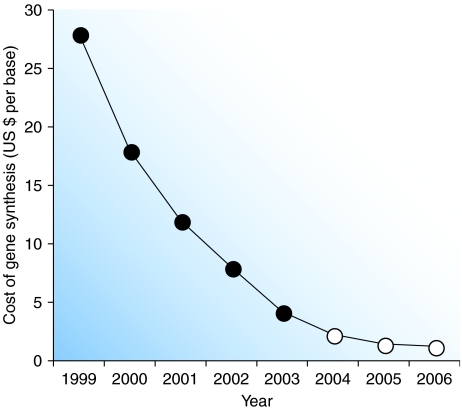
A paradigm shift is underway within the methodology of heterologous protein expression. Specifically, researchers are moving away from conventional techniques of cloning genes from cDNA libraries and moving toward the rational design and de novo synthesis of entire protein-coding sequences from pre-annealed oligonucleotides (Libertini and Di Donato, 1992; Gustafsson et al, 2004). It was the invention of polymerase chain reaction (PCR) that allowed efficient construction of synthetic genes. Since then, the steadily increasing accuracy and decreasing cost of oligonucleotide synthesis (now as low as $0.10 per base; Carlson, 2003; Carr et al, 2004; Kong et al, 2007, see Figure 1) has created a research environment in which gene synthesis offers three main advantages over molecular cloning: cost efficiency, scope and flexibility of redesign (Libertini and Di Donato, 1992). As a result, the emerging field of synthetic biology is highly motivated to improve this approach, as it seeks to expand the sophistication of human-engineered genetic architectures, leading ultimately to the synthesis of entire genomes (Yount et al, 2000; Smith et al, 2003).
Figure 1.
The cost, per base, of commercial oligonucleotide assembly from 1999 to 2006. The price of gene synthesis has decreased almost 30-fold in the past 7 years (data for years 1999–2003 are taken from Carlson, 2003). Data for years 2004–2006 reflect the lowest price found in advertisements placed within Science magazine).
Current research into synthetic gene construction has focused largely on improving PCR-based methods. Areas under active investigation include the following: increasing the accuracy of gene products by reducing errors in oligonucleotide construction and PCR synthesis/amplification (Ciccarelli et al, 1991; Young and Dong, 2004), reducing the relatively high cost of post-synthesis sequencing (Young and Dong, 2004), increasing the length of genes that can be synthesized (Kodumal et al, 2004), developing microchip-based technology and/or microfluidic devices that allow for the simultaneous assembly of multiple genes (Tian et al, 2004; Zhou et al, 2004; Kong et al, 2007), and automating the whole pipeline from gene design to synthetic gene screening (Cox et al, 2007). All frontiers show signs of rapid improvement (e.g., Xiong et al, 2004; Engels, 2005; Wu et al, 2006a), therefore the current challenges for gene synthesis are essentially optimizations of existing concepts.
In stark contrast, it appears that we have much left to learn when it comes to the conceptual design of gene sequences. A significant fraction of the biologically and commercially important genes that have been redesigned report little or no success in increasing protein expression (e.g., see Alexeyev and Winkler, 1999; Flick et al, 2004; Wu et al, 2004; Hillier et al, 2005). More surprising, some of these ‘improvements' have led to a direct and observable reduction in protein production (Griswold et al, 2003). Even those that do report increased protein yield require careful scrutiny, because many have not controlled for altered mRNA levels in their system (e.g., Deng, 1997; Alexeyev and Winkler, 1999; Feng et al, 2000; Humphreys et al, 2000; Nalezkova et al, 2005). Thus, although excellent progress in the practice of gene synthesis enables experimental implementation of the technique, the scientific community remains far from a complete understanding of what constitutes a rational design strategy for a protein-coding gene. Instead, the very concept of a ‘translationally optimal codon' has grown to incorporate dimensions of translational speed, translational accuracy and sustainability of yield that could vary from one experiment to another. Meanwhile, we have learned that a codon's position within a coding sequence, its ‘neighborhood' of other codons, its structural role within the mRNA sequence and the nature of the genomic system in which it is to be expressed can all influence the effects of ‘synonymous' codon choices. Given that we can physically construct any gene, what rules define the appropriate sequence to manufacture? Here, we examine current progress and emerging challenges in both theory and practice, showing how this topic exemplifies the interdisciplinary challenges of 21st century biology.
Why redesign the coding sequence?
Modern expression vectors have undergone extensive manipulation to maximize mRNA transcription. Yet a relatively weak correlation can exist between expression levels of mRNA and those of translated protein products (e.g., Futcher et al, 1999; Nie et al, 2006). Thus, it is now widely understood that persistent poor expression of protein product can result from problems occurring at a post-transcriptional stage, especially at the point of translation (Kurland and Gallant, 1996; Gustafsson et al, 2004). The issue here is that the ‘digital' portrayal of translation found in biology textbooks oversimplifies a bio-mechanical process in which different populations of tRNAs essentially compete to translate an appropriate codon of mRNA within the context of a ribosome (e.g.,Rodnina et al, 2005). Different organisms can vary enough in their relative contents of isoaccepting tNRAs to change the dynamics of this competition, such that different choices from a suite of synonymous codons can influence the speed and accuracy of translation. For this reason it can be useful to redesign a protein-coding sequence to suit its new context when moving it between genomes.
What should we build? The theory of synthetic gene design
The most direct method to find an optimal encoding for heterologous expression would be to comprehensively screen all possible alternative sequences. This is however impractical for sequences of any appreciable length because of the near-infinite encoding possibilities: approximately 3.7 × 1021 different nucleic acid sequences could encode a single peptide comprising 150 amino acids, thus top-down screening procedures must be guided by bottom-up gene design.
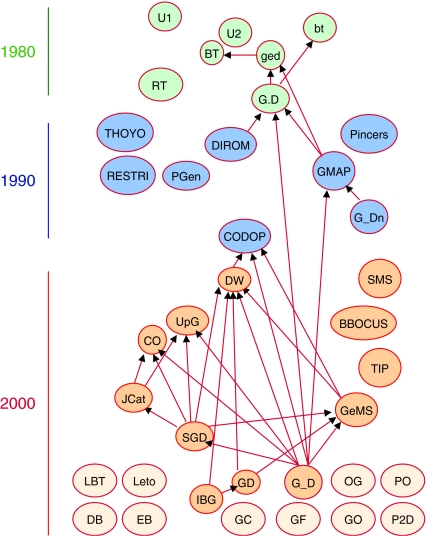
To this end, a wealth of software has been developed to help bench scientists achieve reverse translation (Arentzen and Ripka, 1984; Mount and Conrad, 1984; Danckaert et al, 1987; Pesole et al, 1988; Presnell and Benner, 1988; Weiner and Scheraga, 1989; Bains, 1990; Tamura et al, 1991; Libertini and Di Donato, 1992; Makarova et al, 1992; Nash, 1993; Raghava and Sahni, 1994; Withers-Martinez et al, 1999; Hoover and Lubkowski, 2002; Fuglsang, 2003; Gao et al, 2004; Grote et al, 2005; Jayaraj et al, 2005; Richardson et al, 2006; Villalobos et al, 2006; Wu et al, 2006b; Puigbo et al, 2007). Broadly speaking, this software can be divided into two categories according to algorithmic purpose: one seeking gene designs that facilitate empirical sequence manipulations, the other seeking designs that translate well into protein products. Perhaps the two most salient features of this software are the diversity of opinion as to what rules will optimize translation and a general lack of awareness by each software solution that numerous competitors exist (Figure 2).
Figure 2.
The gene design software shown as a network of citations by date of publication. Each of the 37 nodes represents a specific software application for gene design; arrows indicate acknowledgements (citations) of pre-existing, published software. Appropriate web development would alleviate this patchy awareness of competing efforts, and could eliminate inefficient and needless duplication of effort. Abbreviations: BBOCUS: BackTranslation Based On Codon Usage Strategy; BT: BACKTR (Pesole et al, 1988); bt: backtrans (Mount and Conrad, 1984); CO: Codon Optimizer (Fuglsang, 2003); CODOP: CODon OPtimization (Withers-Martinez et al, 1999); DB: DNA Builder (Pacific Northwest National Laboratory); DIROM (Makarova et al, 1992); DW: DNAWorks (Hoover and Lubkowski, 2002); EB: EasyBack (University of Catania, Italy); G.D: Gene.Design (Weiner and Scheraga, 1989) G_D: Gene Designer (Villalobos et al, 2006); G_Dn, GeneDn (Ju et al, 1998); GC: Gene Composer (by Emerald Biosystems); GD: GeneDesign (Richardson et al, 2006); ged: gene design (Presnell and Benner, 1988); GeMS: Gene Morphing System (Jayaraj et al, 2005); GF: The Gene Forge (by AptaGen LLC); GMAP (Raghava and Sahni, 1994); GO: GeneOptimizer (by GeneArt, Germany); IBG: IBG GeneDesigner (Vogelbacher et al, 2006); JCat: Java Codon Adaptation Tool (Grote et al, 2005); LBT: Locally Sensitive BackTranslation; Leto (by Entelechon Inc.) OG: OptGene (by Ocimum Biosolutions); P2D: Protein2DNA (by DNA 2.0 Inc); PGen: PrimerGen (Nash, 1993); PINCERS (Tamura et al, 1991); PO: Primo Optimum (by Chang Bioscience); RESTRI (Libertini and Di Donato, 1992); RT: Reverse Translate (Danckaert et al, 1987); SGD: Synthetic Gene Designer (Wu et al, 2006a, 2006b); SMS: Sequence Manipulation Suite (Stothard, 2000); THOYO (Bains, 1990); TIP: Traducción Inversa de Proteínas/Protein Backtranslation (Moreira and Maass, 2004); U1: unnamed1 (Arentzen and Ripka, 1984); U2: unnamed2 (Danckaert et al, 1987); UpG: UpGene (Gao et al, 2004). Applications shown in dashed lines indicate software that has never appeared in peer reviewed scientific literature.
So where should we seek guidance as to the rules of optimal encoding? Up until now, the overwhelming majority of synthetic genes that have been reported in peer reviewed literature represent unique attempts to re-engineer different genes. As a result, their collection into a single database (Wu et al, 2007) currently presents a ‘broad and shallow' scatter of isolated points in sequence space. A shift in research emphasis is needed to refocus efforts toward a ‘narrow and deep' systematic comparison of different recoding strategies for a few genes. Meanwhile, the nearest we have to such a dataset are the numerous gene variants produced by evolution. It has been long recognized that codon usage frequency appears to be unequal for most synonymous codons within naturally occurring genomes (Grantham et al, 1980). Much of this bias is a passive reflection of the mutation biases at work in a genome (Sharp et al, 1993; Knight et al, 2001), however it can be tricky to ascertain which features of which sequences have been shaped by natural selection. Not only do precise predictions from evolutionary theory rely on parameters that we may never know with certainty, but the noise to signal ratio implicit within any ‘naturally optimized' sequence can confound the most careful analyses.
Where to next? Specific objectives for future progress
Although the major unknowns of synthetic gene technology are mostly those of design theory, the current problem is an excess, and not deficit, of ideas. Major progress thus seems poised to occur when empirical studies start to compare these ideas systematically.
An important step would be to standardize experimental protocols and reports so that the emerging patchwork of results can be examined as a coherent whole. Specifically, experiments must standardize their measurement of mRNA expression levels for the target genes (as a baseline for interpreting protein yields), and measure protein production in absolute rather than relative terms (e.g., mg/l or percentage of total protein rather than ‘n-fold increase/decrease') if they are to be compared.
A further step would be to identify one or more standardized (model) experimental systems for use by any and all research groups that are willing to share information. An ideal expression system would not be pre-engineered in any way that could confound interpretation of results (e.g., by containing enriched tRNA pools), it would employ a protein product that is amenable to clear, quantitative assay and could include an internal control (such as a dual reporter system in which only one gene has been redesigned) to add further confidence to measurements of protein yields.
The idea of standardization extends into the philosophy of bioinformatics software that predicts gene design. Current software typically requires a combination of logically independent gene optimizing steps as a mandatory, pre-packaged whole. This renders the comparison of results difficult and suggests the need for secondary design algorithms designed to isolate specific gene features (e.g., changing codons while maintaining overall GC content, or varying GC content while maintaining RNA structural motifs).
It is noteworthy that the underlying nature of all gene design software is similar and simple: a user must input a protein sequence and a genetic code. The protein sequence is then reverse translated into a nucleotide sequence using one or more algorithms, and the resulting nucleotide sequence is returned to the user. Independent applications must duplicate at least this much functionality. A promising direction of future software development in this field would be an emphasis on integration into a unified, distributed, modular web service for synthetic gene design. Specifically, programmers could take advantage of purpose-built web technologies, such as XML (a data sharing language) and SOAP (a language for wrapping independent applications), to facilitate interconnection of disparate, pre-existing software. New algorithms could be added as pathways through which a synthetic gene might travel en route to final design. This would provide users with a common interface through which they could choose the specific algorithm(s) to use at each step of synthetic gene design. Far from restricting the diversity of independent ideas for design services offered by different groups (on different web-servers), this type of coordination through a common interface would focus attention where it belongs: on the overlapping (and sometimes directly competing) concepts of how to design genes for optimal expression.
Critical assessment
Our suggested shift in research emphasis toward standardized protocols and integration of competing design strategies would create a foundation with potential that exceeds the capabilities of any one group or traditional collaboration. How then can the diverse interests of those interested in synthetic gene design be harnessed into a common framework for progress?
We advocate the introduction of a competitive model, similar to the CASP approach that has been used within the protein folding research community (Moult, 2005). Given a standardized experimental protocol, it would be possible to pick genes of major research interest that are proving problematic for heterologous expression. For example, a recent study of Plasmodium falciparum, the causative agent of the most deadly form of malaria, reported that ‘12 targets, which did not express in Escherichia coli from the native gene sequence were codon-optimized through whole gene synthesis, resulting in the expression of three of these proteins' (Mehlin et al, 2006). Presumably, malaria researchers would be motivated to call for theoretical predictions of redesign that could help their situation. Theorists and software developers should in turn be motivated to demonstrate their algorithms' worth as the marketplace of redesign ideas becomes increasingly saturated, and those who research the optimization of gene assembly protocols (regardless of sequence content) would be motivated to absorb a significant fraction of the effort required for synthesizing these predictions. The net result would be a distributed (community wide) version of the direct screening approach favored by early pioneers of synthetic gene technology (Stemmer et al, 1993; Humphreys et al, 2000), in which each segment of the community directly benefits from a united focus. If all designs were deposited within the SGDB (Synthetic Gene Database) (Wu et al, 2007), then this could quickly transform the knowledge base for synthetic gene technology. Fortunately, recent advancement in multiplex gene synthesis technology has implied the feasibility of simultaneous synthesis of thousands of genes for large-scale experimental tests (Tian et al, 2004; Zhou et al, 2004; Cox et al, 2007; Kong et al, 2007), so the potential for large-scale comparison of predictions may be nearer than we think.
This is an ambitious vision, but the motivation is strong. Current synthetic gene technology offers the potential to become a foundational tool of systems biology. However, until we know how to optimize coding sequences, we cannot construct a single synthetic gene with confidence, let alone produce a whole synthetic genome.
References
- Alexeyev MF, Winkler HH (1999) Gene synthesis, bacterial expression and purification of the Rickettsia prowazekii ATP/ADP translocase. Biochim Biophys Acta 1419: 299–306 [DOI] [PubMed] [Google Scholar]
- Arentzen R, Ripka WC (1984) Introduction of restriction enzyme sites in protein-coding DNA sequences by site-specific mutagenesis not affecting the amino acid sequence: a computer program. Nucleic Acids Res 12: 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains W (1990) A program to optimize DNA sequences for protein expression. Comput Appl Biosci 6: 399–400 [DOI] [PubMed] [Google Scholar]
- Carlson R (2003) The pace and proliferation of biological technologies. Biosecur Bioterror 1: 203–214 [DOI] [PubMed] [Google Scholar]
- Carr PA, Park JS, Lee YJ, Yu T, Zhang S, Jacobson JM (2004) Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res 32: e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli RB, Gunyuzlu P, Huang J, Scott C, Oakes FT (1991) Construction of synthetic genes using PCR after automated DNA synthesis of their entire top and bottom strands. Nucleic Acids Res 19: 6007–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JC, Lape J, Sayed MA, Hellinga HW (2007) Protein fabrication automation. Protein Sci 16: 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckaert A, Mugnier C, Dessen P, Cohen-Solal M (1987) A computer program for the design of optimal synthetic oligonucleotide probes for protein coding genes. Comput Appl Biosci 3: 303–307 [DOI] [PubMed] [Google Scholar]
- Deng T (1997) Bacterial expression and purification of biologically active mouse c-Fos proteins by selective codon optimization. FEBS Lett 409: 269–272 [DOI] [PubMed] [Google Scholar]
- Engels JW (2005) Gene synthesis on microchips. Angew Chem Int Ed Engl 44: 7166–7169 [DOI] [PubMed] [Google Scholar]
- Feng L, Chan WW, Roderick SL, Cohen DE (2000) High-level expression and mutagenesis of recombinant human phosphatidylcholine transfer protein using a synthetic gene: evidence for a C-terminal membrane binding domain. Biochemistry 39: 15399–15409 [DOI] [PubMed] [Google Scholar]
- Flick K, Ahuja S, Chene A, Bejarano MT, Chen Q (2004) Optimized expression of Plasmodium falciparum erythrocyte membrane protein 1 domains in Escherichia coli. Malar J 3: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang A (2003) Codon optimizer: a freeware tool for codon optimization. Protein Expr Purif 31: 247–249 [DOI] [PubMed] [Google Scholar]
- Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI (1999) A sampling of the yeast proteome. Mol Cell Biol 19: 7357–7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Rzewski A, Sun H, Robbins PD, Gambotto A (2004) UpGene: application of a web-based DNA codon optimization algorithm. Biotechnol Prog 20: 443–448 [DOI] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Gouy M (1980) Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res 8: 1893–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold KE, Mahmood NA, Iverson BL, Georgiou G (2003) Effects of codon usage versus putative 5′-mRNA structure on the expression of Fusarium solani cutinase in the Escherichia coli cytoplasm. Protein Expr Purif 27: 134–142 [DOI] [PubMed] [Google Scholar]
- Grote A, Hiller K, Scheer M, Munch R, Nortemann B, Hempel DC, Jahn D (2005) JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 33: W526–W531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J (2004) Codon bias and heterologous protein expression. Trends Biotechnol 22: 346–353 [DOI] [PubMed] [Google Scholar]
- Hillier CJ, Ware LA, Barbosa A, Angov E, Lyon JA, Heppner DG, Lanar DE (2005) Process development and analysis of liver-stage antigen 1, a preerythrocyte-stage protein-based vaccine for Plasmodium falciparum. Infect Immun 73: 2109–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover DM, Lubkowski J (2002) DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res 30: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys DP, Sehdev M, Chapman AP, Ganesh R, Smith BJ, King LM, Glover DJ, Reeks DG, Stephens PE (2000) High-level periplasmic expression in Escherichia coli using a eukaryotic signal peptide: importance of codon usage at the 5′ end of the coding sequence. Protein Expr Purif 20: 252–264 [DOI] [PubMed] [Google Scholar]
- Jayaraj S, Reid R, Santi DV (2005) GeMS: an advanced software package for designing synthetic genes. Nucleic Acids Res 33: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju LW, Xing LH, Hong PW, Jin WJ (1998) GeneDn: for high-level expression design of heterologous genes in a prokaryotic system. Bioinformatics 14: 884–885 [DOI] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF (2001) A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol 2 RESEARCH0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV (2004) Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc Natl Acad Sci USA 101: 15573–15578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong DS, Carr PA, Chen L, Zhang S, Jacobson JM (2007) Parallel gene synthesis in a microfluidic device. Nucleic Acids Res 35: e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C, Gallant J (1996) Errors of heterologous protein expression. Curr Opin Biotechnol 7: 489–493 [DOI] [PubMed] [Google Scholar]
- Libertini G, Di Donato A (1992) Computer-aided gene design. Protein Eng 5: 821–825 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Mazin AV, Wolf YI, Soloviev VV (1992) DIROM: an experimental design interactive system for directed mutagenesis and nucleic acids engineering. Comput Appl Biosci 8: 425–431 [DOI] [PubMed] [Google Scholar]
- Mehlin C, Boni E, Buckner FS, Engel L, Feist T, Gelb MH, Haji L, Kim D, Liu C, Mueller N, Myler PJ, Reddy JT, Sampson JN, Subramanian E, Van Voorhis WC, Worthey E, Zucker F, Hol WG (2006) Heterologous expression of proteins from Plasmodium falciparum: results from 1000 genes. Mol Biochem Parasitol 148: 144–160 [DOI] [PubMed] [Google Scholar]
- Moreira A, Maass A (2004) TIP: protein backtranslation aided by genetic algorithms. Bioinformatics 20: 2148–2149 [DOI] [PubMed] [Google Scholar]
- Moult J (2005) A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol 15: 285–289 [DOI] [PubMed] [Google Scholar]
- Mount DW, Conrad B (1984) Microcomputer programs for back translation of protein to DNA sequences and analysis of ambiguous DNA sequences. Nucleic Acids Res 12: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalezkova M, de Groot A, Graf M, Gans P, Blanchard L (2005) Overexpression and purification of Pyrococcus abyssi phosphopantetheine adenylyltransferase from an optimized synthetic gene for NMR studies. Protein Expr Purif 39: 296–306 [DOI] [PubMed] [Google Scholar]
- Nash JH (1993) A computer program to calculate and design oligonucleotide primers from amino acid sequences. Comput Appl Biosci 9: 469–471 [DOI] [PubMed] [Google Scholar]
- Nie L, Wu G, Zhang W (2006) Correlation between mRNA and protein abundance in Desulfovibrio vulgaris: a multiple regression to identify sources of variations. Biochem Biophys Res Commun 339: 603–610 [DOI] [PubMed] [Google Scholar]
- Pesole G, Attimonelli M, Liuni S (1988) A backtranslation method based on codon usage strategy. Nucleic Acids Res 16: 1715–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnell SR, Benner SA (1988) The design of synthetic genes. Nucleic Acids Res 16: 1693–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P, Guzman E, Romeu A, Garcia-Vallve S (2007) OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res 35: W126-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghava GP, Sahni G (1994) GMAP: a multi-purpose computer program to aid synthetic gene design, cassette mutagenesis and the introduction of potential restriction sites into DNA sequences. Biotechniques 16: 1116–1123 [PubMed] [Google Scholar]
- Richardson SM, Wheelan SJ, Yarrington RM, Boeke JD (2006) GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res 16: 550–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Gromadski KB, Kothe U, Wieden HJ (2005) Recognition and selection of tRNA in translation. FEBS Lett 579: 938–942 [DOI] [PubMed] [Google Scholar]
- Sharp PM, Stenico M, Peden JF, Lloyd AT (1993) Codon usage: mutational bias, translational selection, or both? Biochem Soc Trans 21: 835–841 [DOI] [PubMed] [Google Scholar]
- Smith HO, Hutchison CA III, Pfannkoch C, Venter JC (2003) Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci USA 100: 15440–15445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer WP, Morris SK, Kautzer CR, Wilson BS (1993) Increased antibody expression from Escherichia coli through wobble-base library mutagenesis by enzymatic inverse PCR. Gene 123: 1–7 [DOI] [PubMed] [Google Scholar]
- Stothard P (2000) The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28: 1102, 1104 [DOI] [PubMed] [Google Scholar]
- Tamura T, Holbrook SR, Kim SH (1991) A Macintosh computer program for designing DNA sequences that code for specific peptides and proteins. Biotechniques 10: 782–784 [PubMed] [Google Scholar]
- Tian J, Gong H, Sheng N, Zhou X, Gulari E, Gao X, Church G (2004) Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S (2006) Gene designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics 7: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelbacher R, Angulo D, Koide S (2006) Sequence optimization for synthetic genes using a genetic algorithm. Proceedings of the Midwest Software Engineering Conference/DePaul CTI Research Symposium
- Weiner MP, Scheraga HA (1989) A set of Macintosh computer programs for the design and analysis of synthetic genes. Comput Appl Biosci 5: 191–198 [DOI] [PubMed] [Google Scholar]
- Withers-Martinez C, Carpenter EP, Hackett F, Ely B, Sajid M, Grainger M, Blackman MJ (1999) PCR-based gene synthesis as an efficient approach for expression of the A+T-rich malaria genome. Protein Eng 12: 1113–1120 [DOI] [PubMed] [Google Scholar]
- Wu G, Wolf JB, Ibrahim AF, Vadasz S, Gunasinghe M, Freeland SJ (2006a) Simplified gene synthesis: a one-step approach to PCR-based gene construction. J Biotechnol 124: 496–503 [DOI] [PubMed] [Google Scholar]
- Wu G, Bashir-Bello N, Freeland SJ (2006b) The Synthetic Gene Designer: a flexible web platform to explore sequence manipulation for heterologous expression. Protein Expr Purif 47: 441–445 [DOI] [PubMed] [Google Scholar]
- Wu G, Zheng Y, Qureshi I, Zin HT, Beck T, Bulka B, Freeland SJ (2007) SGDB: a database of synthetic genes redesigned for optimizing protein overexpression. Nucleic Acids Res 35: D76–D79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Jornvall H, Berndt KD, Oppermann U (2004) Codon optimization reveals critical factors for high level expression of two rare codon genes in Escherichia coli: RNA stability and secondary structure but not tRNA abundance. Biochem Biophys Res Commun 313: 89–96 [DOI] [PubMed] [Google Scholar]
- Xiong AS, Yao QH, Peng RH, Li X, Fan HQ, Cheng ZM, Li Y (2004) A simple, rapid, high-fidelity and cost-effective PCR-based two-step DNA synthesis method for long gene sequences. Nucleic Acids Res 32: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Dong Q (2004) Two-step total gene synthesis method. Nucleic Acids Res 32: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B, Curtis KM, Baric RS (2000) Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J Virol 74: 10600–10611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Cai S, Hong A, You Q, Yu P, Sheng N, Srivannavit O, Muranjan S, Rouillard JM, Xia Y, Zhang X, Xiang Q, Ganesh R, Zhu Q, Matejko A, Gulari E, Gao X (2004) Microfluidic PicoArray synthesis of oligodeoxynucleotides and simultaneous assembling of multiple DNA sequences. Nucleic Acids Res 32: 5409–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]