Abstract
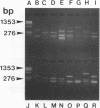
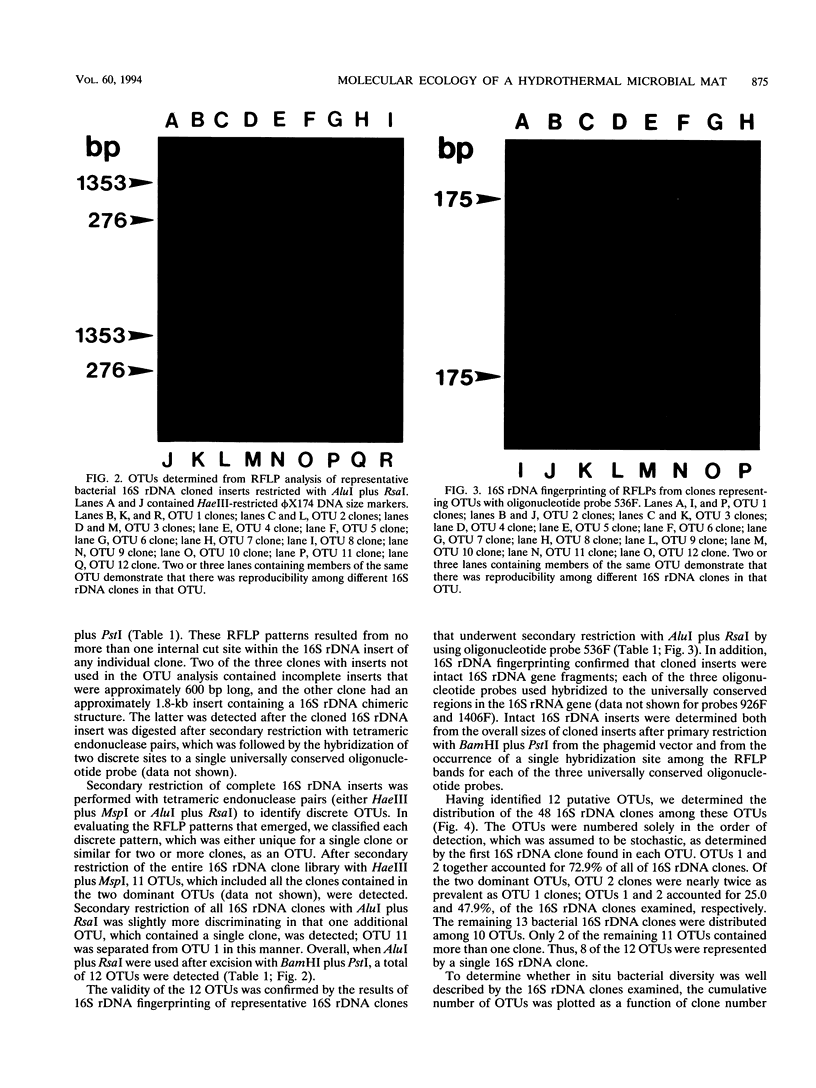
PCR was used to amplify (eu)bacterial small-subunit (16S) rRNA genes from total-community genomic DNA. The source of total-community genomic DNA used for this culture-independent analysis was the microbial mats from a deep-sea, hydrothermal vent system, Pele's Vents, located at Loihi Seamount, Hawaii. Oligonucleotides complementary to conserved regions in the 16S rRNA-encoding DNA (rDNA) of bacteria were used to direct the synthesis of PCR products, which were then subcloned by blunt-end ligation into phagemid vector pBluescript II. Restriction fragment length polymorphism patterns, created by using tandem tetrameric restriction endonucleases, revealed the presence of 12 groups of 16S rRNA genes representing discrete operational taxonomic units (OTUs). The rank order abundance of these putative OTUs was measured, and the two most abundant OTUs accounted for 72.9% of all of the 16S rDNA clones. Among the remaining 27.1% of the 16S rDNA clones, none of the 10 OTUs was represented by more than three individual clones. The cumulative OTU distribution for 48 bacterial 16S rDNA clones demonstrated that the majority of taxa represented in the clone library were detected, a result which we assume to be an estimate of the diversity of bacteria in the native hydrothermal vent habitat. 16S rDNA fingerprinting of individual clones belonging to particular OTUs by using an oligonucleotide probe that binds to a universally conserved region of the 16S rDNA fragments was conducted to confirm OTU specificity and 16S rDNA identity.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E. F. Archaea in coastal marine environments. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Dennis P. P., Bremer H. Macromolecular composition during steady-state growth of Escherichia coli B-r. J Bacteriol. 1974 Jul;119(1):270–281. doi: 10.1128/jb.119.1.270-281.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich H. A., Gelfand D., Sninsky J. J. Recent advances in the polymerase chain reaction. Science. 1991 Jun 21;252(5013):1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- Flärdh K., Cohen P. S., Kjelleberg S. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J Bacteriol. 1992 Nov;174(21):6780–6788. doi: 10.1128/jb.174.21.6780-6788.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman J. A., McCallum K., Davis A. A. Novel major archaebacterial group from marine plankton. Nature. 1992 Mar 12;356(6365):148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- Fuhrman J. A., McCallum K., Davis A. A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993 May;59(5):1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., Britschgi T. B., Moyer C. L., Field K. G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990 May 3;345(6270):60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Kramer J. G., Singleton F. L. Variations in rRNA content of marine Vibrio spp. during starvation-survival and recovery. Appl Environ Microbiol. 1992 Jan;58(1):201–207. doi: 10.1128/aem.58.1.201-207.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer C. L., Morita R. Y. Effect of growth rate and starvation-survival on cellular DNA, RNA, and protein of a psychrophilic marine bacterium. Appl Environ Microbiol. 1989 Oct;55(10):2710–2716. doi: 10.1128/aem.55.10.2710-2716.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer C. L., Morita R. Y. Effect of growth rate and starvation-survival on the viability and stability of a psychrophilic marine bacterium. Appl Environ Microbiol. 1989 May;55(5):1122–1127. doi: 10.1128/aem.55.5.1122-1127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen L. K., Ballard G., Stahl D. A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993 May;59(5):1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosset R., Julien J., Monier R. Ribonucleic acid composition of bacteria as a function of growth rate. J Mol Biol. 1966 Jul;18(2):308–320. doi: 10.1016/s0022-2836(66)80248-6. [DOI] [PubMed] [Google Scholar]
- SCHAECHTER M., MAALOE O., KJELDGAARD N. O. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958 Dec;19(3):592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sarkar G., Sommer S. S. Shedding light on PCR contamination. Nature. 1990 Jan 4;343(6253):27–27. doi: 10.1038/343027a0. [DOI] [PubMed] [Google Scholar]
- Schmidt T. M., DeLong E. F., Pace N. R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991 Jul;173(14):4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. M., Pace B., Pace N. R. Detection of DNA contamination in Taq polymerase. Biotechniques. 1991 Aug;11(2):176–177. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- Ward D. M., Weller R., Bateson M. M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990 May 3;345(6270):63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- Ward D. M., Weller R., Bateson M. M. 16S rRNA sequences reveal uncultured inhabitants of a well-studied thermal community. FEMS Microbiol Rev. 1990 Jun;6(2-3):105–115. doi: 10.1111/j.1574-6968.1990.tb04088.x. [DOI] [PubMed] [Google Scholar]
- Weller R., Bateson M. M., Heimbuch B. K., Kopczynski E. D., Ward D. M. Uncultivated cyanobacteria, Chloroflexus-like inhabitants, and spirochete-like inhabitants of a hot spring microbial mat. Appl Environ Microbiol. 1992 Dec;58(12):3964–3969. doi: 10.1128/aem.58.12.3964-3969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller R., Weller J. W., Ward D. M. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl Environ Microbiol. 1991 Apr;57(4):1146–1151. doi: 10.1128/aem.57.4.1146-1151.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]