Abstract
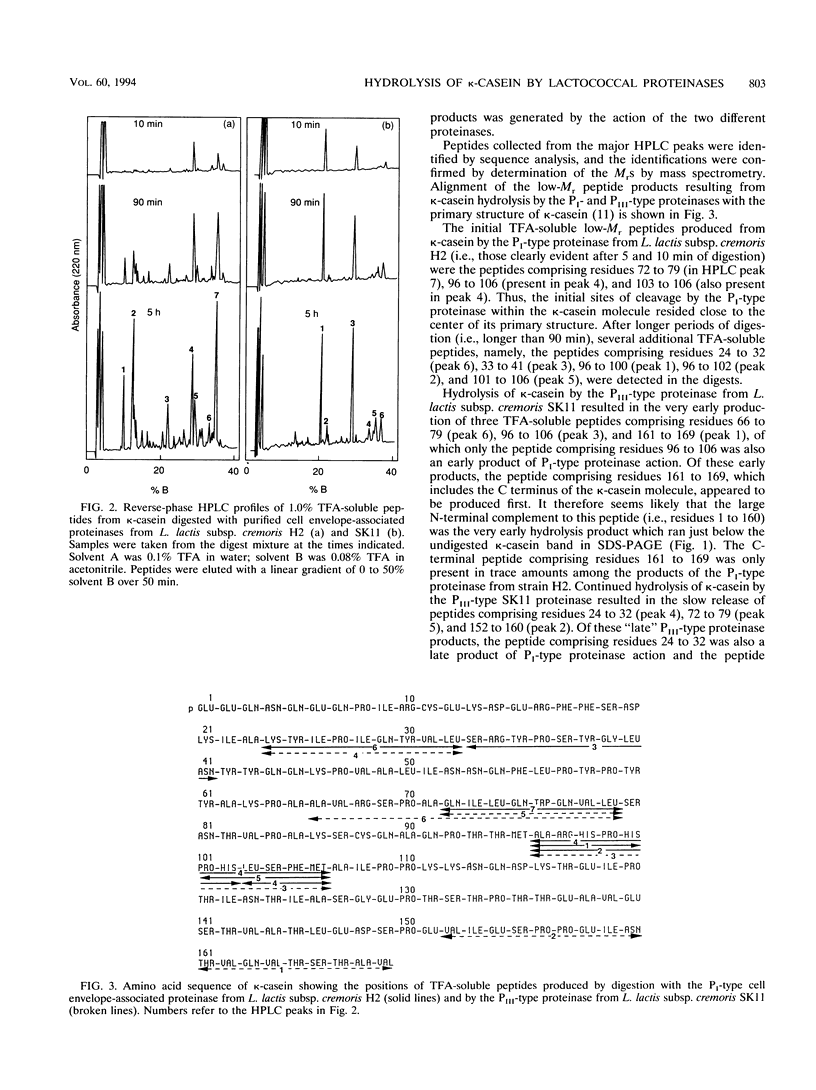
The cell envelope-associated proteinases from Lactococcus lactis subsp. cremoris H2 (a PI-type proteinase-producing strain) and SK11 (a PIII-type proteinase-producing strain) both actively hydrolyze the kappa-casein component of bovine milk but with significant differences in the specificity of peptide bond hydrolysis. The peptide bonds Ala-23-Lys-24, Leu-32-Ser-33, Ala-71-Gln-72, Leu-79-Ser-80, Met-95-Ala-96, and Met-106-Ala-107 were cleaved by both proteinase types, although the relative rates of hydrolysis at some of these sites were quite different for the two proteinases. Small histidine-rich peptides were formed as early products of the action of the cell envelope-associated proteinases on kappa-casein, implicating this casein as a possible significant source of histidine, which is essential for starter growth. The major difference between the two proteinase types in their action on kappa-casein was in their ability to cleave bonds near the C-terminal end of the molecule. The bond Asn-160-Thr-161 and, to a lesser extent, the bond Glu-151-Val-152 were very rapidly cleaved by the PIII-type proteinase, whereas hydrolysis of these bonds by the PI-type proteinase was barely detectable (even after 24 h of digestion). Differential hydrolysis of kappa-casein at these sites by the two different proteinase types resulted in the formation of distinctive, high-M(r) products detectable by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolbear T., Reid J. R., Pritchard G. G. Stability and Specificity of the Cell Wall-Associated Proteinase from Lactococcus lactis subsp. cremoris H2 Released by Treatment with Lysozyme in the Presence of Calcium Ions. Appl Environ Microbiol. 1992 Oct;58(10):3263–3270. doi: 10.1128/aem.58.10.3263-3270.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exterkate F. A., Alting A. C., Slangen C. J. Specificity of two genetically related cell-envelope proteinases of Lactococcus lactis subsp. cremoris towards alpha s1-casein-(1-23)-fragment. Biochem J. 1991 Jan 1;273(Pt 1):135–139. doi: 10.1042/bj2730135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon J. J., Delorme C., Ehrlich S. D., Renault P. Divergence of Genomic Sequences between Lactococcus lactis subsp. lactis and Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1992 Dec;58(12):4045–4047. doi: 10.1128/aem.58.12.4045-4047.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C. Structure and stability of bovine casein micelles. Adv Protein Chem. 1992;43:63–151. doi: 10.1016/s0065-3233(08)60554-9. [DOI] [PubMed] [Google Scholar]
- Mercier J. C., Brignon G., Ribadeau-Dumas B. Structure primaire de la caséine kappa B bovine. Séquence complète. Eur J Biochem. 1973 Jun;35(2):222–235. doi: 10.1111/j.1432-1033.1973.tb02829.x. [DOI] [PubMed] [Google Scholar]
- Monnet V., Ley J. P., Gonzàlez S. Substrate specificity of the cell envelope-located proteinase of Lactococcus lactis subsp. lactis NCDO 763. Int J Biochem. 1992 May;24(5):707–718. doi: 10.1016/0020-711x(92)90004-k. [DOI] [PubMed] [Google Scholar]
- Reid J. R., Moore C. H., Midwinter G. G., Pritchard G. G. Action of a cell wall proteinase from Lactococcus lactis subsp. cremoris SK11 on bovine alpha s1-casein. Appl Microbiol Biotechnol. 1991 May;35(2):222–227. doi: 10.1007/BF00184690. [DOI] [PubMed] [Google Scholar]
- Reid J. R., Ng K. H., Moore C. H., Coolbear T., Pritchard G. G. Comparison of bovine beta-casein hydrolysis by PI and PIII-type proteinases from Lactococcus lactis subsp. cremoris [corrected]. Appl Microbiol Biotechnol. 1991 Dec;36(3):344–351. doi: 10.1007/BF00208154. [DOI] [PubMed] [Google Scholar]
- Smid E. J., Poolman B., Konings W. N. Casein utilization by lactococci. Appl Environ Microbiol. 1991 Sep;57(9):2447–2452. doi: 10.1128/aem.57.9.2447-2452.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tan P. S., Poolman B., Konings W. N. Proteolytic enzymes of Lactococcus lactis. J Dairy Res. 1993 May;60(2):269–286. doi: 10.1017/s0022029900027606. [DOI] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Twining S. S. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal Biochem. 1984 Nov 15;143(1):30–34. doi: 10.1016/0003-2697(84)90553-0. [DOI] [PubMed] [Google Scholar]
- Visser S., Exterkate F. A., Slangen C. J., de Veer G. J. Comparative Study of Action of Cell Wall Proteinases from Various Strains of Streptococcus cremoris on Bovine alpha(s1)-, beta-, and kappa-Casein. Appl Environ Microbiol. 1986 Nov;52(5):1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser S., Robben A. J., Slangen C. J. Specificity of a cell-envelope-located proteinase (PIII-type) from Lactococcus lactis subsp. cremoris AM1 in its action on bovine beta-casein. Appl Microbiol Biotechnol. 1991 Jul;35(4):477–483. doi: 10.1007/BF00169753. [DOI] [PubMed] [Google Scholar]
- Vos P., Boerrigter I. J., Buist G., Haandrikman A. J., Nijhuis M., de Reuver M. B., Siezen R. J., Venema G., de Vos W. M., Kok J. Engineering of the Lactococcus lactis serine proteinase by construction of hybrid enzymes. Protein Eng. 1991 Apr;4(4):479–484. doi: 10.1093/protein/4.4.479. [DOI] [PubMed] [Google Scholar]
- Vos P., Simons G., Siezen R. J., de Vos W. M. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J Biol Chem. 1989 Aug 15;264(23):13579–13585. [PubMed] [Google Scholar]
- Xu F. F., Pearce L. E., Yu P. L. Molecular cloning and expression of a proteinase gene from Lactococcus lactis subsp. cremoris H2 and construction of a new lactococcal vector pFX1. Arch Microbiol. 1990;154(1):99–104. doi: 10.1007/BF00249185. [DOI] [PubMed] [Google Scholar]