Abstract
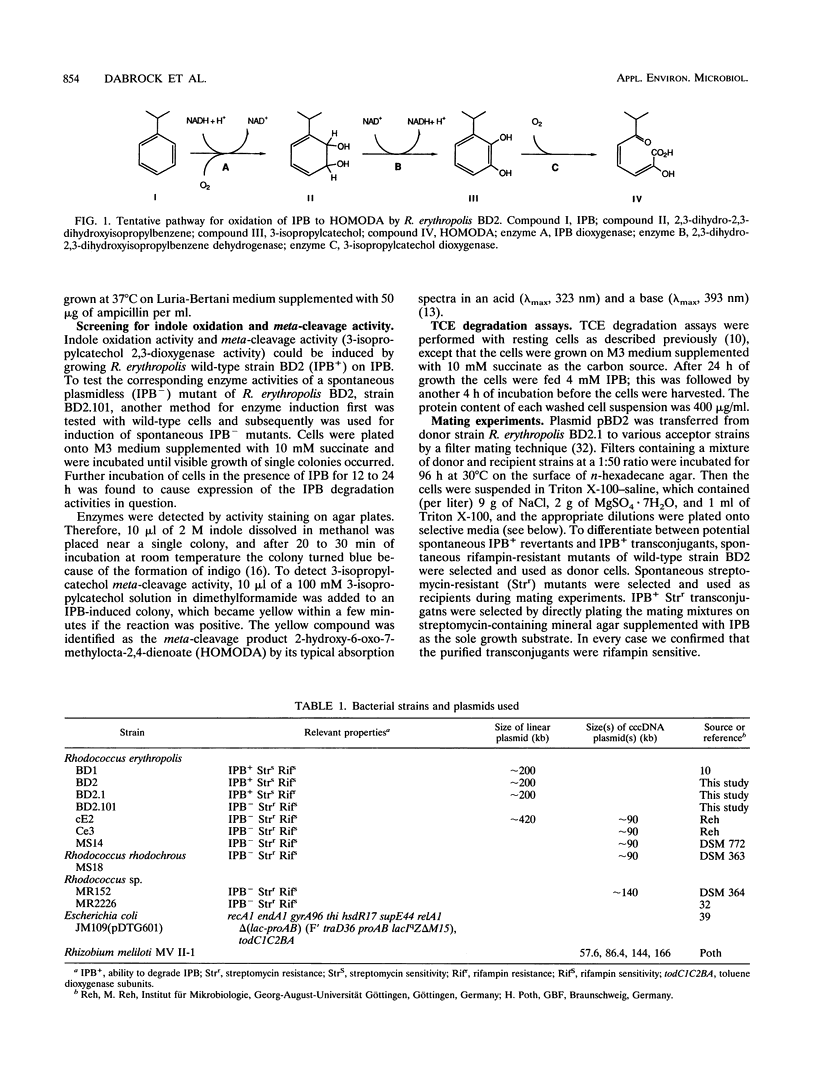
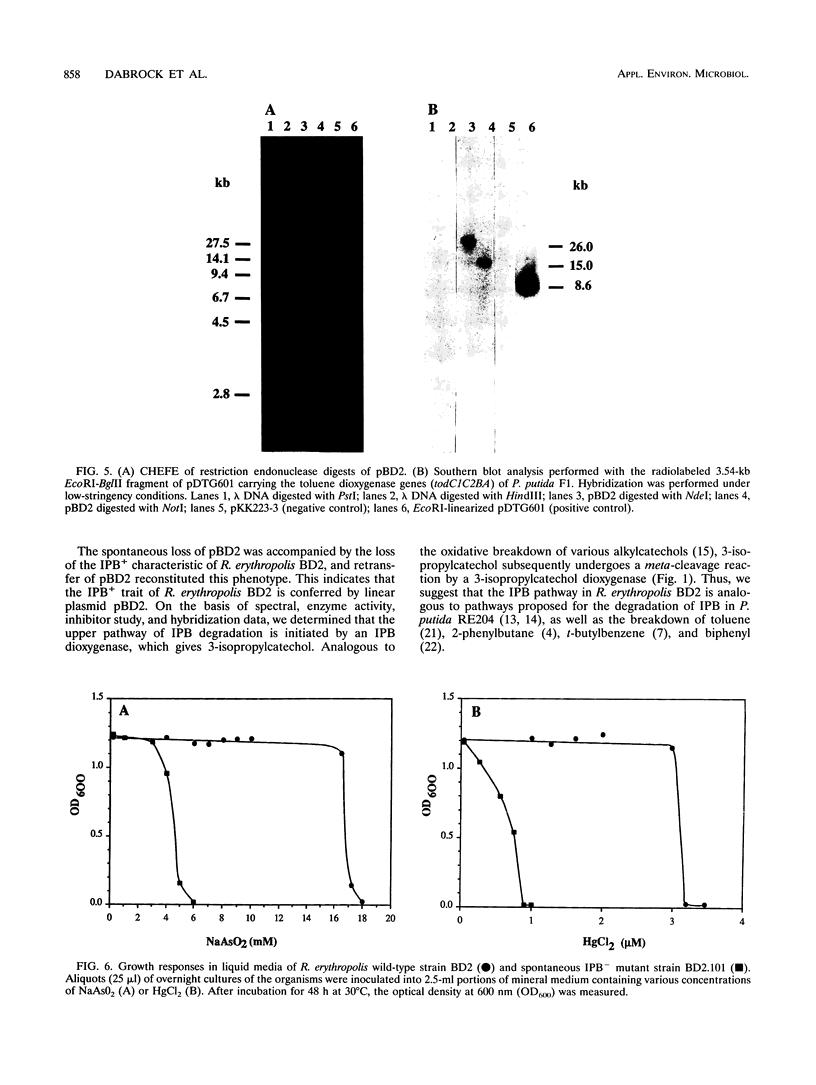
Rhodococcus erythropolis BD2, which is able to utilize isopropylbenzene as a sole carbon and energy source, was shown to contain a conjugative linear plasmid, pBD2. The estimated size of pBD2 is 208 to 212 kb. Linear plasmid-deficient strains had lost both the isopropylbenzene degradation and trichloroethene degradation characteristics, as well as the arsenite resistance and mercury resistance phenotypes. Reintroduction of pBD2 restored all four characteristics. Conjugational transfer of pBD2 to a plasmidless mutant of strain BD2 and other R. erythropolis strains occurred at frequencies between 3.5 x 10(-5) and 2.6 x 10(-3) transconjugants per recipient. R. erythropolis BD2 degrades isopropylbenzene via 3-isopropylcatechol and 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate. Both isopropylbenzene-oxidizing and meta-cleavage activities were shown to correspond with the presence of pBD2. Southern hybridizations with DNA encoding the toluene dioxygenase structural genes (todC1C2BA) from Pseudomonas putida F1 revealed homology to linear plasmid DNA. These results indicate that the isopropylbenzene degradation pathway encoded by linear plasmid pBD2 is initiated by an isopropylbenzene dioxygenase analogous to toluene dioxygenase.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asturias J. A., Timmis K. N. Three different 2,3-dihydroxybiphenyl-1,2-dioxygenase genes in the gram-positive polychlorobiphenyl-degrading bacterium Rhodococcus globerulus P6. J Bacteriol. 1993 Aug;175(15):4631–4640. doi: 10.1128/jb.175.15.4631-4640.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggi G., Catelani D., Galli E., Treccani V. The microbial degradation of phenylalkanes. 2-Phenylbutane, 3-phenylpentane, 3-phenyldodecane and 4-phenylheptane. Biochem J. 1972 Mar;126(5):1091–1097. doi: 10.1042/bj1261091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behki R., Topp E., Dick W., Germon P. Metabolism of the herbicide atrazine by Rhodococcus strains. Appl Environ Microbiol. 1993 Jun;59(6):1955–1959. doi: 10.1128/aem.59.6.1955-1959.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catelani D., Colombi A., Sorlini C., Treccani V. Metabolism of quaternary carbon compounds: 2,2-dimethylheptane and tertbutylbenzene. Appl Environ Microbiol. 1977 Oct;34(4):351–354. doi: 10.1128/aem.34.4.351-354.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbs E. R., Sole G. J. Plasmid-borne resistance to arsenate, arsenite, cadmium, and chloramphenicol in a Rhodococcus species. Mol Gen Genet. 1988 Jan;211(1):148–154. doi: 10.1007/BF00338406. [DOI] [PubMed] [Google Scholar]
- Dabrock B., Riedel J., Bertram J., Gottschalk G. Isopropylbenzene (cumene)--a new substrate for the isolation of trichloroethene-degrading bacteria. Arch Microbiol. 1992;158(1):9–13. doi: 10.1007/BF00249058. [DOI] [PubMed] [Google Scholar]
- Desomer J., Dhaese P., Van Montagu M. Conjugative transfer of cadmium resistance plasmids in Rhodococcus fascians strains. J Bacteriol. 1988 May;170(5):2401–2405. doi: 10.1128/jb.170.5.2401-2405.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton R. W., Timmis K. N. Characterization of a plasmid-specified pathway for catabolism of isopropylbenzene in Pseudomonas putida RE204. J Bacteriol. 1986 Oct;168(1):123–131. doi: 10.1128/jb.168.1.123-131.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton R. W., Timmis K. N. Spontaneous deletion of a 20-kilobase DNA segment carrying genes specifying isopropylbenzene metabolism in Pseudomonas putida RE204. J Bacteriol. 1986 Oct;168(1):428–430. doi: 10.1128/jb.168.1.428-430.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engesser K. H., Rubio M. A., Knackmuss H. J. Bacterial metabolism of side-chain-fluorinated aromatics: unproductive meta-cleavage of 3-trifluoromethylcatechol. Appl Microbiol Biotechnol. 1990 Feb;32(5):600–608. doi: 10.1007/BF00173734. [DOI] [PubMed] [Google Scholar]
- Ensley B. D., Ratzkin B. J., Osslund T. D., Simon M. J., Wackett L. P., Gibson D. T. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science. 1983 Oct 14;222(4620):167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- Ferguson J. A., Korte F. Epoxidation of aldrin to exo-dieldrin by soil bacteria. Appl Environ Microbiol. 1977 Jul;34(1):7–13. doi: 10.1128/aem.34.1.7-13.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerty W. R. The biology and genetics of the genus Rhodococcus. Annu Rev Microbiol. 1992;46:193–218. doi: 10.1146/annurev.mi.46.100192.001205. [DOI] [PubMed] [Google Scholar]
- Gibson D. T., Koch J. R., Kallio R. E. Oxidative degradation of aromatic hydrocarbons by microorganisms. I. Enzymatic formation of catechol from benzene. Biochemistry. 1968 Jul;7(7):2653–2662. doi: 10.1021/bi00847a031. [DOI] [PubMed] [Google Scholar]
- Gibson D. T., Roberts R. L., Wells M. C., Kobal V. M. Oxidation of biphenyl by a Beijerinckia species. Biochem Biophys Res Commun. 1973 Jan 23;50(2):211–219. doi: 10.1016/0006-291x(73)90828-0. [DOI] [PubMed] [Google Scholar]
- Harayama S., Kok M., Neidle E. L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- Kalkus J., Reh M., Schlegel H. G. Hydrogen autotrophy of Nocardia opaca strains is encoded by linear megaplasmids. J Gen Microbiol. 1990 Jun;136(6):1145–1151. doi: 10.1099/00221287-136-6-1145. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Nagasawa T., Yamada H. Enzymatic synthesis of acrylamide: a success story not yet over. Trends Biotechnol. 1992 Nov;10(11):402–408. doi: 10.1016/0167-7799(92)90283-2. [DOI] [PubMed] [Google Scholar]
- Martin R. R., Marshall V. D., Sokatch J. R., Unger L. Common enzymes of branched-chain amino acid catabolism in Pseudomonas putida. J Bacteriol. 1973 Jul;115(1):198–204. doi: 10.1128/jb.115.1.198-204.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J. R., Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- Rosenstein R., Peschel A., Wieland B., Götz F. Expression and regulation of the antimonite, arsenite, and arsenate resistance operon of Staphylococcus xylosus plasmid pSX267. J Bacteriol. 1992 Jun;174(11):3676–3683. doi: 10.1128/jb.174.11.3676-3683.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallis P. J., Armfield S. J., Bull A. T., Hardman D. J. Isolation and characterization of a haloalkane halidohydrolase from Rhodococcus erythropolis Y2. J Gen Microbiol. 1990 Jan;136(1):115–120. doi: 10.1099/00221287-136-1-115. [DOI] [PubMed] [Google Scholar]
- Sensfuss C., Reh M., Schlegel H. G. No correlation exists between the conjugative transfer of the autotrophic character and that of plasmids in Nocardia opaca strains. J Gen Microbiol. 1986 Apr;132(4):997–1007. doi: 10.1099/00221287-132-4-997. [DOI] [PubMed] [Google Scholar]
- Silver S., Budd K., Leahy K. M., Shaw W. V., Hammond D., Novick R. P., Willsky G. R., Malamy M. H., Rosenberg H. Inducible plasmid-determined resistance to arsenate, arsenite, and antimony (III) in escherichia coli and Staphylococcus aureus. J Bacteriol. 1981 Jun;146(3):983–996. doi: 10.1128/jb.146.3.983-996.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. E., Finnerty W. R. Construction of an Escherichia coli-Rhodococcus shuttle vector and plasmid transformation in Rhodococcus spp. J Bacteriol. 1988 Feb;170(2):638–645. doi: 10.1128/jb.170.2.638-645.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Straube G., Hensel J., Niedan C., Straube E. Kinetic studies of phenol degradation by Rhodococcus sp. P1. I. Batch cultivation. Antonie Van Leeuwenhoek. 1990 Jan;57(1):29–32. doi: 10.1007/BF00400332. [DOI] [PubMed] [Google Scholar]
- Wheatcroft R., Williams P. A. Rapid methods for the study of both stable and unstable plasmids in Pseudomonas. J Gen Microbiol. 1981 Jun;124(2):433–437. doi: 10.1099/00221287-124-2-433. [DOI] [PubMed] [Google Scholar]
- Zylstra G. J., Gibson D. T. Aromatic hydrocarbon degradation: a molecular approach. Genet Eng (N Y) 1991;13:183–203. doi: 10.1007/978-1-4615-3760-1_8. [DOI] [PubMed] [Google Scholar]
- Zylstra G. J., Gibson D. T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989 Sep 5;264(25):14940–14946. [PubMed] [Google Scholar]