Abstract
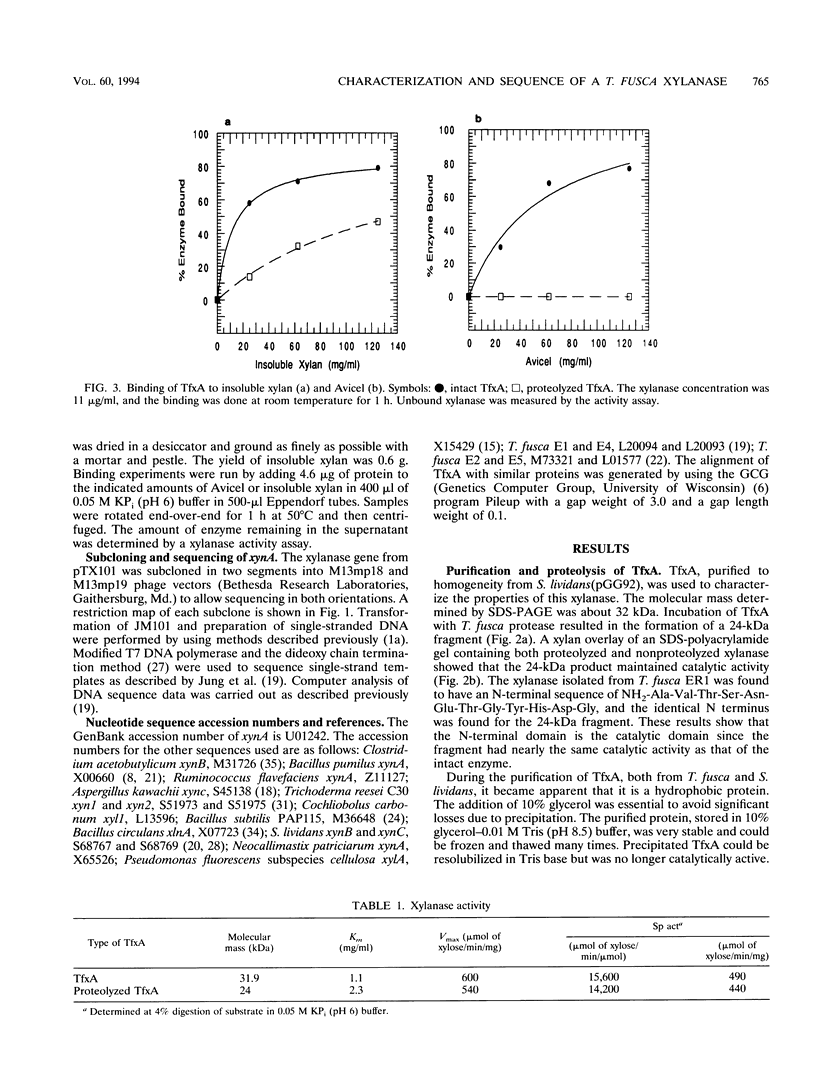
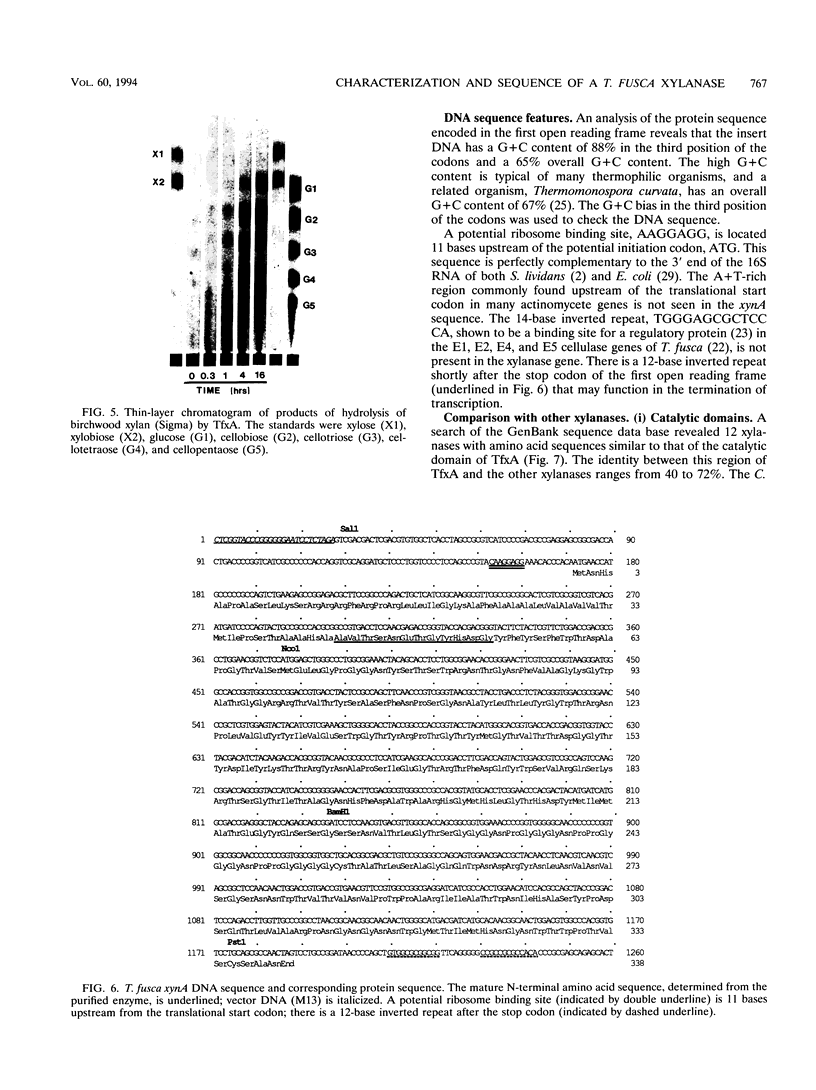
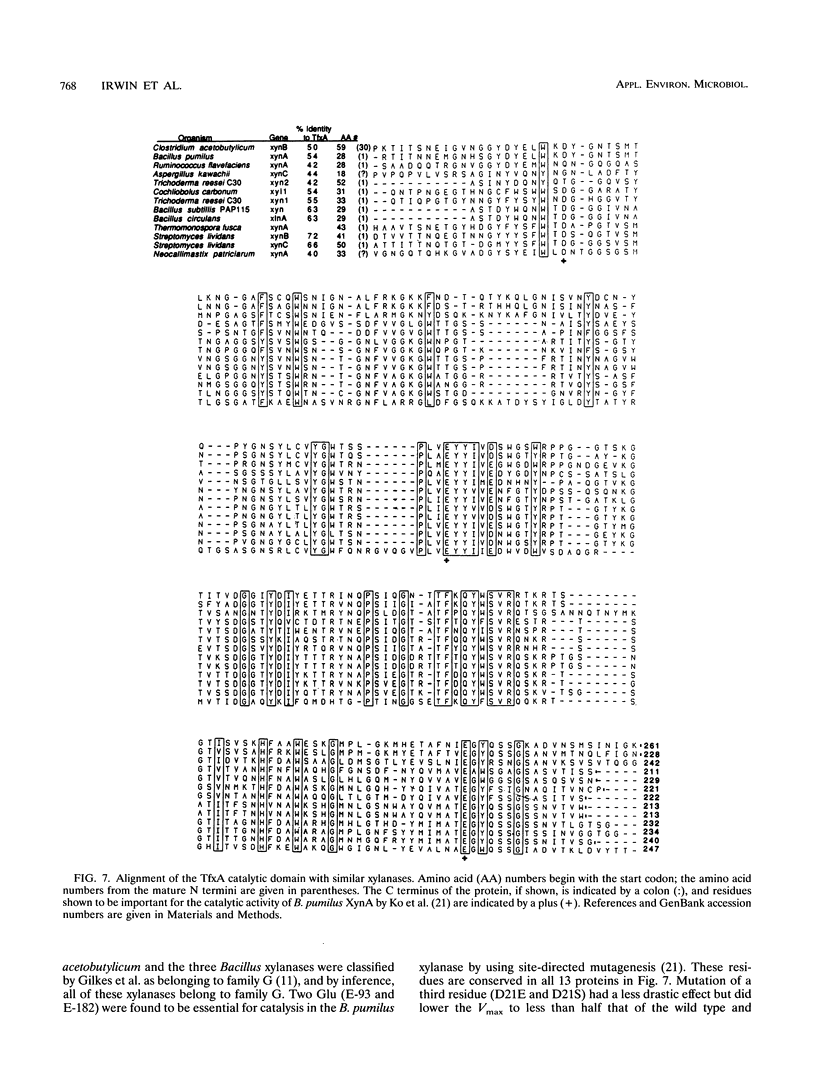
TfxA is a thermostable xylanase produced by the thermophilic soil bacterium Thermomonospora fusca. The enzyme was purified to homogeneity from the culture supernatant of Streptomyces lividans transformed by plasmid pGG92, which carries the gene for TfxA, xynA. The molecular mass of TfxA by sodium dodecyl sulfate-polyacrylamide gel electrophoresis is 32 kDa. TfxA is extremely stable, retaining 96% of its activity after 18 h at 75 degrees C. It has a broad pH optimum around pH 7 and retains 80% of its maximum activity between pH 5 and 9. The native enzyme binds strongly to both cellulose and insoluble xylan even though it has no activity on cellulose. Treatment of TfxA with a T. fusca protease produced a 24-kDa catalytically active fragment that had the same N-terminal sequence as TfxA. The fragment does not bind to cellulose and binds weakly to xylan. The Vmax values for TfxA and the fragment are 600 and 540 mumol/min/mg, respectively, while the Kms are 1.1 and 2.3 mg of xylan per ml, respectively. The DNA sequence of the xynA gene was determined, and it contains an open reading frame that codes for a 42-amino-acid (42-aa) actinomycete signal peptide followed by the 32-kDa mature protein. There is a 21-aa Gly-Pro-rich region that separates the catalytic domain from an 86-aa C-terminal binding domain. The amino acid sequence of the catalytic domain of TfxA has from 40 to 72% identity with the sequence of 12 other xylanases from seven different organisms and belongs to family G.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L. M., Durrant A. J., Hall J., Hazlewood G. P., Gilbert H. J. Spatial separation of protein domains is not necessary for catalytic activity or substrate binding in a xylanase. Biochem J. 1990 Jul 1;269(1):261–264. doi: 10.1042/bj2690261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghangas G. S., Hu Y. J., Wilson D. B. Cloning of a Thermomonospora fusca xylanase gene and its expression in Escherichia coli and Streptomyces lividans. J Bacteriol. 1989 Jun;171(6):2963–2969. doi: 10.1128/jb.171.6.2963-2969.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grépinet O., Chebrou M. C., Béguin P. Nucleotide sequence and deletion analysis of the xylanase gene (xynZ) of Clostridium thermocellum. J Bacteriol. 1988 Oct;170(10):4582–4588. doi: 10.1128/jb.170.10.4582-4588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusek T. W., Kinsella J. E. Purification and characterization of the heat-stable serine proteinase from Thermomonospora fusca YX. Biochem J. 1987 Sep 1;246(2):511–517. doi: 10.1042/bj2460511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Hazlewood G. P., Huskisson N. S., Durrant A. J., Gilbert H. J. Conserved serine-rich sequences in xylanase and cellulase from Pseudomonas fluorescens subspecies cellulosa: internal signal sequence and unusual protein processing. Mol Microbiol. 1989 Sep;3(9):1211–1219. doi: 10.1111/j.1365-2958.1989.tb00271.x. [DOI] [PubMed] [Google Scholar]
- Hägerdal B. G., Ferchak J. D., Pye E. K. Cellulolytic Enzyme System of Thermoactinomyces sp. Grown on Microcrystalline Cellulose. Appl Environ Microbiol. 1978 Oct;36(4):606–612. doi: 10.1128/aem.36.4.606-612.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Iwashita K., Iwano K. Cloning and sequencing of the xynC gene encoding acid xylanase of Aspergillus kawachii. Biosci Biotechnol Biochem. 1992 Aug;56(8):1338–1340. doi: 10.1271/bbb.56.1338. [DOI] [PubMed] [Google Scholar]
- Jung E. D., Lao G., Irwin D., Barr B. K., Benjamin A., Wilson D. B. DNA sequences and expression in Streptomyces lividans of an exoglucanase gene and an endoglucanase gene from Thermomonospora fusca. Appl Environ Microbiol. 1993 Sep;59(9):3032–3043. doi: 10.1128/aem.59.9.3032-3043.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko E. P., Akatsuka H., Moriyama H., Shinmyo A., Hata Y., Katsube Y., Urabe I., Okada H. Site-directed mutagenesis at aspartate and glutamate residues of xylanase from Bacillus pumilus. Biochem J. 1992 Nov 15;288(Pt 1):117–121. doi: 10.1042/bj2880117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao G., Ghangas G. S., Jung E. D., Wilson D. B. DNA sequences of three beta-1,4-endoglucanase genes from Thermomonospora fusca. J Bacteriol. 1991 Jun;173(11):3397–3407. doi: 10.1128/jb.173.11.3397-3407.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E. S., Wilson D. B. Identification of a celE-binding protein and its potential role in induction of the celE gene in Thermomonospora fusca. J Bacteriol. 1988 Sep;170(9):3843–3846. doi: 10.1128/jb.170.9.3843-3846.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrícek M., Stajner K., Tichý P. Cloning of a thermostable alpha-amylase gene from Thermomonospora curvata and its expression in Streptomyces lividans. J Gen Microbiol. 1989 Dec;135(12):3303–3309. doi: 10.1099/00221287-135-12-3303. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareck F., Roy C., Yaguchi M., Morosoli R., Kluepfel D. Sequences of three genes specifying xylanases in Streptomyces lividans. Gene. 1991 Oct 30;107(1):75–82. doi: 10.1016/0378-1119(91)90299-q. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. A. Molecular biology of xylan degradation. FEMS Microbiol Rev. 1993 Jan;10(1-2):65–82. doi: 10.1111/j.1574-6968.1993.tb05864.x. [DOI] [PubMed] [Google Scholar]
- Törrönen A., Mach R. L., Messner R., Gonzalez R., Kalkkinen N., Harkki A., Kubicek C. P. The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Biotechnology (N Y) 1992 Nov;10(11):1461–1465. doi: 10.1038/nbt1192-1461. [DOI] [PubMed] [Google Scholar]
- Yang R. C., MacKenzie C. R., Narang S. A. Nucleotide sequence of a Bacillus circulans xylanase gene. Nucleic Acids Res. 1988 Jul 25;16(14B):7187–7187. doi: 10.1093/nar/16.14.7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappe H., Jones W. A., Woods D. R. Nucleotide sequence of a Clostridium acetobutylicum P262 xylanase gene (xynB). Nucleic Acids Res. 1990 Apr 25;18(8):2179–2179. doi: 10.1093/nar/18.8.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]





