down).
RESULTS
GCRMA was used to compare GeneChips from detrusor strips from STZ-DM rats after 1 week of STZ-DM to similar data from age-matched controls. Expression analysis identified 145 genes that were up-regulated and 175 genes that were down-regulated in STZ-DM bladder relative to controls. Tables 1 and 2 summarize changes that are likely to be relevant to DM bladder disease. For up-regulated genes, GO analysis using EASE revealed predominant genetic themes involved in ‘cell proliferation’, ‘sugar, protein and macromolecule catabolism/metabolism’, ‘actin cytoskeleton’ and ‘myosin’. The GO analysis and the number of ‘hits’ in each category are summarized in Table 3. These included matrix metalloproteinase 7 (MMP-7, 612-fold), myosin regulatory light chain (3.4-fold), calmodulin (2.2-fold), calreticulin (2 Affymetrix probe sets, 6–14-fold), tropomyosin 4 (5.5-fold) and the Na+/K+ ATPase β1 subunit (29-fold). Prominent biological themes in the down-regulated genes were ‘cytoskeletal organization and biogenesis’, ‘cell motility’, ‘muscle contraction’ and ‘regulation of muscle contraction’ (Table 3). Among the down-regulated genes were neurotrophin-3 (3.5-fold), synuclein-γ (96-fold), neurofilament 3 (3.2-fold), neurofilament light polypeptide (20-fold), myosin phosphatase, target subunit 1 (7.8 fold), and the tachykinin receptor 2 (8.5-fold), and the M3 muscarinic cholinergic receptor. In short, biological themes in the up-regulated category were consistent with genes that would be expected to be involved in altered detrusor myocyte contractility, while alterations in gene expression in the down-regulated data set were associated with alterations that might account for neurogenic and myogenic mechanisms of bladder dysfunction.
TABLE 1.
Up-regulation in expression of individual genes in detrusor myocytes/bladder wall and corporal myocytes
| Affymetrix probesets | Fold change | Gene name (UP) | Gene symbol |
|---|---|---|---|
| Detrusor myocytes | |||
| L24374_at | 612 | MMP-7 | Mmp7 |
| RC_AI230614_S_AT | 29 | ATPase Na+/K+ transporting β1 polypeptide | Atp1b1 |
| S57478CDS_S_AT | 26 | Annexin 1 | Anxa1 |
| RC_AI105348_I_AT | 20.9 | Cofilin 1 | Cfl1 |
| X53363CDS_S_AT | 14 | Calreticulin | Calr |
| X53363cds_s_at | 13 | TGFβ1 | Tgfb1 |
| U31463_AT | 11 | Myosin, heavy polypeptide 9 | Myh9 |
| U39875_AT | 10.3 | Ca2+ binding protein p22 | Chp |
| L13039_S_AT | 6.2 | Calpactin I heavy chain | Anxa2 |
| D78308_G_AT | 6.1 | Calreticulin | Calr |
| J02780_AT | 5.5 | Tropomyosin 4 | Tpm4 |
| X68199_AT | 5.3 | Myosin Ib | Myo1b |
| D14568_AT | 4.6 | Protein phospatase 3, regulatory subunit B, α isoform, type 1 | Ppp3r1 |
| M86870_AT | 4.5 | Protein disulphide isomerase related protein (Ca2+-binding protein, intestinal-related) | Erp70 |
| RC_AA817887_AT | 3.6 | Profilin | Pfn1 |
| D14688_S_AT | 3.4 | Myosin regulatory light chain | Mrlcb |
| X14265_AT | 2.2 | Calmodulin 3 | Calm3 |
| J03627_AT | 2.0 | S-100 related protein, clone 42C | S100a10 |
| Corporal myocytes | |||
| rc_AA818144_at | 2.3 | C-reactive protein, petaxin related | Crp |
| M15191_s_at | 2.0 | Tachykinin 1 | Tac1 |
| J05189_at | 1.7 | Tachykinin receptor 3 | Tacr3 |
| AF087454_at | 1.6 | K+ voltage-gated channel, subfamily Q, member 3 | Kcnq3 |
| U92279_at | 1.5 | Regulator of G-protein signalling 14 | Rgs14 |
| L13606_at | 1.5 | Myosin, heavy polypeptide 4 | Myh4 |
| AF055477_at | 1.5 | Ca2+ channel, voltage-dependent, N type, α1B subunit | Cacna1b |
| AF029310_at | 1.4 | Transient receptor potential cation channel, subfamily V, member 1 | Trpv1 |
| D17764_at | 1.4 | Synuclein-β | Sncb |
| U09243_at | 1.4 | K+ inwardly rectifying channel, subfamily J, member 3 | Kcnj3 |
| U16802_at | 1.4 | Ca2+-dependent activator protein | Caps |
| J04629_at | 1.4 | ATPase, Na+/K+ transporting, β2 polypeptide | Atp1b2 |
| rc_AI144709_s_at | 1.3 | Regulator of G-protein signalling 8 | Rgs8 |
| AF016191_at | 1.3 | K+ channel erg3 | erg3 |
| X55519_at | 1.3 | Cyclic nucleotide gated channel 4 | Cncg4 |
| D13927_at | 1.3 | Cl- channel K1 | Clcnk1 |
| D31962cds_at | 1.2 | Rho GTPase activating protein 7 | Arhgap7 |
TABLE 2.
Down-regulation in expression of individual genes in detrusor myocytes/bladder wall and corporal myocytes
| Affymetrix probesets | Fold change | Gene name (DOWN) | Gene symbol |
|---|---|---|---|
| Detrusor myocytes | |||
| X86789_at | 96 | Synuclein-γ | Sncg |
| J05592_g_at | 62 | Protein phosphatase 1, regulatory (inhibitor) subunit 1A | Ppp1r1a |
| X65036_at | 62 | Integrin-α7 | Itga7 |
| rc_AA925248_at | 25.3 | Na+ channel, voltage-gated, type 6, α polypeptide | Scn6a |
| AF031880_AT | 20 | Neurofilament, light polypeptide | Nfl |
| D45920_at | 9.7 | 130 kDa-Ins(1,4,5)P3 binding protein | LOC84587 |
| rc_AA892251_at | 9.6 | Arginine vasopressin receptor 1A | Avpr1a |
| L03382_at | 9.4 | Phospholamban | Plm |
| X04139_s_at | 9.4 | Protein kinase C, β1 | Prkcb1 |
| M31838_at | 8.5 | Tachykinin receptor 2 | Tacr2 |
| U50185_AT | 7.8 | Myosin phosphatase, target subunit 1 | Mypt1 |
| rc_AI230211_s_at | 7.2 | K+ voltage gated channel, Shal-related family, member 3 | Kcnd3 |
| AA799276_AT | 6.6 | ATPase, Ca2+ transporting, cardiac muscle, slow twitch 2 | Atp2a2 |
| D85435_at | 6.4 | Protein kinase C, δ binding protein | Prkcdbp |
| M17526_G_AT | 6.2 | Guanine nucleotide binding protein, α o | Gnao |
| U21101_at | 4.7 | cGMP-stimulated phosphodiesterase | Pde2a |
| M34643_AT | 3.5 | Neurotrophin-3 (HDNF/NT-3) | Ntf3 |
| Z12152_AT | 3.2 | Neurofilament 3, medium | Nef3 |
| RC_AI145367_AT | 1.7 | Adenylyl cyclase-associated protein 2 | Cap2 |
| U17971_AT | 1.5 | Protein tyrosine phosphatase 2E | Ptp2E |
| M16407_AT | 1.4 | Cholinergic receptor, muscarinic 3 | Chrm3 |
| Corporal myocytes | |||
| rc_AI231472_s_at | 380 | Collagen, type 1, α1 | Col1a1 |
| rc_AA900769_s_at | 70.8 | Smooth muscle α-actin | Acta2 |
| Z78279_g_at | 36 | Collagen, type 1, α1 | Col1a1 |
| Z78279_at | 18.2 | Collagen, type 1, α1 | Col1a1 |
| rc_AA944422_at | 14.6 | Calponin 3, acidic | Cnn3 |
| X70369_s_at | 10.8 | Collagen, type III, α1 | Col3a1 |
| rc_AI176170_at | 6.9 | FK506-binding protein 1a | Fkbp1a |
| rc_AI228738_s_at | 5.7 | FK506-binding protein 1a | Fkbp1a |
TABLE 3.
GOs/biological themes associated with alterations in gene expression in detrusor and corporal myocytes after 1 week of STZ-DM
| System | Gene category | List hits | EASE score |
|---|---|---|---|
| Detrusor myocytes | |||
| Up-regulation: | |||
| GO cellular component | Actin cytoskeleton | 7 | 1.16E-02 |
| GO cellular component | Myosin | 4 | 2.29E-02 |
| GO biological process | Cell proliferation | 16 | 1.48E-03 |
| GO biological process | Catabolism | 17 | 1.90E-03 |
| GO biological process | Protein catabolism | 11 | 1.17E-02 |
| GO biological process | Macromolecule catabolism | 11 | 1.38E-02 |
| GO biological process | Protein metabolism | 26 | 1.44E-02 |
| GO biological process | Glycolysis | 4 | 4.01E-02 |
| GO biological process | Monosaccharide catabolism | 4 | 4.39E-02 |
| GO biological process | Hexose catabolism | 4 | 4.39E-02 |
| GO biological process | Alcohol catabolism | 4 | 4.39E-02 |
| GO biological process | Glucose catabolism | 4 | 4.39E-02 |
| GO molecular function | Calcium ion binding | 11 | 1.61E-02 |
| GO molecular function | Peptidase activity | 10 | 2.31E-02 |
| GO molecular function | Actin binding | 6 | 2.69E-02 |
| GO molecular function | Transferase activity | 18 | 4.37E-02 |
| GO molecular function | Enzyme inhibitor activity | 6 | 4.43E-02 |
| Down-regulation: | |||
| GO biological process | Cytoskeleton organization and biogenesis | 7 | 3.44E-03 |
| GO biological process | Cell motility | 8 | 3.90E-03 |
| GO biological process | Cell organization and biogenesis | 9 | 7.03E-03 |
| GO biological process | Cytoplasm organization and biogenesis | 8 | 8.01E-03 |
| GO biological process | Muscle contraction | 5 | 1.40E-02 |
| GO biological process | Regulation of muscle contraction | 3 | 3.20E-02 |
| Corporal myocytes | |||
| Up-regulation: | |||
| GO biological process | Ion transport | 21 | 3.21E-04 |
| GO biological process | Metal ion transport | 15 | 4.62E-04 |
| GO biological process | Cation transport | 16 | 4.99E-04 |
| GO biological process | Monovalent inorganic cation transport | 12 | 3.34E-03 |
| GO biological process | Sodium ion transport | 7 | 4.87E-03 |
| GO biological process | Potassium ion transport | 7 | 4.40E-02 |
| GO biological process | Transport | 30 | 4.88E-02 |
| GO cellular component | Voltage-gated sodium channel complex | 3 | 3.30E-02 |
| GO molecular function | Ion channel activity | 12 | 3.09E-02 |
| GO molecular function | Cation channel activity | 9 | 4.09E-02 |
| GO molecular function | ATPase activity\, coupled to transmembrane movement of ions | 4 | 4.87E-02 |
| GO molecular function | ATPase activity\, coupled to transmembrane movement of ions\, phosphorylative mechanism | 4 | 4.87E-02 |
| Down-regulation: | |||
| GO cellular component | Collagen | 2 | 3.41E-02 |
| GO molecular function | Cyclin-dependent protein kinase regulator activity | 2 | 3.27E-02 |
The use of GCRMA to analyse GeneChips from corporal tissue of STZ-DM rats identified 269 genes that were up-regulated and 44 that were down-regulated relative to age-matched controls. Tables 1 and 2 summarize pertinent changes in gene expression that would be expected to be of interest to erectile physiology/function. GO analysis for the up-regulated genes revealed prominent biological themes for ‘ion channel transport’ and ‘ion channel activity’ (Table 3). The changes in expression were relatively modest and were represented by the tachykinin 1 and 3 receptor subtypes (1.4 and 2.0-fold, respectively), the transient receptor potential cation channel, subfamily V, member 1 (Trpv1) receptor (1.4-fold), as well as 3 distinct K+ channel subtypes, e.g. Kcnq3 voltage-gated K+ channel (1.6-fold), Kcnj3, an inwardly rectifying K+ channel (1.4-fold) and erg3 K+ channel (1.3-fold), as well as the Na+/K+ ATPase β1 subunit (1.4-fold). However, as there were fewer down-regulated genes, the main theme identified by GO analysis involved ‘collagen’ genes (Table 3). Three different probe sets (i.e. distinct Affymetrix gene identifications) probing different parts of collagen type 1 α1 were identified with changes of 380-fold, 36-fold and 18.2-fold, respectively. Furthermore, smooth muscle α-actin was decreased 70-fold, as was expression of the actin binding protein calponin (14.6-fold). In addition, FK506 binding protein 12 was determined to be decreased 6–7 fold with two distinct probes.
To further show the potential impact of STZ-DM induced alterations in smooth muscle gene expression on bladder and erectile function, we created several data sets and imported them into pre-existing GenMapp pathways. A ‘colour template’ was then created by assigning each data set a colour, and this colour template was overlaid on existing pathways. No pathway currently exists in GenMapp for corporal or detrusor smooth muscle cells, so to investigate the physiological relevance of the gene expression analysis, as well as its implications to urological disease initiation and progression, we used four existing pathways with common features for the over-represented biological themes in our dataset. The GenMapp pathways revealed significant involvement of the following analogous pathways: (i) myometrial relaxation and contraction pathways (Fig. 2); (ii) calcium regulation in the cardiac cell (Fig. 3), (iii) regulation of actin cytoskeleton (Fig. 4), and (iv) cytokines and inflammatory response pathway (Fig. 5).
FIG. 2.
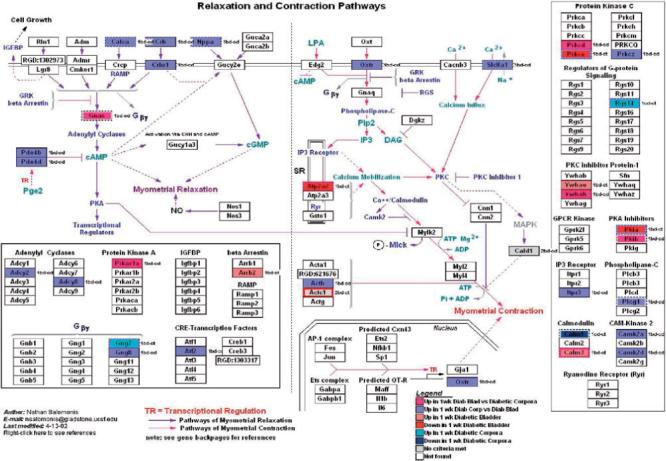
GenMapp pathway showing the impact of 1 week of STZ-DM on transcription in pathways involved in myocyte contraction and relaxation. Although this pathway was specifically developed for the myometrium, there is enough transcriptional overlap in the relevant pathways with the detrusor and corporal myocyte to make this a useful illustrative tool.
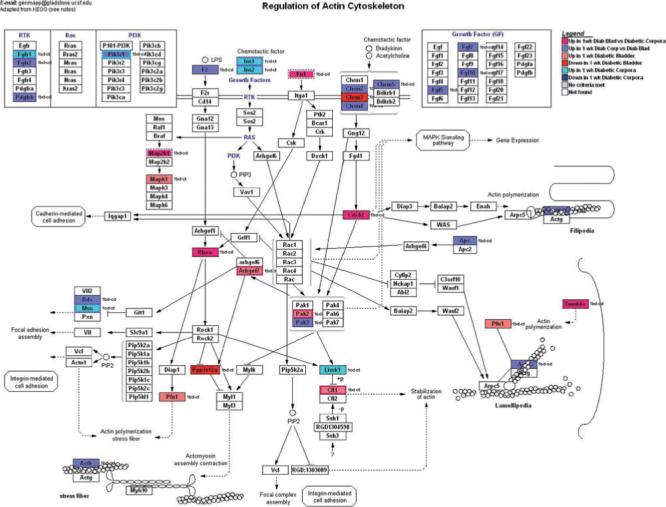
FIG. 3.
GenMapp pathway showing the impact of 1 week of STZ-DM on transcription in pathways involved in regulation of the actin cytoskeleton. Again, although this pathway was not specifically developed for the detrusor or corporal myocyte, there is enough transcriptional overlap to make this a useful illustrative tool.
FIG. 4.
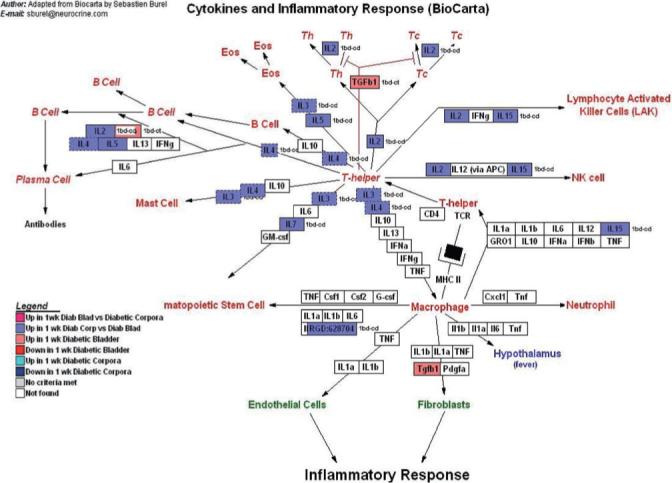
GenMapp pathway showing the impact of 1 week of STZ-DM on transcription in pathways related to cytokines and the inflammatory response. Again, although this pathway was not specifically developed for the detrusor or corporal myocyte, there is enough transcriptional overlap to make this a useful illustrative tool.
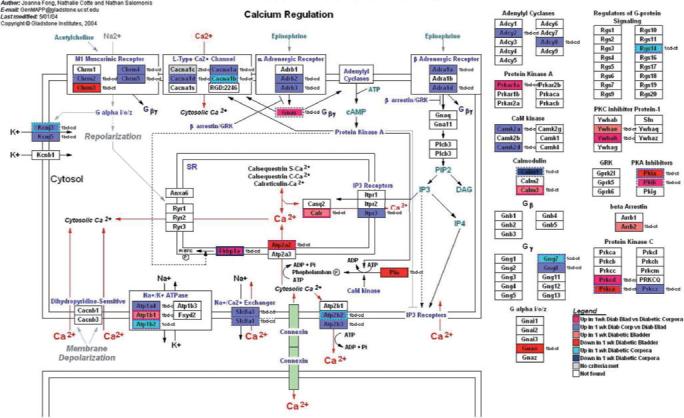
FIG. 5.
GenMapp pathway showing the impact of 1 week of STZ-DM on transcription in pathways involved in calcium regulation. Although this pathway was specifically developed for the myocardium, there is enough transcriptional overlap in the relevant pathways with the detrusor and corporal myocyte to make this a useful illustrative tool.
DISCUSSION
DM is a multifactorial disease with a major impact on lower urinary tract function. Thus, it is not surprising that the expression of many smooth muscle genes is altered upon induction of STZ-DM, and changes of a similar magnitude were recently reported in the corporal tissue of 10 week-STZ-DM rats [30]. We recently reported that at least one gene, Vcsa1, undergoes a temporal change with the onset of DM, which correlates with the development of ED [31]. To begin to improve our understanding of the initiation, development and progression of the diabetic disease process, we examined the impact of STZ-DM on smooth muscle cell gene expression in two organ systems at one sample time (1 week). This time was selected because it is after the onset of DM, but before evidence of organ dysfunction in vivo. This approach is based on the premise that genomic studies of progressive diseases such as DM, whose clinical sequelae depend on both the duration and severity of disease, would provide insights into molecular changes that are harbingers of organ failure (i.e. diagnostic and preventative), as well as molecular alterations that represent mechanisms of organ failure (i.e. therapeutic targets). This approach would be part of a strategy aimed at preventing complications for chronic disease manifestations, such as the end-organ complications of DM. The critical importance of such an approach was emphasized by Dr E.A. Zerhouni, Director of the USA National Institutes of Health (NIH; http://appropriations.senate.gov/hearmarkups/Zerhouni_sStatementfinalFY2006.htm).
In the present study, we used established, conservative analytical methods that are freely available, to estimate changes in gene expression in bladder and corporal smooth muscle in the STZ-DM rat (i.e. GCRMA; Fig. 1). The resulting gene sets were subsequently coupled to two distinct, but complementary (and also publicly available), software programs for statistical interpretation of common biological themes in the data sets (i.e. EASE), as well as to visualize the results of those analyses using physiologically relevant GenMapp pathways. In short, we grouped genes by biology/function to relate them to previously observed changes in physiology (i.e. bladder dysfunction and ED). Below is a description of how this approach can identify organ- and tissue-specific responses to DM, and to interpret these results in the context of their physiological relevance. The present analyses were sufficiently sensitive to identify many neuromuscular genes, which can better explain the causes and/or compensation mechanisms of the neuropathy associated with DM-bladders and DM-ED, thus providing therapeutic targets for the clinicians. This is reflected by the detection of altered expression of neuronal-specific genes in each tissue (i.e. neurofilaments, neurotrophins, synuclein, Trpv1, FK506 binding proteins, etc.). Nonetheless, most of the genes detected reflect the major myocyte component of the tissues studied, but also indicate that the neuronal genes detected are still reflective of the overall pathophysiology. To make the essential points that the present analytical approach can yield, we confine our analysis and the detailed descriptions below to only a few salient comparisons.
BIOLOGICAL THEMES AND THE PHYSIOLOGY/PATHOPHYSIOLOGY OF CHANGES IN GENE EXPRESSION IN DETRUSOR MYOCYTES
As illustrated for the up-regulated genes, GO analysis revealed predominant genetic themes involved in ‘cell proliferation’, ‘sugar, protein and macromolecule catabolism/metabolism’, ‘actin cytoskeleton’ and ‘myosin’ (Table 3). This included increased expression of myosin, as well as myosin regulatory and myosin binding proteins (Table 1). Increased expression of tropomyosin, myosin regulatory light chain, calmodulin 3 and calreticulin were also detected. These increases in thin filament proteins are consistent with previous reports in the bladder of diabetic rabbits [52], where such changes were proposed to contribute to the altered contractile and cytoskeletal structure in bladder myocytes (in this case decreased contractility). In addition, there was a 612-fold increase in MMP-7, a protease involved in the hypoxia-induced proliferation of rat bladder smooth muscle cells [53]. These are the types of alterations are as expected if cell hypertrophy and/or proliferation, and altered detrusor myocyte contractility are ongoing processes that contribute to the remodelling of the enlarged bladder in diabetic animals/patients.
Consistent with these observations, recent digital imaging microscopy studies showed significant remodelling of the bladder of STZ-DM rats at 2–3 weeks after induction of DM [34]. Together, such observations suggest that some of these targets might be harbingers of bladder dysfunction. The decreases in gene expression are also consistent with impending alterations in bladder function, e.g. increased myosin phosphatase, target subunit 1, phospholamban, and 130 kDa-Ins(1,4,5)P3 binding protein, are consistent with increased contractility (Table 2). There was also a decrease in detrusor myocyte M3 receptor subtype gene expression; this might seem counterintuitive given reported increases in cholinergic sensitivity in detrusor strips from diabetic animals [54]. However, Stevens et al. [55] reported that the increased sensitivity to carbachol-induced contraction of detrusor strips from 1 week STZ-DM rats was associated with only a small, albeit significant, increase in the total muscarinic receptor population (≈ 40%). This is a modest change relative to the magnitude required (i.e. 6-fold) for changes in receptor density alone to provide the mechanistic basis for the increased carbachol contractility, especially as only a fraction of the total muscarinic receptor population in rat bladder is M3 receptors. Given the many steps between receptor binding and smooth muscle cell contraction, such reports indicate that the DM-related enhancement in carbachol-induced detrusor contractility occurs elsewhere in the intracellular processes that regulate contraction; perhaps as a result of some of the changes identified in the present report. The decreases in synuclein-γ, neurofilament, light polypeptide, neurotrophin-3, neurofilament 3, medium (Table 2) are what one might expect to be associated with the development of neuropathy or neurodegeneration [56-59]. Finally, the increase in Na+/K+ ATPase β1 subunit is also consistent with altered bladder function.
Overall, the present data indicate that there are a host of changes in gene expression that are consistent with the development of increased detrusor contractility in the short term (i.e. detrusor overactivity), perhaps decreased detrusor contractility in the long term, and decreased innervation (i.e. neuropathy) [27,54,55]. It is possible that the magnitude and duration of the changes in gene expression of some of these targets (and others that might be detected on a different chip or with an increased sample size, etc.) at this time point could further influence the course of disease, and might account for some of the phenotypic diversity seen in STZ-DM induced changes in bladder function. Nonetheless, the 6 × 3 analysis allowed for the identification of hundreds of statistically significant differentially expressed genes.
BIOLOGICAL THEMES AND THE PHYSIOLOGY/PATHOPHYSIOLOGY OF CHANGES IN GENE EXPRESSION IN CORPORAL MYOCYTES
GO analysis for up-regulated genes in corporal myocytes revealed prominent biological themes for ‘ion channel transport’ and ‘ion channel activity’ (Table 1). Alterations in gene expression were observed in three distinct K+ channel subtypes (Kcnq3, voltage-gated K+ channel, Kcnj3, an inwardly rectifying K+ channel, and erg3 K+ channel, as well as the Na+/K+ ATPase β1 subunit; Table 2). Most of these changes are modest and have not yet been characterized or pursued in corporal smooth muscle/erectile tissue, but given the importance of membrane potential to the regulation of corporal smooth muscle cell tone, these alterations are consistent with altered ionic mechanisms in diabetic corporal smooth muscle cells, and thus could lead to altered corporal myocyte excitability/function. Also, there was an increase in gene expression for both the tachykinin 1 and 3 receptor subtypes. The influence of tachykinins on corporal myocyte function has not been extensively studied, and there are species differences in tachykinin-induced effects on these receptor subtypes and their effect on corporal myocyte contractility [60,61]. Nonetheless, increased expression of these might correlate with altered corporal myocyte function, and perhaps, altered erectile function leading to ED.
There was also an increase in Trpv1 receptor expression (Table 1). Stein et al. [62] documented the presence of the Trpv1 receptor in the rat corpora [62], raising the possibility of an interaction between tachykinins and Trpv1 in the corpora, as shown in the bladder [63]. Specifically, they proposed that capsaicin-induced activation of Trpv1 on sensory nerves results in release of tachykinins and cyclooxygenase metabolites, thus evoking myocyte contraction. If a similar link exists in corporal tissue, the increased expression of Trpv1 and tachykinin receptor subtypes might affect corporal smooth muscle contractility.
There were fewer down-regulated genes (Table 2), and the main theme identified by GO analysis involved ‘collagen’ genes (Table 3). The decrease in collagen type 1 α1 expression detected with three distinct Affymetrix probe sets ranged from 18.2- to 360-fold. A similar pattern of reduction in collagen synthesis appears to be conserved during identifiable ED, as reported by Sullivan et al. [30]. Furthermore, smooth muscle α-actin was decreased, as was expression of the actin binding protein calponin. FK506 binding protein 12 was also decreased using two distinct probes; this is particularly interesting given reports of the importance of this gene to recovery of nerve function after cavernosal nerve injury [64,65]. If this gene is down-regulated throughout DM, the use of FK506 and other immunophilin ligands might be less effective for treating DM-neuropathy (as opposed to the neuropathy that results from radical prostate surgery).
USING GENMAPP TO ILLUSTRATE BIOLOGICAL PATHWAYS
The implications of the points discussed above are visible in the few selected GenMapp pathways (Figs 2-5). Not surprisingly, DM-related transcriptional alterations were detected at many points in the signal transduction pathways that govern myocyte contraction and relaxation (Fig. 2), calcium regulation (Fig. 3), as well as the regulation of the actin cytoskeleton (Fig. 4). Interestingly, up-regulation of various cytokines and inflammatory mediators was also detected in both tissues (Fig. 5), although more predominantly in corporal tissue. The presence of this early inflammatory component of DM-related changes in gene expression is worth further exploration. Inflammation might affect not only myocyte phenotype and survival, but perhaps also myocyte contractility (e.g. tachykinin-mediated effects on corporal tissue), and thus tissue/organ function as well.
The present initial results suggest that transcriptional profiling 1 week after the onset of DM can ‘predict’ the advent of some of the pathophysiological processes reported in these organ systems at later times. In pursuing this strategy in a longitudinal fashion throughout the duration of DM, these profiles might help to determine the mechanistic basis for the severity and duration of DM in a tissue- and organ-specific manner, as well as to identify biomarkers of disease progression. While these changes are not fully characterized at the protein level, they are consistent with the findings of others and with the pathophysiology of this disease process in the selected organ systems. Also of interest is that there was little overlap in the GOs and genes of interest between these two urological tissues. Thus, while both myogenic and neurogenic mechanisms contribute to organ dysfunction in the bladder and penis, the transcriptional changes associated with these changes appear to be distinct. Even when there was overlapping gene alterations in both detrusor and corporal myocytes (i.e. the Na+/K+ ATPase β1 subunit), this change differed substantially in magnitude between the two tissue/cell types (29-fold increase in bladder and 1.4-fold increase in corpora). Thus, intervening at the transcriptional (i.e. siRNA) or pharmacological levels (e.g. oral therapy) might selectively target the deficits in the corpora or bladder, offering novel possibilities for treating these debilitating diabetic complications.
In summary, our conservative analytical approach was capable of detecting alterations in gene expression, as well as biological themes/pathways involved in the causes of the organ dysfunction eventually seen in the diabetic bladder and corpora. Further investigation, with larger sample sizes, more time points, complemented by confirmation of the suspected changes in gene (i.e. PCR) and/or protein expression are required. Nonetheless, these initial observations and methods provide a realistic approach for revealing molecular correlates of the initiation, development and progression of common urological complications of DM. This approach has the potential for identifying organ-specific, smooth muscle responses to DM that might provide valuable diagnostic and therapeutic targets.
Finally, these genomic data might have applications for several progressive urological diseases/conditions, e.g. in men with BOO secondary to BPH, identifying molecular milestones for the initiation, development and progression of the inevitable bladder complications would be valuable in trying to delay the onset of symptoms as well as slowing or preventing the progression of bladder dysfunction. A molecular ‘roadmap’ indicative of disease onset and progression for ED would be similarly beneficial to the patient and clinician. Given the promise of this approach, further investigation is warranted.
ACKNOWLEDGEMENTS
This work was supported in part by NIH Grants DK60037 and DK70229, and an NIAAA Pre-Doctoral Fellowship (F30AA014299) to JH. KPD was supported by K01-DK67270 from the NIH, NIDDK and received grants from the International Society for Sexual and Impotence Research and Kidney and Urology Foundation of America (E. Darracott Vaughan Award) to support this research.
Glossary
Abbreviations
- GO
gene ontology
- DM
diabetes mellitus
- ED
erectile dysfunction
- STZ
streptozotocin
- LIMMA
Linear Models for Microarray Data
- GUI
Graphical User Interface
- EASE
Expression Analysis Systematic Explorer
- CEL
cell intensity (file)
- GenMapp
Gene Map Annotator and Pathway Profiler
- MMP-7
matrix metalloproteinase 7
- NIH
National Institutes of Health
Footnotes
JDH and KPD contributed equally to this work.
CONFLICT OF INTEREST
None declared.
REFERENCES
- 1.Bonow RO, Gheorghiade M. The diabetes epidemic: a national and global crisis. Am J Med. 2004;116(Suppl 5A):2S–10S. doi: 10.1016/j.amjmed.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Engelgau MM, Narayan KM, Saaddine JB, Vinicor F. Addressing the burden of diabetes in the 21st century: better care and primary prevention. J Am Soc Nephrol. 2003;14(Suppl 2):S88–91. doi: 10.1097/01.asn.0000070143.71933.b0. [DOI] [PubMed] [Google Scholar]
- 3.Permutt MA, Wasson J, Cox N. Genetic epidemiology of diabetes. J Clin Invest. 2005;115:1431–9. doi: 10.1172/JCI24758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–53. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 5.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–7. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 6.Alloussi S, Mast GJ, Kopper B, Ziegler M. [Micturition disorder as a sequela of sacral autonomic diabetic neuropathy]. Urologe A. 1985;24:291–5. [PubMed] [Google Scholar]
- 7.Brown JS, Wessells H, Chancellor MB, et al. Urologic complications of diabetes. Diabetes Care. 2005;28:177–85. doi: 10.2337/diacare.28.1.177. [DOI] [PubMed] [Google Scholar]
- 8.Costabile RA. Optimizing treatment for diabetes mellitus induced erectile dysfunction. J Urol. 2003;170:S35–9. doi: 10.1097/01.ju.0000077448.31995.99. [DOI] [PubMed] [Google Scholar]
- 9.Faerman I, Maler M, Jadzinsky M, et al. Asymptomatic neurogenic bladder in juvenile diabetics. Diabetologia. 1971;7:168–72. doi: 10.1007/BF01212549. [DOI] [PubMed] [Google Scholar]
- 10.Frimodt-Moller C, Mortensen S. Treatment of diabetic cystopathy. Ann Intern Med. 1980;92:327–8. doi: 10.7326/0003-4819-92-2-327. [DOI] [PubMed] [Google Scholar]
- 11.Guay AT. Sexual dysfunction in the diabetic patient. Int J Impot Res. 2001;13(Suppl 5):S47–50. doi: 10.1038/sj.ijir.3900779. [DOI] [PubMed] [Google Scholar]
- 12.Kaplan SA, Te AE, Blaivas JG. Urodynamic findings in patients with diabetic cystopathy. J Urol. 1995;153:342–4. doi: 10.1097/00005392-199502000-00013. [DOI] [PubMed] [Google Scholar]
- 13.Melman A, Gingell JC. The epidemiology and pathophysiology of erectile dysfunction. J Urol. 1999;161:5–11. [PubMed] [Google Scholar]
- 14.Menendez V, Cofan F, Talbot-Wright R, Ricart MJ, Gutierrez R, Carretero P. Urodynamic evaluation in simultaneous insulin-dependent diabetes mellitus and end stage renal disease. J Urol. 1996;155:2001–4. [PubMed] [Google Scholar]
- 15.Yoshimura N, Chancellor MB, Andersson KE, Christ GJ. Recent advances in understanding the biology of diabetes-associated bladder complications and novel therapy. BJU Int. 2005;95:733–8. doi: 10.1111/j.1464-410X.2005.05392.x. [DOI] [PubMed] [Google Scholar]
- 16.Benko R, Lazar Z, Porszasz R, Somogyi GT, Bartho L. Effect of experimental diabetes on cholinergic, purinergic and peptidergic motor responses of the isolated rat bladder to electrical field stimulation or capsaicin. Eur J Pharmacol. 2003;478:73–80. doi: 10.1016/j.ejphar.2003.08.035. [DOI] [PubMed] [Google Scholar]
- 17.Bezuijen MW, Levendusky MC, Longhurst PA. Functional response of bladder strips from streptozotocin diabetic rats depends on bladder mass. J Urol. 2003;169:2397–401. doi: 10.1097/01.ju.0000060120.47657.8a. [DOI] [PubMed] [Google Scholar]
- 18.Latifpour J, Gousse A, Kondo S, Morita T, Weiss RM. Effects of experimental diabetes on biochemical and functional characteristics of bladder muscarinic receptors. J Pharmacol Exp Ther. 1989;248:81–8. [PubMed] [Google Scholar]
- 19.Lincoln J, Crockett M, Haven AJ, Burnstock G. Rat bladder in the early stages of streptozotocin-induced diabetes: adrenergic and cholinergic innervation. Diabetologia. 1984;26:81–7. doi: 10.1007/BF00252269. [DOI] [PubMed] [Google Scholar]
- 20.Liu G, Daneshgari F. Alterations in neurogenically mediated contractile responses of urinary bladder in rats with diabetes. Am J Physiol Renal Physiol. 2005;288:F1220–6. doi: 10.1152/ajprenal.00449.2004. [DOI] [PubMed] [Google Scholar]
- 21.Nakahara T, Mitani A, Kubota Y, et al. MaxiK channel-triggered negative feedback system is preserved in the urinary bladder smooth muscle from streptozotocin-induced diabetic rats. J Smooth Muscle Res. 2004;40:97–109. doi: 10.1540/jsmr.40.97. [DOI] [PubMed] [Google Scholar]
- 22.Rizk DE, Padmanabhan RK, Tariq S, Shafiullah M, Ahmed I. Ultra-structural morphological abnormalities of the urinary bladder in streptozotocin-induced diabetic female rats. Int Urogynecol J Pelvic Floor Dysfunct. 2006;17:143–54. doi: 10.1007/s00192-005-1359-5. [DOI] [PubMed] [Google Scholar]
- 23.Santicioli P, Gamse R, Maggi CA, Meli A. Cystometric changes in the early phase of streptozotocin-induced diabetes in rats: evidence for sensory changes not correlated to diabetic neuropathy. Naunyn Schmiedebergs Arch Pharmacol. 1987;335:580–7. doi: 10.1007/BF00169128. [DOI] [PubMed] [Google Scholar]
- 24.Waring JV, Wendt IR. Effects of streptozotocin-induced diabetes mellitus on intracellular calcium and contraction of longitudinal smooth muscle from rat urinary bladder. J Urol. 2000;163:323–30. [PubMed] [Google Scholar]
- 25.Watanabe T, Miyagawa I. Characteristics of detrusor contractility during micturition in diabetics. Neurourol Urodyn. 1999;18:163–71. doi: 10.1002/(sici)1520-6777(1999)18:3<163::aid-nau2>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 26.Christ GJ, Day N, Santizo C, et al. Intracorporal injection of hSlo cDNA restores erectile capacity in STZ-diabetic F-344 rats in vivo. Am J Physiol Heart Circ Physiol. 2004;287:H1544–53. doi: 10.1152/ajpheart.00792.2003. [DOI] [PubMed] [Google Scholar]
- 27.Christ GJ, Hsieh Y, Zhao W, et al. Effects of streptozotocin-induced diabetes on bladder and erectile (dys)function in the same rat in vivo. BJU Int. 2006;97:1076–82. doi: 10.1111/j.1464-410X.2006.06058.x. [DOI] [PubMed] [Google Scholar]
- 28.El-Sakka AI, Lin CS, Chui RM, Dahiya R, Lue TF. Effects of diabetes on nitric oxide synthase and growth factor genes and protein expression in an animal model. Int J Impot Res. 1999;11:123–32. doi: 10.1038/sj.ijir.3900392. [DOI] [PubMed] [Google Scholar]
- 29.Rehman J, Chenven E, Brink P, et al. Diminished neurogenic but not pharmacological erections in the 2- to 3-month experimentally diabetic F-344 rat. Am J Physiol. 1997;272:H1960–71. doi: 10.1152/ajpheart.1997.272.4.H1960. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan CJ, Teal TH, Luttrell IP, Tran KB, Peters MA, Wessells H. Microarray analysis reveals novel gene expression changes associated with erectile dysfunction in diabetic rats. Physiol Genomics. 2005;23:192–205. doi: 10.1152/physiolgenomics.00112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong Y, Tar M, Davelman F, Christ G, Melman A, Davies KP. Variable coding sequence protein A1 as a marker for erectile dysfunction. BJU Int. 2006;98:396–401. doi: 10.1111/j.1464-410X.2006.06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin RM, Wein AJ, Buttyan R, Monson FC, Longhurst PA. Update on bladder smooth-muscle physiology. World J Urol. 1994;12:226–32. doi: 10.1007/BF00191201. [DOI] [PubMed] [Google Scholar]
- 33.Daneshgari F, Liu G, Imrey PB. Time dependent changes in diabetic cystopathy in rats include compensated and decompensated bladder function. J Urol. 2006;176:380–6. doi: 10.1016/S0022-5347(06)00582-9. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, Daneshgari F. Temporal diabetes-and diuresis-induced remodeling of the urinary bladder in the rat. Am J Physiol Regul Integr Comp Physiol. 2006;291:R837–43. doi: 10.1152/ajpregu.00917.2005. [DOI] [PubMed] [Google Scholar]
- 35.Brink PR, Valiunas V, Wang HZ, Zhao W, Davies K, Christ GJ. Experimental diabetes alters connexin43 derived gap junction permeability in short-term cultures of rat corporeal vascular smooth muscle cells. J Urol. 2006;175:381–6. doi: 10.1016/S0022-5347(05)00007-8. [DOI] [PubMed] [Google Scholar]
- 36.Hipp J, Atala A. Genechips in Regenerative Medicine. Elsevier Academic Press; New York: 2006. [Google Scholar]
- 37.Wettenhall JM. 2004. affylmGUI Package Vignette.
- 38.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–6. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 39.Bolstad BM, Collin F, Simpson KM, Irizarry RA, Speed TP. Experimental design and low-level analysis of microarray data. Int Rev Neurobiol. 2004;60:25–58. doi: 10.1016/S0074-7742(04)60002-X. [DOI] [PubMed] [Google Scholar]
- 40.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 41.Cope LM, Irizarry RA, Jaffee HA, Wu Z, Speed TP. A benchmark for Affymetrix GeneChip expression measures. Bioinformatics. 2004;20:323–31. doi: 10.1093/bioinformatics/btg410. [DOI] [PubMed] [Google Scholar]
- 42.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucl Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 44.Wu Z, Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol. 2005;12:882–93. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- 45.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 46.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry RA, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 47.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 48.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 51.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mannikarottu AS, Changolkar AK, Disanto ME, Wein AJ, Chacko S. Over expression of smooth muscle thin filament associated proteins in the bladder wall of diabetics. J Urol. 2005;174:360–4. doi: 10.1097/01.ju.0000161602.18671.c7. [DOI] [PubMed] [Google Scholar]
- 53.Sabha N, Aitken K, Lorenzo AJ, Szybowska M, Jairath A, Bagli DJ. Matrix metalloproteinase-7 and epidermal growth factor receptor mediate hypoxia-induced extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase activation and subsequent proliferation in bladder smooth muscle cells. In Vitro Cell Dev Biol Anim. 2006;42:124–33. doi: 10.1290/0510070.1. [DOI] [PubMed] [Google Scholar]
- 54.Uchiyama T, Chess-Williams R. Muscarinic receptor subtypes of the bladder and gastrointestinal tract. J Smooth Muscle Res. 2004;40:237–47. doi: 10.1540/jsmr.40.237. [DOI] [PubMed] [Google Scholar]
- 55.Stevens LA, Sellers DJ, McKay NG, Chapple CR, Chess-Williams R. Muscarinic receptor function, density and G-protein coupling in the overactive diabetic rat bladder. Auton Autacoid Pharmacol. 2006;26:303–9. doi: 10.1111/j.1474-8673.2006.00371.x. [DOI] [PubMed] [Google Scholar]
- 56.Iwai A. Properties of NACP/alpha-synuclein and its role in Alzheimer's disease. Biochim Biophys Acta. 2000;1502:95–109. doi: 10.1016/s0925-4439(00)00036-3. [DOI] [PubMed] [Google Scholar]
- 57.Pittenger G, Vinik A. Nerve growth factor and diabetic neuropathy. Exp Diabesity Res. 2003;4:271–85. doi: 10.1155/EDR.2003.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Bohlen Und Halbach O. Synucleins and their relationship to Parkinson's disease. Cell Tissue Res. 2004;318:163–74. doi: 10.1007/s00441-004-0921-7. [DOI] [PubMed] [Google Scholar]
- 59.Yasuda H, Terada M, Maeda K, et al. Diabetic neuropathy and nerve regeneration. Prog Neurobiol. 2003;69:229–85. doi: 10.1016/s0301-0082(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 60.Patacchini R, Barbagli G, Palminteri E, Lazzeri M, Turini D, Maggi CA. Tachykinin NK1 and NK2 receptors mediate inhibitory vs excitatory motor responses in human isolated corpus cavernosum and spongiosum. Br J Pharmacol. 2002;135:1351–4. doi: 10.1038/sj.bjp.0704650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi R, Nishimura J, Hirano K, Naito S, Kanaide H. The mechanisms for tachykinin-induced contractions of the rabbit corpus cavernosum. Br J Pharmacol. 2002;137:845–54. doi: 10.1038/sj.bjp.0704938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stein RJ, Santos S, Nagatomi J, et al. Cool (TRPM8) and hot (TRPV1) receptors in the bladder and male genital tract. J Urol. 2004;172:1175–8. doi: 10.1097/01.ju.0000134880.55119.cf. [DOI] [PubMed] [Google Scholar]
- 63.Andrade EL, Ferreira J, Andre E, Calixto JB. Contractile mechanisms coupled to TRPA1 receptor activation in rat urinary bladder. Biochem Pharmacol. 2006;72:104–14. doi: 10.1016/j.bcp.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 64.Burnett AL, Becker RE. Immunophilin ligands promote penile neurogenesis and erection recovery after cavernous nerve injury. J Urol. 2004;171:495–500. doi: 10.1097/01.ju.0000089775.88825.ec. [DOI] [PubMed] [Google Scholar]
- 65.Sezen SF, Blackshaw S, Steiner JP, Burnett AL. FK506 binding protein 12 is expressed in rat penile innervation and upregulated after cavernous nerve injury. Int J Impot Res. 2002;14:506–12. doi: 10.1038/sj.ijir.3900919. [DOI] [PubMed] [Google Scholar]