Abstract
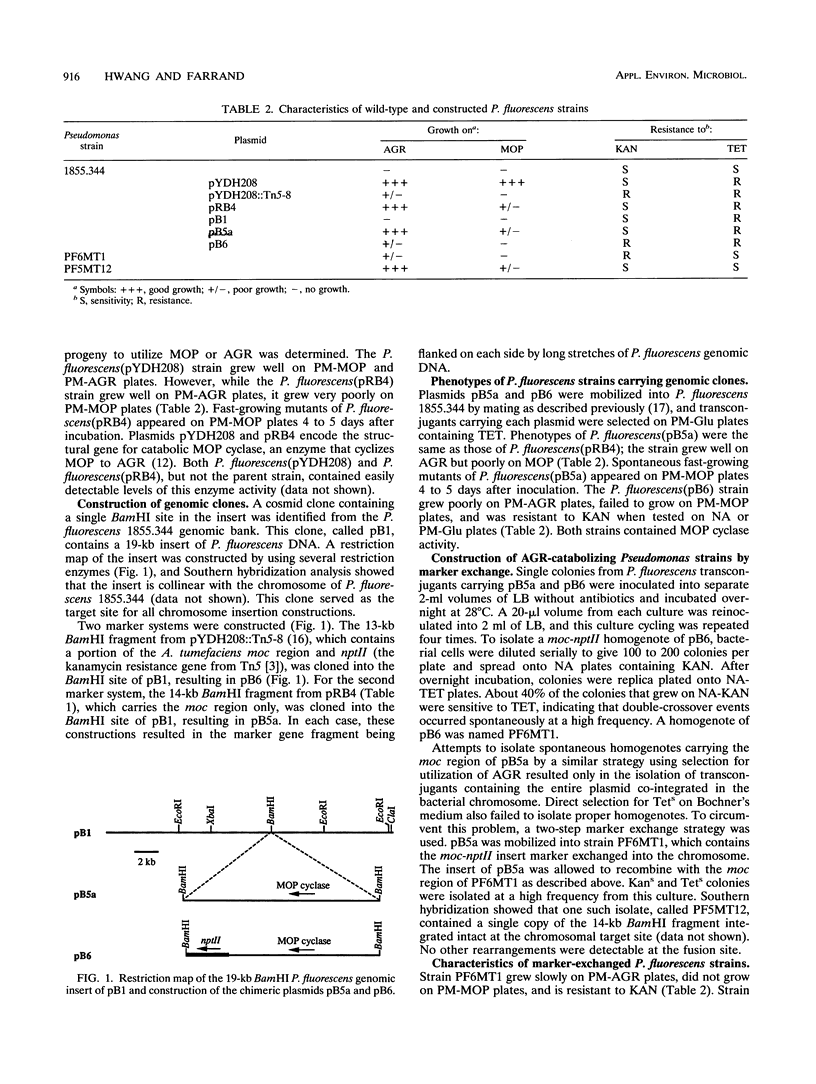
A novel method using a moc (mannityl opine catabolism) region from the Agrobacterium tumefaciens Ti plasmid pTi15955 was developed as a tag to identify genetically modified microorganisms released into the environment. Pseudomonas fluorescens 1855.344, a plant-growth-promoting rhizosphere bacterium, was chosen as the organism in which to develop and test the system. moc genes carried by pYDH208, a cosmid clone containing a 20-kb segment of the octopine-mannityl opine-type Ti plasmid, conferred on P. fluorescens strains the capacity to utilize mannopine and agropine (AGR) as a sole source of carbon and energy. Modified P. fluorescens strains containing moc or moc::nptII inserted into a chromosomal site were constructed by marker exchange. One such modified strain, PF5MT12, utilized AGR as a sole carbon source and contained detectable levels of mannopine cyclase, an easily assayable enzyme encoded by the moc region. Catabolism of AGR could be used to recover selectively the marked strain from mixed populations containing a large excess of closely related bacteria. Nucleic acid-based detection strategies were developed on the basis of the unique fusion region between Agrobacterium DNA and Pseudomonas DNA in strain PF5MT12. The specificity and sensitivity of detection of PF5MT12 were enhanced by amplifying the fused DNA region by using PCR. The target fragment could be detected at levels of sensitivity comparable to those of other described PCR-based gene tags, even in the presence of high levels of Agrobacterium, Pseudomonas, or Escherichia coli DNA. This gene tag strategy gives a method for direct selection and enumeration of the marked strain from mixtures containing a large excess of closely related bacteria and a sensitive and highly specific system for detection by PCR amplification of the target fragment even in the presence of large amounts of DNA from related or unrelated organisms.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck E., Ludwig G., Auerswald E. A., Reiss B., Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982 Oct;19(3):327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Bej A. K., Mahbubani M. H. Applications of the polymerase chain reaction in environmental microbiology. PCR Methods Appl. 1992 Feb;1(3):151–159. doi: 10.1101/gr.1.3.151. [DOI] [PubMed] [Google Scholar]
- Bej A. K., Mahbubani M. H., Dicesare J. L., Atlas R. M. Polymerase chain reaction-gene probe detection of microorganisms by using filter-concentrated samples. Appl Environ Microbiol. 1991 Dec;57(12):3529–3534. doi: 10.1128/aem.57.12.3529-3534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej A. K., Steffan R. J., DiCesare J., Haff L., Atlas R. M. Detection of coliform bacteria in water by polymerase chain reaction and gene probes. Appl Environ Microbiol. 1990 Feb;56(2):307–314. doi: 10.1128/aem.56.2.307-314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesney R. H., Scott J. R., Vapnek D. Integration of the plasmid prophages P1 and P7 into the chromosome of Escherichia coli. J Mol Biol. 1979 May 15;130(2):161–173. doi: 10.1016/0022-2836(79)90424-8. [DOI] [PubMed] [Google Scholar]
- Darzins A., Chakrabarty A. M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984 Jul;159(1):9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaux Y., Guyon P., Farrand S. K., Petit A., Tempé J. Agrobacterium Ti and Ri plasmids specify enzymic lactonization of mannopine to agropine. J Gen Microbiol. 1986 Sep;132(9):2549–2559. doi: 10.1099/00221287-132-9-2549. [DOI] [PubMed] [Google Scholar]
- Dessaux Y., Tempé J., Farrand S. K. Genetic analysis of mannityl opine catabolism in octopine-type Agrobacterium tumefaciens strain 15955. Mol Gen Genet. 1987 Jun;208(1-2):301–308. doi: 10.1007/BF00330457. [DOI] [PubMed] [Google Scholar]
- Guyon P., Chilton M. D., Petit A., Tempé J. Agropine in "null-type" crown gall tumors: Evidence for generality of the opine concept. Proc Natl Acad Sci U S A. 1980 May;77(5):2693–2697. doi: 10.1073/pnas.77.5.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. B., Dessaux Y., Chilton W. S., Farrand S. K. Organization and regulation of the mannopine cyclase-associated opine catabolism genes in Agrobacterium tumefaciens 15955. J Bacteriol. 1993 Jan;175(2):401–410. doi: 10.1128/jb.175.2.401-410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Lim S. M., Shaw P. D. Cloning and characterization of pathogenicity genes from Xanthomonas campestris pv. glycines. J Bacteriol. 1992 Mar;174(6):1923–1931. doi: 10.1128/jb.174.6.1923-1931.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. W., Edlin G. Expression of tetracycline resistance in pBR322 derivatives reduces the reproductive fitness of plasmid-containing Escherichia coli. Gene. 1985;39(2-3):173–180. doi: 10.1016/0378-1119(85)90311-7. [DOI] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal C. S., Dion P. Characterization of the Opine-Utilizing Microflora Associated with Samples of Soil and Plants. Appl Environ Microbiol. 1990 Aug;56(8):2576–2579. doi: 10.1128/aem.56.8.2576-2579.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obukowicz M. G., Perlak F. J., Bolten S. L., Kusano-Kretzmer K., Mayer E. J., Watrud L. S. IS50L as a non-self transposable vector used to integrate the Bacillus thuringiensis delta-endotoxin gene into the chromosome of root-colonizing pseudomonads. Gene. 1987;51(1):91–96. doi: 10.1016/0378-1119(87)90478-1. [DOI] [PubMed] [Google Scholar]
- Olexy V. M., Bird T. J., Grieble H. G., Farrand S. K. Hospital isolates of Serratia marcescens transferring ampicillin, carbenicillin, and gentamicin resistance to other gram-negative bacteria including Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1979 Jan;15(1):93–100. doi: 10.1128/aac.15.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P. E., Rice W. A. Rhizobium strain identification and quantification in commercial inoculants by immunoblot analysis. Appl Environ Microbiol. 1989 Feb;55(2):520–522. doi: 10.1128/aem.55.2.520-522.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S. D., Josephson K. L., Bailey R. L., Gerba C. P., Pepper I. L. Rapid method for processing soil samples for polymerase chain reaction amplification of specific gene sequences. Appl Environ Microbiol. 1991 Aug;57(8):2283–2286. doi: 10.1128/aem.57.8.2283-2286.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. J., Dane F., Geiger D., Kloepper J. W. Use of bioluminescence for detection of genetically engineered microorganisms released into the environment. Appl Environ Microbiol. 1992 Jan;58(1):267–273. doi: 10.1128/aem.58.1.267-273.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- Tempé J., Petit A., Holsters M., Montagu M., Schell J. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: Possible relation to transformation in crown gall. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2848–2849. doi: 10.1073/pnas.74.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992 Feb;58(2):754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson B., Currier T. C., Gordon M. P., Chilton M. D., Nester E. W. Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1975 Jul;123(1):255–264. doi: 10.1128/jb.123.1.255-264.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]