Abstract
Antisense oligonucleotides are being evaluated in clinical trials as novel therapeutic agents. To further improve the properties of antisense oligonucleotides, we have designed mixed-backbone oligonucleotides (MBOs) that contain phosphorothioate segments at the 3′ and 5′ ends and have a modified oligodeoxynucleotide or oligoribonucleotide segment located in the central portion of the oligonucleotide. Some of these MBOs indicate improved properties compared with phosphorothioate oligodeoxynucleotides with respect to affinity to RNA, RNase H activation, and anti-HIV activity. In addition, more acceptable pharmacological, in vivo degradation and pharmacokinetic profiles were obtained with these MBOs.
Keywords: pharmacokinetics, anti-HIV agents, coagulation, hemolytic complement
Synthetic oligonucleotides (oligos) complementary to messenger RNA have shown promise as therapeutic agents for various diseases (1). Phosphorothioate oligodeoxynucleotides (PS-oligos) have been studied intensively in both in vitro and in vivo models, and clinical trials in humans are underway (2).
Although PS-oligos continue to show promising results as first generation antisense oligos, they have certain limitations (2). Toxicity studies of PS-oligos in mice, rats, monkeys, and humans have shown some dose-dependent side effects (2). In mice and rats, these side effects include thrombocytopenia, elevation of liver transaminases, hyperplasia of reticuloendothelial cells in various organs, and renal tubular changes (2, 3). In monkeys, the side effects observed are activation of complement (4) and prolongation of activated partial thromboplastin time (aPTT; refs. 2 and 5). These side effects may be caused by the PS-oligos or by their in vivo metabolites. Because similar side effects have been observed after administration of dextran sulfate (2, 6), the inference is that these side effects are caused by the polyanionic nature of PS-oligos and are not nucleotide-sequence-specific. In addition, certain conditions (e.g., splenomegaly) may be caused by a sequence-dependent mitogenic response leading to cytokine induction (2, 7). The aim of the present study was to design less polyanionic oligos that would still be biologically active as antisense agents but less immune stimulatory than the original oligos. A related aim was to retain control of degradation in vivo and therefore generate a favorable pharmacological and safety profile.
MATERIALS AND METHODS
Synthesis and Analysis of Oligonucleotides.
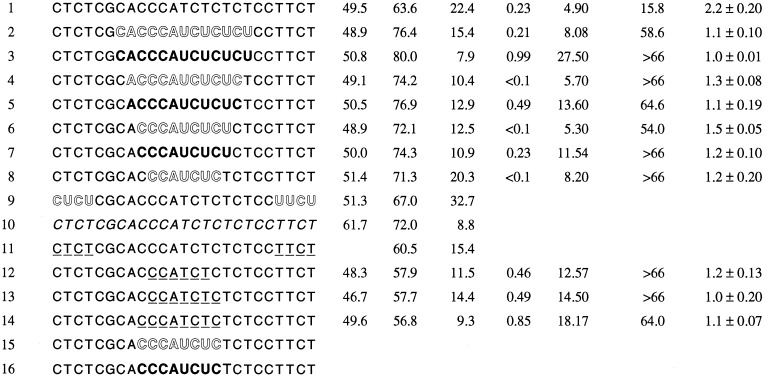
The structures of oligos used in the present study are illustrated in Table 1. The synthesis of oligos was carried out on a 15-μmol scale (Biosearch 8900 DNA Synthesizer) or on a 1-mmol scale (Pharmacia OligoPilot II Synthesizer) using appropriate nucleoside phosphoramidites (8). Oligos 1, 9, 10, and 11 were synthesized and purified as reported (8). Oligos 2, 4, 6, 8, and 15 were synthesized in a three-step reaction sequence. First, the 3′ end was prepared using deoxynucleoside phosphoramidites. The central region was next synthesized using 2′-O-methylribonucleoside phosphoramidites, and finally, the 5′ end was prepared from deoxynucleoside phosphoramidites. Oxidation following each coupling was carried out using 3H-1,2-benzodithiol-3-one-1,1-dioxide (9) to generate phosphorothioate linkages. Oligos 3, 5, 7, and 16 were synthesized by the same procedure used for oligos 2, 4, 6, 8, and 15 except that, after 2′-O-methylribonucleosides couplings, oxidation was carried out using iodine reagent to generate phosphodiester linkages. Oligos 12–14 were prepared by the same procedure used above, except that the middle segment of the oligo was synthesized using nucleoside methylphosphonamidites (10), followed by oxidation with the iodine reagent. Deprotection of oligos 2–8 was completed in two steps using concentrated ammonia as reported (11). Oligos 12–14 were also deprotected in two steps using concentrated ammonia followed by ethylenediamine:ethanol treatment (10).
Table 1.
Structures of oligos used in this study and their various parameters
| Oligo no. | Sequence | Melting temperature, °C |
RNase H t1/2, min | HIV IC90, μM | aPTT 50% conc., μM | Complement lysis | Lymphocyte stimulation index | |
|---|---|---|---|---|---|---|---|---|
| DNA | RNA | |||||||
Lightface type, deoxynucleoside phosphorothioate; openface type, 2′-O-methylribonucleoside phosphorothioate; boldface type, 2′-O-methylribonucleoside phosphodiester; italic type, deoxynucleoside phosphodiester; underline, deoxynucleoside methylphosphonate.
Purification of oligos followed a published procedure (8). Analysis of the purified oligos was carried out using PAGE and capillary gel electrophoresis. The percentage of the desired length of each oligo was greater than 90, and the nature and ratio of internucleotide linkages were confirmed by 31P NMR.
35S-Labeled Oligos.
Synthesis of 35S-labeled oligos was carried out using a published procedure (12). Oligos 14–16 were each labeled with 35S at five internucleotide linkages at the 5′ end. The specific activities of the PAGE-purified products were 0.62, 0.75, and 0.27 μCi/μg (1 Ci = 37 GBq) for oligos 14–16, respectively.
Melting Temperature.
Melting temperatures (Tm) were recorded using a GBC 920 Spectrophotometer (GBC Scientific Equipment, Victoria, Australia) with six cuvettes, and the temperature was controlled by a Peltier-effect temperature controller. Oligos were mixed with complementary oligodeoxynucleotide (35-mer) or oligoribonucleotide (25-mer) in a buffer containing a 1:1 ratio to obtain an overall concentration of 0.2 A260 units in a buffer containing 10 mM Pipes, 1 mM EDTA, and 100 mM NaCl (pH 7). The samples were heated to 90°C and cooled to room temperature. The samples were then heated at a rate of 0.5°C/min, and A260 versus time was recorded. The Tm values (temperatures at which the oligos were half-dissociated) were obtained from the first derivative plots.
RNase H Cleavage and Kinetics.
RNase H kinetics data were obtained using a GBC 920 Spectrophotometer at 259 nm. Oligos (0.05 A260 units) were mixed with 0.1 A260 units of complementary oligoribonucleotide (35-mer) in a buffer containing 20 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 100 mM KCl, 2% glycerol, and 0.1 mM DTT. The mixture was placed in a cuvette and heated to 95°C for 5 min, then gradually cooled to room temperature, and incubated at 37°C for 10 min, and the absorbance was recorded. Five units of RNase H (Boehringer Mannheim) was added and incubated at 37°C for 3 hr, and A260 versus time was recorded. From the curve, t1/2 (time at which 50% absorbance change was observed) was calculated.
Nuclease Stability.
32P end-labeled oligos (0.05 A260 units) were suspended in 39 μl of buffer containing 10 mM Tris (pH 8) and 10 mM MgCl2. A 10-μl aliquot was removed at time zero. To the remaining sample, 0.003 units of phosphodiesterase from Crotalus durissus (Boehringer Mannheim) was added, and the solution was incubated at 37°C. At 30, 60, and 120 min, 10-μl aliquots were removed and mixed with 10 μl of formamide. The resultant samples were then analyzed by PAGE (20% acrylamide, 8.3 M urea) and subjected to autoradiography.
Anti-HIV-1.
The biological activity of oligos against HIV-1 was studied in cell culture assays (13, 14). Molt 3 cells (4 × 105 cells per ml) were infected with HIV-1 suspension (tissue culture 50% infective dose per milliliter = 10−5.3) and cultured in the presence of oligos. Cell concentrations were adjusted to 4 × 105 cells per ml on days 3 and 6, and the cells were treated with oligos at their initial concentrations. On day 9, HIV-1 replication was monitored by the p24 antigen capture assay (Coulter). The inhibitory concentration to suppress HIV-1 replication by 90% (IC90) was determined from the concentration versus percent inhibition plots.
Coagulation and Hemolytic Complement Assays.
Coagulation, which was measured photometrically, was assessed by the aPTT, as performed with citrated normal donor plasma on an Electra 1000C (Medical Laboratory Automation, Mount Vernon, NY) according to the recommended clinical procedures (Actin FSL, Baxter Dade, Miami), and 25 mM calcium was used to initiate clot formation. Various concentrations of oligos were tested with parallel controls of buffer alone. Control plasma aPTT values ranged from 27 to 39 sec. The 50% prolongation doses were extrapolated from plots of oligo concentration versus percent prolongation of the clotting time compared with the appropriate buffer control.
Activation of serum complement by oligos was determined indirectly by quantitation of residual hemolytic activity (5). Briefly, fresh normal donor serum was mixed with various concentrations of oligos, with parallel controls of serum mixed with buffer alone. After incubation at 37°C for 10 min, serum samples were diluted and assayed with opsonized red cell targets for determination of hemolysis by standard methods. Data are presented as the oligo dose producing 50% reduction of hemolytic activity, determined from plots of oligo concentration versus percentage of control hemolysis.
In Vitro Cell Proliferation Studies.
In vitro cell proliferation studies were carried out using mouse spleen cells (15). Spleen cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum/50 μM 2-mecaptoethanol/100 units/ml penicillin/100 μg/ml streptomycin/2 mM l-glutamate. The cells were plated in 96-well dishes at a density of 106 cells per ml in a final volume of 100 μl. Mitogen (lipopolysaccharide, 10 μg/ml) or oligos were added to the cell culture in 10 μl of TE buffer (10 mM Tris·HCl, pH 7.5/1 mM EDTA). After 44 hr, 1 μCi of [3H]thymidine (Amersham) in 20 μl of RPMI 1640 medium was added to the culture, the cells were harvested by an automatic cell harvester (Skatron, Sterling, VA), and the filters were counted using a scintillation counter. Each experiment was completed in triplicate.
Pharmacokinetic Studies.
Pharmacokinetic studies were carried out using a protocol similar to that previously described (16, 17). Male Sprague–Dawley rats (120 ± 10 g, Harlan Laboratories, Indianapolis) were fed with commercial diet and water ad libitum for 1 week before the study. Thirty rats were used over 10 time points: 3 animals each at 5, 30, and 60 min, and 1, 3, 6, 12, 24, 48 and 72 hr. Animals received a single bolus injection into the tail vein at a dose of 30 mg per kilogram of body weight. Unlabeled and 35S-labeled oligos were dissolved in physiological saline (0.9% NaCl) at a concentration of 10 mg/ml (15 μCi/ml). After i.v. injection, each animal was placed in a metabolism cage and fed with commercial diet and water ad libitum. Blood samples were collected in heparinized tubes, and plasma was separated by centrifugation. Total voided urine and excreted feces were collected using a rat metabolism cage (Vanguard International, Neptune, NJ).
At scheduled times, animals were killed by exsanguination under sodium pentobarbital anesthesia. Various tissues were collected from each animal, immediately blotted on Whattman No. 1 filter paper, trimmed of extraneous fat or connective tissue, emptied, cleaned of all contents, weighed, and homogenized in 0.9% NaCl saline (5 ml per gram of wet weight). The resultant homogenates were stored at a temperature of −70°C or greater until further analysis. The total radioactivity in tissues and body fluids was determined by liquid scintillation spectrometry (Beckman spectrometer, model LS 6000TA; refs. 16 and 17).
Oligonucleotide Extraction from Plasma and Kidney.
Plasma (180 μl) and kidney homogenate (180 μl) were incubated with proteinase K (100 μg, Boehringer Mannheim) in extraction buffer (0.5% SDS/10 μM NaCl/20 μM Tris·HCl, pH 7.6/10 μM EDTA) for 2 hr at 60°C. The samples were extracted twice with phenol/chloroform (1:1, vol/vol) and precipitated with ethanol. Samples extracted from the kidney homogenate were analyzed by electrophoresis on a 20% polyacrylamide gel containing 8.3 M urea. The gels were fixed in 10% acetic acid/10% methanol solution and dried before autoradiography.
Samples obtained from plasma were 5′ end-labeled with [γ-32P]ATP (Amersham) using T4 polynucleotide kinase (10 units, New England Biolabs). The samples were then treated with RNase A (10 μg, Boehringer Mannheim) for 20 min at 37°C. Each sample was purified on a TE select G25 column (5′3′) before gel electrophoresis and autoradiography.
Toxicity of Oligonucleotides.
To assess the comparative safety profiles, oligos were administered i.v. to Fisher 344 rats (3 per sex per group) at a dose of 3, 10, or 30 mg/kg daily for 7 days. On day 8, animals were killed for clinical chemistry and histopathology of various organs.
RESULTS
Biophysical Properties.
Thermodynamic stability of an oligo to the target RNA and activation of RNase H are important parameters for its antisense activity. MBOs 2–8 containing 2′-O-methylribonucleoside in the central region of the oligos and phosphorothioate linkages throughout have Tm values of 7–13°C greater than the PS-oligo counterpart (oligo 1, Table 1). The increase in Tm correlated with the number of 2′-O-methylribonucleosides and the nature of the internucleotide linkages. MBOs 3, 5, and 7, containing 2′-O-methylribonucleoside phosphodiester (2′-OMePO) linkages had higher Tm values than oligonucleotides 2, 4, and 6, which contained 2′-O-methylribonucleoside phosphorothioate (2′-OMePS) linkages. MBOs 12–14, which contained methylphosphonate (PM) linkages, had lower Tm values than the PS-oligo (oligo 1).
Duplex of PS-oligos and RNA stimulate RNase H, which then excises RNA (18); however, PS-oligos have been shown to be inhibitors of RNase H activity at high concentrations (19). The t1/2 for catalyzing RNA excission by PS-oligo 1 in the presence of RNase H was about three times slower than phosphodiester oligo 10 (ref. 19 and Table 1). Antisense oligo-assisted cleavage of RNA by RNase H may depend upon the on and off rate of oligo binding to RNA. Cleavage rates may also depend on RNase H binding to the modified duplex. Experiments with end-modified MBOs (oligos 9 and 11) showed that the rate of cleavage of RNA by RNase H may also depend on the nature of the oligo modifications as oligo 11 was a better substrate than oligo 9 (Table 1).
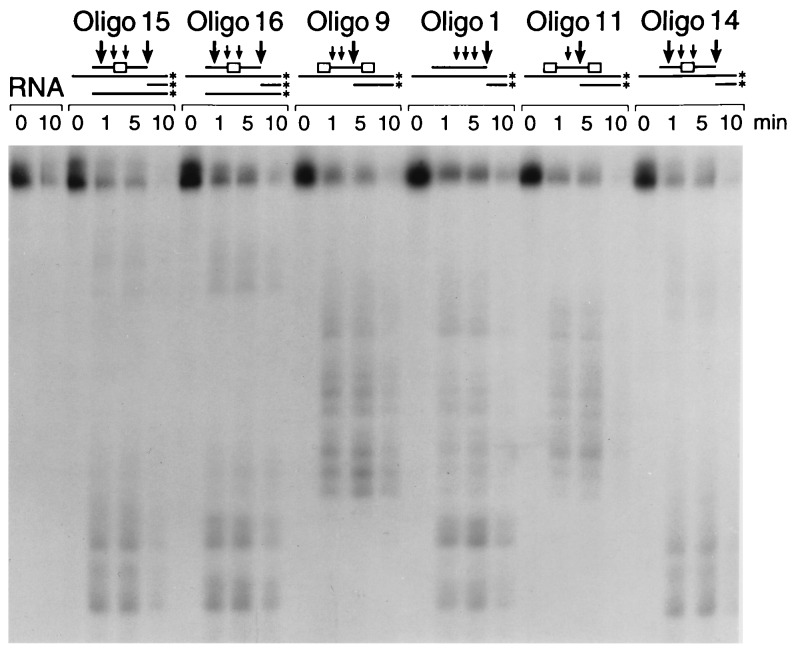
Cleavage of RNA in the presence of centrally modified MBOs (oligos 2–8 and 12–14) was faster than cleavage in the presence of PS-oligo or end-modified MBOs (Table 1). Analysis of the cleavage products by gel electrophoresis showed that in the presence of centrally modified MBOs, the RNA was cleaved on both sides, unlike RNA in the presence of PS-oligo and end-modified MBOs (Fig. 1).
Figure 1.
RNase H cleavage pattern of oligos on target sequence. The RNA was labeled at the 3′ end and incubated with the oligos as described in Materials and Methods. The samples were analyzed by gel electrophoresis. Arrows indicate the approximate locations of the major (large arrow) and minor (small arrow) excission sites of the RNA by RNase H in the presence of oligos 1, 9, 11, and 14–16. The box in the oligo represents the position of the modified region.
Stability Toward Nucleases.
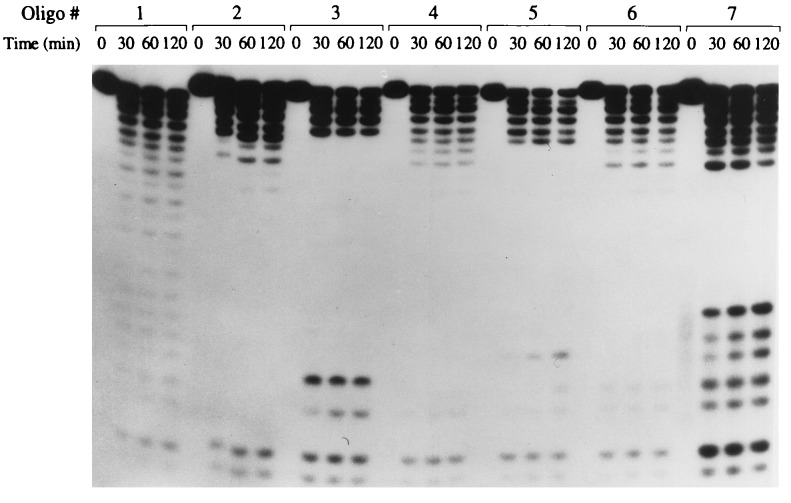
The stabilities of PS-oligo 1 and centrally modified MBOs 2–7 against snake venom phosphodiesterase were very similar, but the degradation profiles were quite different (Fig. 2). Digestion of PS-oligo generated a ladder of oligos of shorter lengths. Digestion of oligos 2, 4, and 6 generated a ladder of up to only 18- to 20-mer, and no further degradation products were observed. This lack of degradation products is because of the increased nuclease resistance of 2′-OMePS linkages. Similar results were obtained for oligos 12–14 (data not shown). Digestion of oligos 3, 5, and 7 showed a ladder of digestion products like that of the PS-oligo, but degradation products in the central region of the ladder were not observed. The degradation profiles of MBOs 3, 5, and 7 suggest that, after digestion of the PS-oligo segment, 2′-OMePO linkages were digested very rapidly, and the remaining PS-oligo segment of the oligo was digested slowly (Fig. 2).
Figure 2.
Stability of oligos against snake venom phosphodiesterase. 5′ end-labeled oligos were incubated with snake venom phosphodiesterase as described in Materials and Methods. At the indicated time, an aliquot was analyzed by gel electrophoresis.
Anti-HIV-1 Activity.
The centrally modified MBOs listed in Table 1 were studied for their inhibitory activity against HIV-1 replication using a long-term infected cell culture system. All oligos inhibited HIV-1 replication in a dose-dependent fashion, with the IC90 ranging from <0.1 to 0.99 μM (Table 1). Oligos 4, 6, and 8, containing 2′-OMePS segments, were more effective than PS-oligo 1. Oligos containing 2′-OMePO (oligos 3, 5, and 7) and PM linkages (oligos 12–14) were 2- to 3-fold less effective than PS-oligo 1. No mismatched oligo was included in the present study, but we have shown that in the present assay system inhibition of HIV-1 replication occurs primarily by the sequence-specific mechanism described in our previous studies (13, 14).
Prolongation of aPTT.
Administration of PS-oligos in monkeys and humans has been shown to prolong the aPTT, a clinical parameter measuring blood coagulation (2, 5). In in vitro studies, PS-oligos also prolong aPTT in a dose-dependent manner that is not sequence-dependent but is dependent on the nature of the internucleotide linkages in the oligos. In the present study, all oligos prolonged aPTT, but the concentration of the oligos required to prolong the aPTT by 50% was greater than the concentration of PS-oligo 1 (Table 1). Oligos 3, 5, 7, and 12–14, which contained 2′-OMePO or PM linkages, showed less of an effect on aPTT than oligos 2, 4, 6, and 8, which contained 2′-OMePS linkages.
Lymphocyte Proliferation.
PS-oligos containing a specific nucleotide motif (e.g., CG) are known to induce lymphocyte proliferation that depends on the nature of internucleotide linkages (2, 7). In the present study, all MBOs showed reduction in lymphocyte proliferation compared with PS-oligo 1 (Table 1).
Complement Activation.
In addition to prolonging the aPTT, the other short-term side effect observed in monkeys was complement activation and reduction of serum hemolytic activity (2, 4, 5). All oligos showed concentration-dependent reduction of complement hemolysis in in vitro assays. PS-oligo 1 at 15 μM reduced hemolysis by 50%, whereas all other oligos required concentrations greater than 60 μM (Table 1).
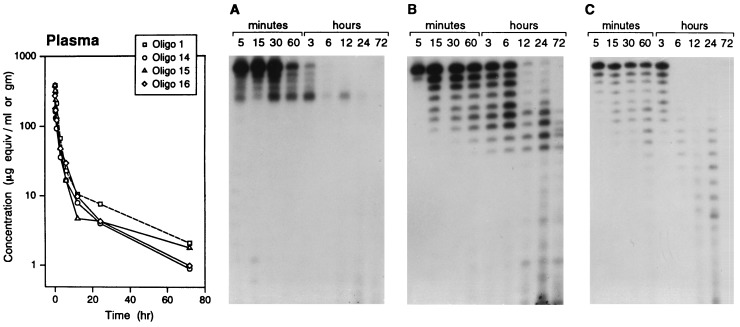
Plasma Clearance and in Vivo Stability.
After i.v. administration of a 30 mg/kg dose of oligos 14–16, plasma clearance was rapid, as had been observed earlier with PS-oligo 1 (ref. 16 and Fig. 3). The half-lives of distribution (t1/2α) and elimination (t1/2β) were in the range of 0.32–0.79 hr and 8.5–12.7 hr, respectively. Analysis of the extracted oligo from the plasma showed the presence of both intact and degraded oligo (Fig. 3). The degradation pattern depended on the oligo modification. Oligo 15 (Fig. 3C) showed time-dependent degradation, and a continuous ladder of degradation products was generated, presumably after degradation of the PS-oligo segment from the 3′ end. No major digestion products were observed at 6 and 12 hr. At 24 hr, however, degradation products of even shorter lengths, presumably generated in tissues after degradation of the 2′-OMePS segment, were observed. In the case of oligo 16 (Fig. 3B), eight degradation products, representing the length of PS-oligo segment, were observed, but shorter metabolites were not observed. The mobility of the bands on the gel suggests that degradation of oligos 15 and 16 was primarily by 3′ exonuclease. The degradation pattern suggested that the segment of oligo 16 containing 2′-OMePO linkages remained stable toward endonucleases. In contrast to band patterns for oligos 15 and 16, the pattern of bands for oligo 14 (Fig. 3A) on the gel suggests that only the PS-oligo portion was degraded; further degradation of the oligo slowed significantly at the site of PM linkages.
Figure 3.
Plasma clearance and stability of oligos 1 and 14–16. A plot of plasma concentration versus the time course of oligo-derived radioactivity (35S) is shown at the left. The plasma samples were extracted, and the oligos were analyzed after 5′ end-labeling with 32P by gel electrophoresis and were autoradiographed. (A) Oligo 14. (B) Oligo 16. (C) Oligo 15. (The data for oligo 1 are derived from a previous study, ref. 16.)
Tissue Disposition.
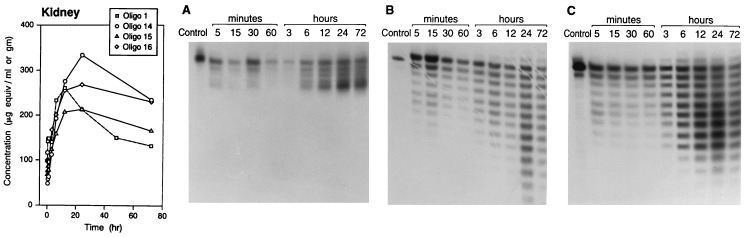
Tissue disposition of oligos 14–16 was similar to that observed for PS-oligo 1 as reported earlier (16). While the initial concentrations (based on radioactivity equivalents) of these oligos in various tissues were quite similar, at 48–72 hr after dosing, oligos 15 and 16 were present at lower concentrations in tissues than the concentration of oligo 14, except in the kidneys. The disposition of oligos 14–16 in kidneys as compared with that of PS-oligo 1 is shown in Fig. 4. At the 72-hr time point, all three oligos were present in kidney at higher concentrations than that of PS-oligo 1. The degradation of oligos 15 and 16 showed a very similar profile as observed in plasma (Fig. 4 B and C). In the case of oligo 14, degradation after digestion of PS-oligo linkages seemed to cease at the site of the PM linkages (Fig. 4A).
Figure 4.
Kidney disposition and stability of oligos 1 and 14–16. A plot of kidney concentration versus the time course of oligo-derived radioactivity (35S) is shown at the left. The kidney samples were extracted, and the oligos were analyzed by gel electrophoresis and autoradiographed. (A) Oligo 14. (B) Oligo 15. (C) Oligo 16. (The data for oligo 1 are derived from a previous study, ref. 16.)
Urinary Excretion.
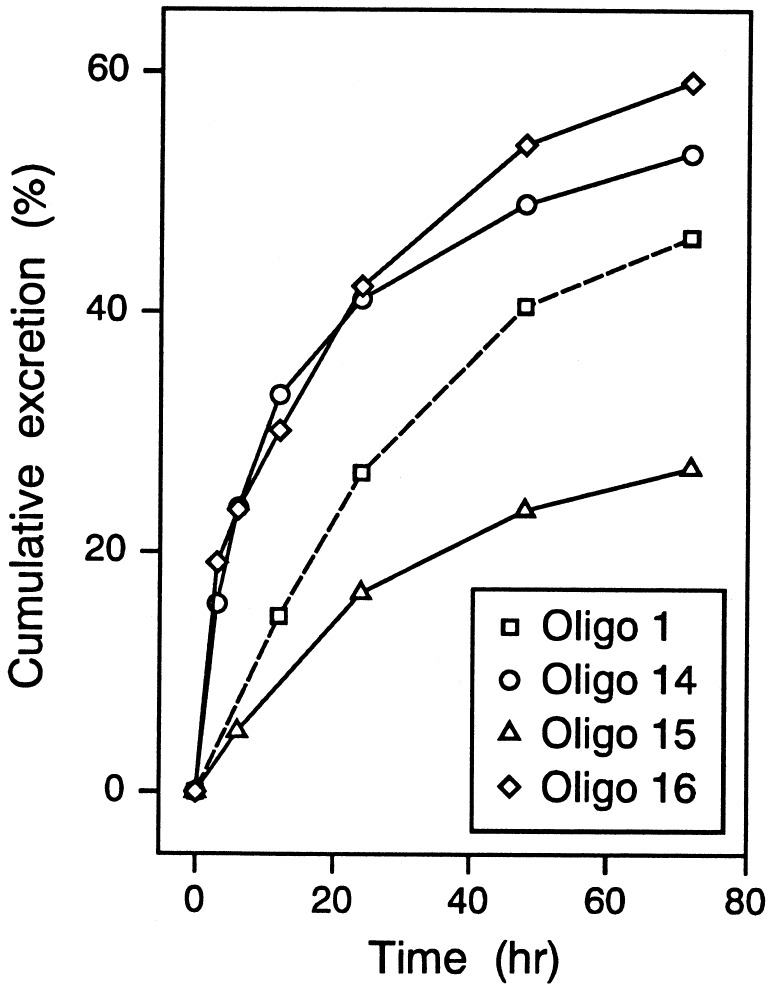
The major elimination route for PS-oligo from the body is through urinary excretion. Oligo 15 was excreted up to 30% in 72 hr, whereas oligos 14 and 16 were excreted in urine more than 50% (Fig. 5). In comparison, the excretion of PS-oligo 1 during the same time period was about 40% (16). These data suggest that elimination of oligos in urine depends on the nature and stability of the degradation products. Oligo 16, which contained degradable 2′-OMePO linkages, and oligo 14, which contained PM linkages, were excreted more than oligo 15, which contained 2′-OMePS linkages.
Figure 5.
Cumulative urinary excretion of oligos 1 and 14–16. (The data from oligo 1 are derived from a previous study, ref. 16.)
Toxicity of Oligos.
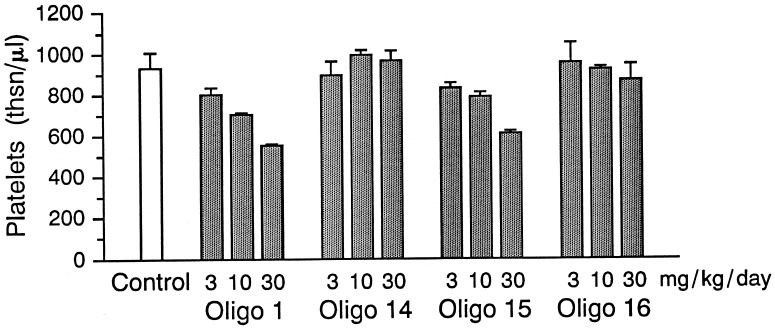
Oligos 14–16 were studied for their safety profile in rats after i.v. administration of 3, 10, and 30 mg/kg daily for 7 days. In general, PS-oligos in mice and rats cause dose-dependent side effects, including thrombocytopenia, elevation of transaminases, splenomegaly, hyperplasia of reticuloendothelial cells, and renal tubular changes (2, 3). Similar side effects were also observed with PS-oligo 1, in a dose-dependent fashion. Oligos 14–16 also produced similar side effects, but at much lower rates. Oligos 14 and 16 caused no significant thrombocytopenia compared with control animals, whereas oligos 1 and 15 showed dose-dependent side effects (Fig. 6). Similarly, no significant changes in levels of alanine amino transferase and aspartate amino transferase were observed with oligos 14 and 16 (data not shown). Histopathology of kidneys showed dose-dependent effects on tubular degeneration with oligos 1 and 15, but the severity of the tubular degeneration was significantly lower with oligos 14 and 16.
Figure 6.
Comparative dose-dependent effect of oligos 1 and 14–16 on thrombocytopenia in rats after i.v. administration daily for 7 days.
DISCUSSION
The potential of antisense oligos as therapeutic agents depends on many parameters. In cell culture, the biological activity of an oligo may depend on its length, modification, nuclease stability, cellular uptake, and intracellular localization, and the target mRNA sequences, whereas in vivo biological activity depends on in vivo stability, pharmacokinetics, tissue disposition, and the safety/toxicity profile. PS-oligos have proven to be very effective antisense oligonucleotides, in both in vitro and in vivo studies, and a number of human clinical trials are underway for various disease indications (2). While studies to establish the therapeutic potential of PS-oligos are ongoing, newer generations of oligos are being synthesized and studied for their antisense properties (2). In our earlier studies, the properties of antisense oligos were studied by designing MBOs containing a segment of PS-oligo and segments of other oligodeoxynucleotide analogs (e.g., methylphosphonate, phosphoramidates, etc.; ref. 19) or oligoribonucleotide (e.g., 2′-O-alkyl) analogs (20). Incorporation of these modified segments at the ends of PS-oligos (oligos 9 and 11) significantly improved their in vivo stability, safety profile, and biological activity as an antisense agent, and the oligos were absorbed after oral administration in rats (2, 5, 15, 17, 21–23).
In the present work, additional modifications of PS-oligos were studied to further understand the issues underlying the oligos’ improved biological activity as antisense agents, in vivo metabolism, and safety profile. These oligos incorporated a segment of modified oligonucleotide in the center. Such a modification was proposed as an attempt to (i) reduce the contiguous length of PS-oligo linkages, thereby reducing the polyanionic-related side effects; (ii) modulate the mitogenic response; (iii) control the rate of in vivo degradation and nature of metabolites, thereby reducing the side effects; and (iv) improve the affinity to RNA and RNase H activity, which should improve antisense properties of PS-oligos. To carry out these studies, the PS-oligo 1 GEM 91, which is complementary to the initiation site of the gag gene of HIV-1, was used as these parameters have previously been studied in detail with this sequence (2, 4, 5, 8, 12, 14–16).
Incorporation of the 2′-OMe segment in the center of PS-oligo 1 produced increases in Tm (7–13°C) and the rate of cleavage of RNA by RNase H, compared with PS-oligo 1. The changes in Tm values and RNase H activation depended on the nature of the internucleotide linkage and the length of 2′-OMe segment (oligos 2–8). Incorporation of PM linkages in the center reduced the Tm (5–6°C) but increased the rate of cleavage of RNA by RNase H. The general increase in the rate of cleavage of RNA may be caused by the cleavage of the RNA substrate at two sites rather than cleavage at one site, which is the case with oligos 1, 9, and 11 (Fig. 1).
Digestion of oligos by snake venom phosphodiesterase confirmed that after degradation of PS-oligo, further degradation of the oligo depended on the nature of the central segments. Further degradation was slowed in oligos containing nuclease-resistant 2′-OMePS or PM linkages, whereas increased degradation was observed in oligos containing 2′-OMePO linkages.
These centrally modified oligos were also studied for their anti-HIV-1 activity in HIV-1 infected cell culture systems. The IC90 of the oligos ranged from <0.1 to 0.99 μM. Oligos 4, 6, and 8 showed greater inhibitory activity than oligo 1. Oligos 14–16, containing eight PM, 2′-OMePS, and 2′-OMePO linkages, respectively, were used to study the pharmacokinetics, in vivo stability, and safety profile. The pharmacokinetics of these three oligos after i.v. administration in rats showed a profile for plasma clearance similar to that for PS-oligo 1, but analysis of the extracted oligo from the plasma by PAGE showed different degradation profiles. In the case of oligo 14, degradation almost ceased, possibly after digestion of the PS-oligo segment. With oligo 15, a continuous ladder of degradation products was obtained, whereas with oligo 16, only eight major degradation products were observed. Disposition of the oligonucleotide in kidney and liver was quite different. Oligo 15, which contains all PS-linkages, had a concentration in kidney similar to that of oligo 1. Oligos 14 and 16 (eight PM and eight 2′-OMePO linkages, respectively) had a higher concentration in kidney at 24 hr or later. Analysis of the extracted material from kidney homogenate by PAGE showed a profile similar to the one observed in plasma. Digestion of oligo 14 almost ceased at the site of PM linkages, whereas oligo 15 slowed but continued to be degraded at the site of 2′-OMePS linkages. In the case of oligo 16, no digestion products were observed beyond 16- to 17-mer.
The disposition of oligos in liver was also different. Oligo 15 was present in higher concentrations than oligos 14 and 16, but all three oligos had lower concentrations at 72 hr than oligo 1 (data not shown). Elimination of the oligos in urine also depended on the nature of the central segment. The percentage of the administered dose eliminated in urine for oligos 14 and 16 was higher than that of oligo 1, and lowest for oligo 15.
In vitro and in vivo studies were carried out to understand the general safety profile of these oligos. In in vitro studies, higher concentrations of centrally modified oligos than of oligo 1 were required to affect complement and coagulation assays, which could be due to a reduction in contiguous length of PS-oligo. Similarly, centrally modified oligos had significantly less of an impact on lymphocyte proliferation than did oligo 1, thereby reducing the mitogenic responses to oligos. Safety studies in vivo after daily i.v. administration in rats showed that oligos 14 and 16 produced significantly less thrombocytopenia and elevation of aspartate amino transferase and alanine amino transferase than did oligos 1 and 15. Similarly, histopathology of kidneys showed reduced renal tubule degeneration in kidney in animals receiving oligos 14 and 16, compared with oligos 1 and 15.
From the results discussed above, it is evident that the antisense activity, pharmacokinetics and in vivo degradation, and safety profile of an oligo can be modulated by combination of appropriate oligo segments and backbone at defined sites, which may lead to better design of novel antisense agents with improved therapeutic effectiveness.
Acknowledgments
We thank Y. Li, B. Chambless, Yufeng Li, and L. High for technical assistance. The safety studies of oligonucleotides were carried out at Frederick Research Center, Frederick, MD.
ABBREVIATIONS
- oligo
oligonucleotide
- PS-oligo
phosphorothioate oligodeoxynucleotide
- aPTT
activated partial thromboplastin time
- 2′-OMePO
2′-O-methylribonucleoside phosphodiester
- 2′-OMePS
2′-O-methylribonucleoside phosphorothioate
- PM
methylphosphonate
References
- 1.Zamecnik P C. In: Antisense Therapeutics. Agrawal S, editor. Totowa, NJ: Humana; 1996. pp. 1–11. [Google Scholar]
- 2.Agrawal S. Trends Biotechnol. 1996;14:376–387. doi: 10.1016/0167-7799(96)10053-6. [DOI] [PubMed] [Google Scholar]
- 3.Sarmiento U M, Perez J R, Becker J M, Narayanan R. Antisense Res Dev. 1994;4:99–107. doi: 10.1089/ard.1994.4.99. [DOI] [PubMed] [Google Scholar]
- 4.Galbraith W M, Hobson W C, Giclas P C, Schechter P J, Agrawal S. Antisense Res Dev. 1994;4:201–206. doi: 10.1089/ard.1994.4.201. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal S, Rustagi P K, Shaw D. Toxicol Lett. 1996;82/83:431–434. doi: 10.1016/0378-4274(95)03573-7. [DOI] [PubMed] [Google Scholar]
- 6.Flexner C, Barditch-Crovo P A, Kornhauser D M, Farzadegan H, Nerhood L J, Chaisson R E, Bell K M, Lorensten K J, Hendrix C W, Petty B G, Lietman P S. Antimicrob Agents Chemother. 1991;35:2544–2550. doi: 10.1128/aac.35.12.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krieg A M, Yi A E, Matson S, Waldschmidt T S, Bishop G A, Teasdale R, Koretzky G A, Klinman D M. Nature (London) 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 8.Padmapriya A P, Tang J Y, Agrawal S. Antisense Res Dev. 1994;4:185–199. doi: 10.1089/ard.1994.4.185. [DOI] [PubMed] [Google Scholar]
- 9.Iyer R P, Egan W, Regan J B, Beaucage S L. J Am Chem Soc. 1990;112:1253–1254. [Google Scholar]
- 10.Agrawal S, Goodchild J. Tetrahedron Lett. 1987;28:3539–3542. [Google Scholar]
- 11.Iyer R P, Yu D, Jiang Z, Agrawal S. Nucleosides Nucleotides. 1995;14:1349–1357. [Google Scholar]
- 12.Agrawal S, Temsamani J, Tang J-Y. Proc Natl Acad Sci USA. 1991;88:7595–7599. doi: 10.1073/pnas.88.17.7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lisziewicz J, Sun D, Klotman M, Agrawal S, Zamecnik P C, Gallo R. Proc Natl Acad Sci USA. 1992;89:11209–11213. doi: 10.1073/pnas.89.23.11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lisziewicz J, Sun D, Weichold F F, Thierry A R, Lusso P, Tang J-Y, Gallo R C, Agrawal S. Proc Natl Acad Sci USA. 1994;91:7942–7946. doi: 10.1073/pnas.91.17.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J, Temsamani J, Iadarola P, Jiang Z, Agrawal S. Biochem Pharmacol. 1996;51:173–182. doi: 10.1016/0006-2952(95)02177-9. [DOI] [PubMed] [Google Scholar]
- 16.Zhang R, Diasio R B, Lu Z, Liu T, Jiang Z, Galbraith W M, Agrawal S. Biochem Pharmacol. 1995;49:929–939. doi: 10.1016/0006-2952(95)00010-w. [DOI] [PubMed] [Google Scholar]
- 17.Zhang R, Lu Z, Liu T, Zhao H, Zhang X, Diasio R, Habus I, Jiang Z, Iyer R P, Yu D, Agrawal S. Biochem Pharmacol. 1995;50:545–556. doi: 10.1016/0006-2952(95)00159-w. [DOI] [PubMed] [Google Scholar]
- 18.Agrawal S, Mayrand S M, Zamecnik P C, Pederson T. Proc Natl Acad Sci USA. 1990;87:1401–1405. doi: 10.1073/pnas.87.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao W-Y, Han F-S, Storm C, Egan W, Cheng Y-C. Mol Pharmacol. 1992;41:223–229. [PubMed] [Google Scholar]
- 20.Metelev V, Lisziewicz J, Agrawal S. Bioorg Med Chem Lett. 1994;4:2929–2934. [Google Scholar]
- 21.Zhang R, Iyer R, Tan W, Yu D, Zhang Z, Lu Z, Zhao H, Agrawal S. J Pharmacol Exp Ther. 1996;278:971–979. [PubMed] [Google Scholar]
- 22.Shaw, D. R., Rustagi, P. K., Kandimalla, E. R., Manning, A. M., Jiang, Z. & Agrawal S. (1997) Biochem. Pharmacol., in press. [DOI] [PubMed]
- 23.Agrawal S, Zhang X, Zhao H, Lu Z, Yan J, Cai H, Diasio R B, Habus I, Jiang Z, Iyer R P, Yu D, Zhang R. Biochem Pharmacol. 1995;50:571–576. doi: 10.1016/0006-2952(95)00160-2. [DOI] [PubMed] [Google Scholar]