Abstract
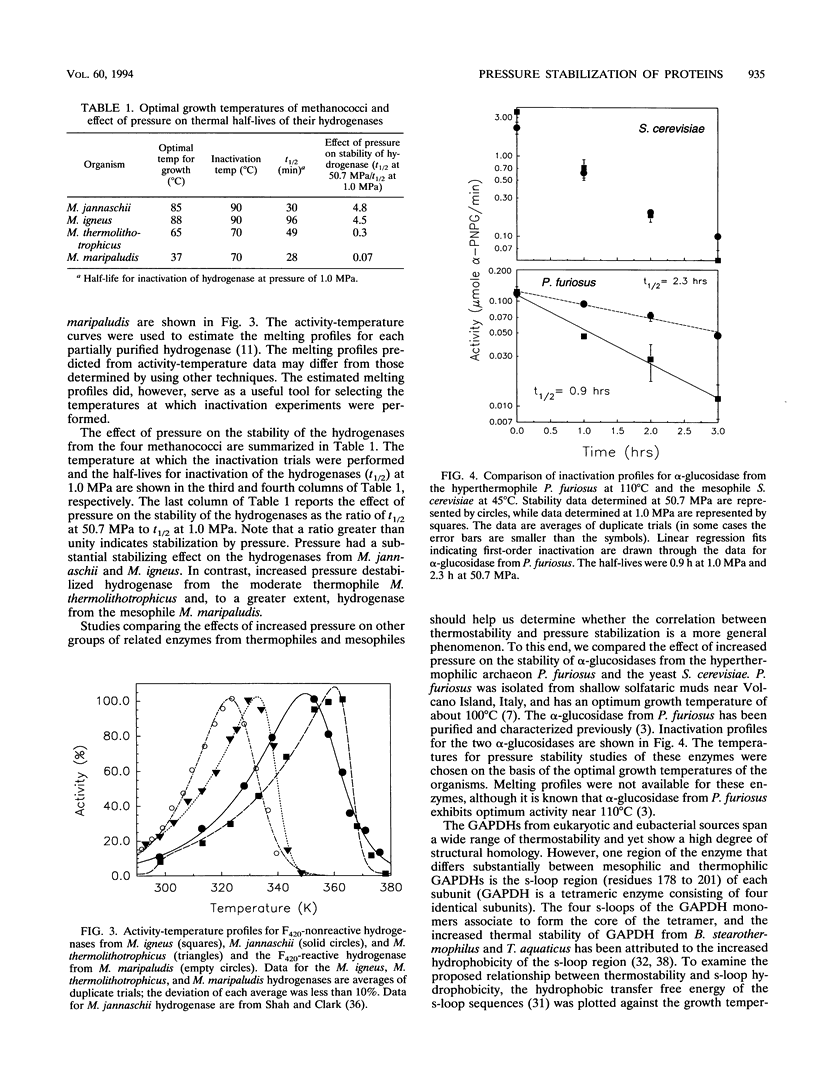
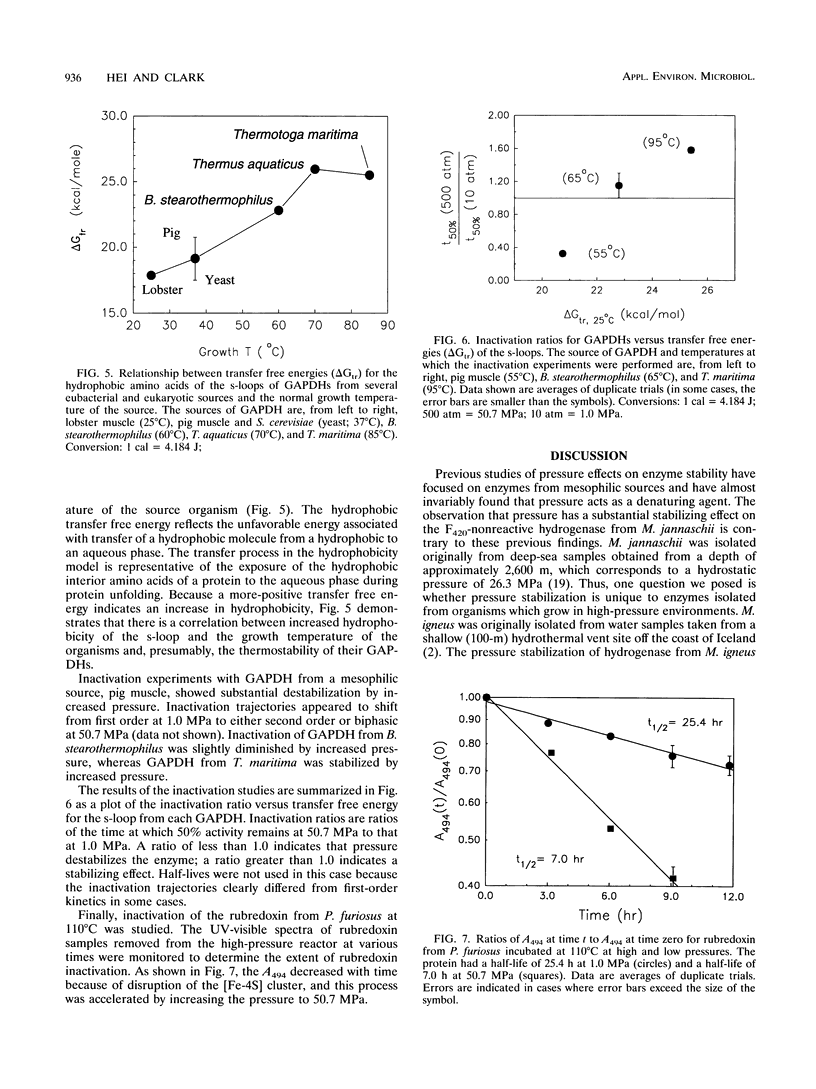
We describe the stabilization by pressure of enzymes, including a hydrogenase from Methanococcus jannaschii, an extremely thermophilic deep-sea methanogen. This is the first published report of proteins from thermophiles being stabilized by pressure. Inactivation studies of partially purified hydrogenases from an extreme thermophile (Methanococcus igneus), a moderate thermophile (Methanococcus thermolithotrophicus), and a mesophile (Methanococcus maripaludis), all from shallow marine sites, show that pressure stabilization is not unique to enzymes isolated from high-pressure environments. These studies suggest that pressure stabilization of an enzyme may be related to its thermophilicity. Further experiments comparing the effects of increased pressure on the stability of α-glucosidases from the hyperthermophile Pyrococcus furiosus and Saccharomyces cerevisiae support this possibility. We have also examined pressure effects on several highly homologous glyceraldehyde-3-phosphate dehydrogenases from mesophilic and thermophilic sources and a rubredoxin from P. furiosus. The results suggest that hydrophobic interactions, which have been implicated in the stabilization of many thermophilic proteins, contribute to the pressure stabilization of enzymes from thermophiles.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blake P. R., Park J. B., Bryant F. O., Aono S., Magnuson J. K., Eccleston E., Howard J. B., Summers M. F., Adams M. W. Determinants of protein hyperthermostability: purification and amino acid sequence of rubredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus and secondary structure of the zinc adduct by NMR. Biochemistry. 1991 Nov 12;30(45):10885–10895. doi: 10.1021/bi00109a012. [DOI] [PubMed] [Google Scholar]
- Burggraf S., Fricke H., Neuner A., Kristjansson J., Rouvier P., Mandelco L., Woese C. R., Stetter K. O. Methanococcus igneus sp. nov., a novel hyperthermophilic methanogen from a shallow submarine hydrothermal system. Syst Appl Microbiol. 1990;13:263–269. doi: 10.1016/s0723-2020(11)80197-9. [DOI] [PubMed] [Google Scholar]
- Costantino H. R., Brown S. H., Kelly R. M. Purification and characterization of an alpha-glucosidase from a hyperthermophilic archaebacterium, Pyrococcus furiosus, exhibiting a temperature optimum of 105 to 115 degrees C. J Bacteriol. 1990 Jul;172(7):3654–3660. doi: 10.1128/jb.172.7.3654-3660.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton T. E. Protein folding. Biochem J. 1990 Aug 15;270(1):1–16. doi: 10.1042/bj2700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B. E., Sajgò M., Noller H. F., Harris J. I. Amino-acid sequence of glyceraldehyde 3-phosphate dehydrogenase from lobster muscle. Nature. 1967 Dec 23;216(5121):1181–1185. doi: 10.1038/2161181a0. [DOI] [PubMed] [Google Scholar]
- Harris J. I., Perham R. N. Glyceraldehyde 3-phosphate dehydrogenase from pig muscle. Nature. 1968 Sep 7;219(5158):1025–1028. doi: 10.1038/2191025a0. [DOI] [PubMed] [Google Scholar]
- Hocking J. D., Harris J. I. D-glyceraldehyde-3-phosphate dehydrogenase. Amino-acid sequence of the enzyme from the extreme thermophile Thermus aquaticus. Eur J Biochem. 1980 Jul;108(2):567–579. doi: 10.1111/j.1432-1033.1980.tb04752.x. [DOI] [PubMed] [Google Scholar]
- Holland J. P., Holland M. J. The primary structure of a glyceraldehyde-3-phosphate dehydrogenase gene from Saccharomyces cerevisiae. J Biol Chem. 1979 Oct 10;254(19):9839–9845. [PubMed] [Google Scholar]
- Hvidt A. A discussion of pressure-volume effects in aqueous protein solutions. J Theor Biol. 1975 Mar;50(1):245–252. doi: 10.1016/0022-5193(75)90035-1. [DOI] [PubMed] [Google Scholar]
- Jaenicke R. Protein stability and molecular adaptation to extreme conditions. Eur J Biochem. 1991 Dec 18;202(3):715–728. doi: 10.1111/j.1432-1033.1991.tb16426.x. [DOI] [PubMed] [Google Scholar]
- Jannasch H. W., Wirsen C. O., Molyneaux S. J., Langworthy T. A. Extremely thermophilic fermentative archaebacteria of the genus desulfurococcus from deep-sea hydrothermal vents. Appl Environ Microbiol. 1988 May;54(5):1203–1209. doi: 10.1128/aem.54.5.1203-1209.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Lin Y. Z., Liang S. J., Zhou J. M., Tsou C. L., Wu P. Q., Zhou Z. K. Comparison of inactivation and conformational changes of D-glyceraldehyde-3-phosphate dehydrogenase during thermal denaturation. Biochim Biophys Acta. 1990 Apr 19;1038(2):247–252. doi: 10.1016/0167-4838(90)90212-x. [DOI] [PubMed] [Google Scholar]
- Martin W., Cerff R. Prokaryotic features of a nucleus-encoded enzyme. cDNA sequences for chloroplast and cytosolic glyceraldehyde-3-phosphate dehydrogenases from mustard (Sinapis alba). Eur J Biochem. 1986 Sep 1;159(2):323–331. doi: 10.1111/j.1432-1033.1986.tb09871.x. [DOI] [PubMed] [Google Scholar]
- Millar D. B., Grafius M. A., Wild J. R., Palmer D. A. High pressure stabilization of acetylcholinesterase sizeozymes against thermal denaturation. Biophys Chem. 1974 Aug;2(2):189–192. doi: 10.1016/0301-4622(74)80042-6. [DOI] [PubMed] [Google Scholar]
- Miller J. F., Shah N. N., Nelson C. M., Ludlow J. M., Clark D. S. Pressure and Temperature Effects on Growth and Methane Production of the Extreme Thermophile Methanococcus jannaschii. Appl Environ Microbiol. 1988 Dec;54(12):3039–3042. doi: 10.1128/aem.54.12.3039-3042.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki Y., Tanford C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J Biol Chem. 1971 Apr 10;246(7):2211–2217. [PubMed] [Google Scholar]
- Olsen K. W. Structural basis for the thermal stability of glyceraldehyde-3-phosphate dehydrogenases. Int J Pept Protein Res. 1983 Oct;22(4):469–475. doi: 10.1111/j.1399-3011.1983.tb02117.x. [DOI] [PubMed] [Google Scholar]
- Samarasinghe S. D., Campbell D. M., Jonas A., Jonas J. High-resolution NMR study of the pressure-induced unfolding of lysozyme. Biochemistry. 1992 Sep 1;31(34):7773–7778. doi: 10.1021/bi00149a005. [DOI] [PubMed] [Google Scholar]
- Schultes V., Deutzmann R., Jaenicke R. Complete amino-acid sequence of glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic eubacterium Thermotoga maritima. Eur J Biochem. 1990 Aug 28;192(1):25–31. doi: 10.1111/j.1432-1033.1990.tb19190.x. [DOI] [PubMed] [Google Scholar]
- Shah N. N., Clark D. S. Partial Purification and Characterization of Two Hydrogenases from the Extreme Thermophile Methanococcus jannaschii. Appl Environ Microbiol. 1990 Apr;56(4):858–863. doi: 10.1128/aem.56.4.858-863.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Carne A. F., Runswick M. J., Bridgen J., Harris J. I. D-glyceraldehyde-3-phosphate dehydrogenase. Complete amino-acid sequence of the enzyme from Bacillus stearothermophilus. Eur J Biochem. 1980 Jul;108(2):549–565. doi: 10.1111/j.1432-1033.1980.tb04751.x. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Wonacott A. J., Harris J. I. Heat stability of a tetrameric enzyme, D-glyceraldehyde-3-phosphate dehydrogenase. Eur J Biochem. 1980 Jul;108(2):581–586. doi: 10.1111/j.1432-1033.1980.tb04753.x. [DOI] [PubMed] [Google Scholar]
- Weber G., Drickamer H. G. The effect of high pressure upon proteins and other biomolecules. Q Rev Biophys. 1983 Feb;16(1):89–112. doi: 10.1017/s0033583500004935. [DOI] [PubMed] [Google Scholar]
- Wright H. T. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit Rev Biochem Mol Biol. 1991;26(1):1–52. doi: 10.3109/10409239109081719. [DOI] [PubMed] [Google Scholar]



