Abstract
Aims
To evaluate the reliability of the omeprazole hydroxylation index as a marker for polymorphic CYP2C19 activity in a Japanese population of healthy young subjects (n =78) and patients with peptic ulcer (n =72).
Methods
Healthy subjects were administered a single dose of omeprazole (20 mg), whereas patients received 20 mg daily for at least 1 week. The ratio of the serum concentration of omeprazole to hydroxyomeprazole at 3 h postdose was determined and used as a measure of CYP2C19 activity. The CYP2C19 wild type (wt) gene and four mutant alleles associated with the poor metaboliser phenotype of (S)-mephenytoin, CYP2C19*2 in exon 5, CYP2C19*3 in exon 4, CYP2C19m4 in exon 9, and CYP2C19m3 in the initial codon were analysed.
Results
In the healthy volunteer study there was complete concordance between genotype and phenotype. However, eight of the patients who had the EM genotype had a high value for their hydroxylation index, and were classified as phenotypic PMs. No CYP2C19m4 and CYP2C19m3 mutations were detected in the eight mismatched patients. They were all genotypic heterozygous EMs, elderly (≥65 years) and/or had hepatic disease. Therefore, impaired CYP2C19 activity combined with partial saturation of omeprazole metabolism during multiple dosing may have contributed to the discrepancy between CYP2C19 genotyping and phenotyping.
Conclusion
Although omeprazole has been used instead of mephenytoin as a probe for polymorphic CYP2C19, it does not appear to be reliable enough for clinical application in Japanese patients.
Keywords: CYP2C19, omeprazole hydroxylation index, genotype, phenotype, clinical applicability
Introduction
Omeprazole, a proton pump inhibitor, is metabolised by the liver to two major plasma metabolites, hydroxyomeprazole and omeprazole sulphone [1]. In vivo and in vitro studies have indicated that the formation of hydroxyomeprazole is mediated by CYP2C19, with a minor contribution from CYP3A4 [1–4]. Because the mephenytoin S/R enantiomeric ratio is significantly correlated with the omeprazole hydroxylation index (the ratio of omeprazole to hydroxyomeprazole in serum 3 h post-dose) [5], omeprazole is used by some to assess CYP2C19 phenotype [6–8]. Although the reliability of the omeprazole hydroxylation index for phenotyping healthy volunteers has been demonstrated, its clinical application has not been investigated. In the present study, we compared CYP2C19 phenotype using the omeprazole hydroxylation index to genotype in populations of healthy volunteers and patients with peptic ulcer.
Methods
Subjects and study protocol
Seventy eight unrelated healthy Japanese subjects participated in the first study. The subjects ranged in age from 20 to 47 years (median with 25% and 75% quartiles 23.0, 21.0 and 30.0), and 58 of them were male. Each subject had no antecedent history of significant illness or medication, or hypersensitivity to any drugs. A physical examination, blood chemistry screening, a complete blood count, and urinalysis, were performed before the subjects were admitted to the study. They were asked to refrain from taking any medication, including alcohol and over-the-counter drugs, for at least 1 week before and then throughout the study. The participants came to the clinic after overnight fasting and received an oral dose of 20 mg omeprazole (Omepral, Fujisawa Co. Ltd, Osaka, Japan) with 150 ml water. Lunch was served 4 h after drug ingestion. In the second study, 72 in-patients with peptic ulcer were recruited from the Internal Medicine Ward of St Mary’s Hospital. All had been receiving an oral dose of 20 mg omeprazole each morning for at least 1 week. The patients ranged in age from 19 to 82 years (median with 25% and 75% quartiles 47.0, 31.5 and 60.5), and 53 of them were male. Information was recorded on gender, age, and liver and renal function. Patients who were taking a drug known to be metabolised by or to inhibit CYP2C19 were not included in the study. The subjects were informed both verbally and in writing of the experimental procedure and the purpose of the study. Each subject gave their written consent before the study, and the protocol was approved by the Institutional Review Board of the Clinical Pharmacology Center, Medical Co.Ltd. Venous blood samples (5 ml) were collected into Vacutainer tubes (Becton Dickinson, Franklin Lakes, N.J.) from an antecubital vein 3 h after drug administration. Serum was separated after centrifugation and stored at −20 °C until analysis.
Phenotyping
The concentration of omeprazole and 5-hydroxyomeprazole in serum was measured by h.p.l.c. [8]. Samples were analysed in duplicate and standard curves were included in each analysis run. The lower limit of sensitivity of the omeprazole assay was 10 ng ml−1; the coefficient of the intra-assay variation was 3.5% and that of the inter-assay variation was 4.0%. The corresponding values were 10 ng ml−1, 3.1%, and 4.3% for the 5-hydroxyomeprazole assay. The recovery of omeprazole and 5-hydroxyomeprazole ranged from 90% to 100%. The ratio of the serum concentration of omeprazole and hydroxyomeprazole 3 h post-dose (the omeprazole hydroxylation index) was used as the index of CYP2C19 activity.
Genotyping
Blood samples (10 ml) were obtained from all subjects, and genomic deoxyribonucleic acid (DNA) was isolated from the peripheral lymphocytes using an extraction kit (Genomix; Talent srl, Trieste, Italy). The CYP2C19 wt gene (CYP2C19*1) and two mutant alleles associated with the poor metaboliser phenotype of (S)-mephenytoin, CYP2C19*2 in exon 5 and CYP2C19*3 in exon 4, were identified according to the methods of de Morais et al. [9, 10] with minor modifications [11]. A new mutant CYP2C19 allele (m4), which contains a single amino acid substitution of Arg433 for Trp433 in exon 9, was analysed according to the method of Xiao et al. [12] with a minor modification, namely the use of hot start PCR. Another new mutant CYP2C19 allele (m3), which contains an A→G mutation in the initial codon, was analysed according to the method of Ferguson et al. [13].
Results
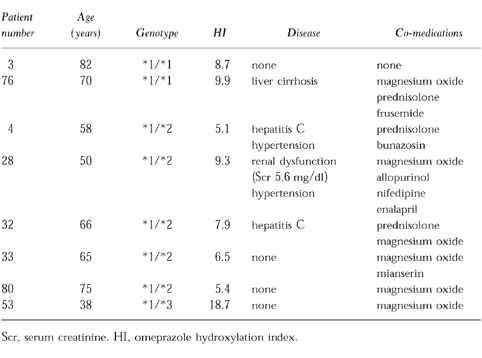
Figure 1 shows the probit plot and frequency distribution of the omeprazole hydroxylation index for the combined healthy subject and patient populations. Using an antimode of 5.1, 21 of the 78 healthy volunteers (27%) and 25 of the 72 patients (35%) were phenotyped as PMs. In the healthy volunteers the omeprazole hydroxylation index ranged from 0.02 to 5.0 in those genotyped as EM, and from 5.1 to 12.3 in those genotyped as PM. There was no overlap in the metabolic ratio of the two groups, thus giving complete concordance between genotype and phenotype. In patients omeprazole hydroxylation index values ranged from 0.05 to 18.7 in the EM genotype group, and from 5.1 to 17.7 in the PM genotype group. As shown in Figure 1, 8 out of 55 who had the EM genotype clearly had a high hydroxylation index (mean±s.d.=8.9±4.3), and were phenotypically classified as a PM. Six of these eight genotypic EMs were heterozygous for either the CYP2C19*2 or CYP2C19*3 mutant allele but no CYP2C19m4 or CYP2C19m3 alleles were detected. Five of these patients were elderly (mean age; 72 years), three had hepatic dysfunction, and one had renal failure. Concomitant drugs used included magnesium oxide in six patients, prednisolone in three, allopurinol in one, frusemide in one, nifedipine in one, enalapril in one, and mianserin in one (Table 1).
Figure 1.
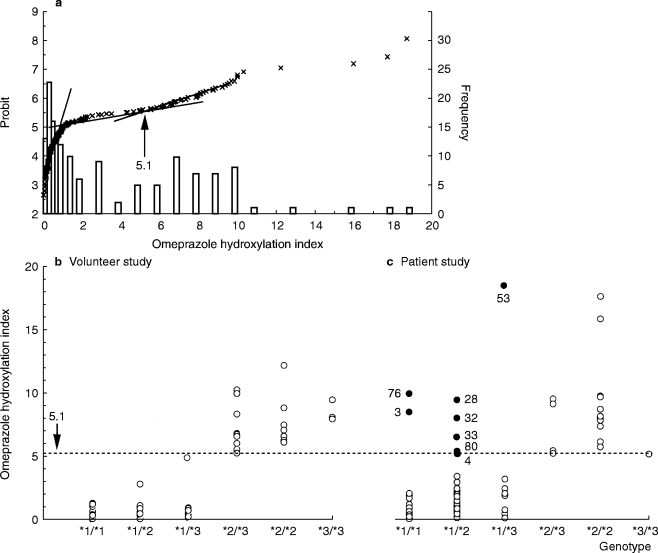
a) Probit plot and frequency distribution of the omeprazole hydroxylation index in the combined population of healthy subjects and patients, the omeprazole hydroxylation index in relation to genotype in healthy volunteers b) and peptic ulcer patients c). The closed circles are from eight EM patients who showed discrepant results between genotyping and phenotyping. CYP2C19*1, *2 and *3 (new nomenclature) represent CYP2C19wt, CYP2C19m1, and CYP2C19m2 (old nomenclature), respectively.
Table 1.
Characteristics of the patients whose CYP2C19 genotype and phenotype results were discrepant.
Discussion
Although mephenytoin, an antiepileptic drug, is well established as an index of CYP2C19 activity in humans, serious side effects have been reported [14]. Therefore, omeprazole, a drug with a high therapeutic index, has been proposed as an alternative to mephenytoin. The ratio of the 3 h plasma concentration of omeprazole to hydroxyomeprazole allows the population to be identified as EMs or PMs, and may enable the detection of homozygous and heterozygous EMs [6–8]. However, in the present study we observed a discrepancy between phenotype and genotype in 14.5% of genotypic EM patients being treated with omeprazole for a gastric ulcer. Furthermore, some patients had hydroxylation indices that were very close to the antimode, leaving their phenotypic status unclear. In contrast, there was no discrepancy between genotype and phenotype in the healthy subjects.
Factors affecting the metabolism of omeprazole and which may have contributed to the discrepancy between genotype and phenotype include gender [15], concomitant use of other drugs [16, 17], liver disease [18], age [19] and length of therapy [6]. The proportion of males to females was similar in patients and healthy subjects, making gender an unlikely cause. None of the mismatched EMs was taking substrates that interact with CYP2C19, although inhibition of activity by a metabolite of a co-administered drug cannot be ruled out. Impaired omeprazole metabolism as a result of liver disease or old age could be a basis for the phenotypic discrepancy in six out of the eight EMs. Indeed liver disease and age has been shown to cause phenocopying, the apparent conversion of an EM into a PM [18–20]. An alternative explanation is saturation of CYP2C19 activity in the patients. The healthy subjects were given single doses of omeprazole, whereas the patients had been receiving the drug daily for at least 1 week. Chang et al. [6] have shown that the omeprazole metabolic ratio increases from the first to the eighth dose in heterozygote but not in homozygote EMs. In the present study, six of the eight patients who were phenotyped incorrectly were heterozygote EMs. Unfortunately, these patients could not be retested using a single dose of omeprazole.
In conclusion, the use of omeprazole instead of mephenytoin as an index of polymorphic CYP2C19 activity may be unreliable particularly in elderly patients, those with liver disease and those on chronic therapy. Because it is not influenced by exogenous factors, genotyping gives unequivocal assignment of CYP2C19 PM and EM status. However, it cannot be used as a quantitative measure of CYP2C19 activity. A combination of genotyping and phenotyping is recommended for the full characterisation of individual CYP2C19 behaviour.
References
- 1.Andersson T, Regardh CG, Lou YC, Zhang Y, Dahl ML, Bertilsson L. Polymorphic hydroxylation of S-mephenytoin and omeprazole metabolism in Caucasian and Chinese subjects. Pharmacogenetics. 1992;2:25–31. doi: 10.1097/00008571-199202000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Andersson T, Miners JO, Veronese M, et al. Identification of human liver cytochrome P450 isoforms mediating omeprazole metabolism. Br J Clin Pharmacol. 1993;36:521–530. doi: 10.1111/j.1365-2125.1993.tb00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balian JD, Sukhova N, Harris JW, et al. The hydroxylation of omeprazole correlates with S-mephenytoin metabolism: a population study. Clin Pharmacol Ther. 1995;57:662–669. doi: 10.1016/0009-9236(95)90229-5. [DOI] [PubMed] [Google Scholar]
- 4.Curi-Pedrosa R, Pichard L, Bonfils C, Jacoz-Aigrain E, Maurel P. Major implication of cytochrome P4503A4 in the oxidative metabolism of the antisecretory drugs omeprazole and lansoprazole in human liver microsomes and hepatocytes. Br J Clin Pharmacol. 1993;36 [Google Scholar]
- 5.Chang M, Dahl M-L, Tybring G, Gotharson E, Bertilsson L. Use of omeprazole as a probe drug for CYP2C19 phenotype in Swedish Caucasians: comparison with S-mephenytoin hydroxylation phenotype and CYP2C19 genotype. Pharmacogenetics. 1995;5:358–363. doi: 10.1097/00008571-199512000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Chang M, Tybring G, Dahl M-L, et al. Interphenotype differences in disposition and effect on gastrin levels of omeprazole—suitability of omeprazole as a probe for CYP2C19. Br J Clin Pharmacol. 1995;39:511–518. doi: 10.1111/j.1365-2125.1995.tb04488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roh H-K, Dahl M-L, Tybring G, Yamada H, Cha Y-N, Bertilsson L. CYP2C19 genotype and phenotype determined by omeprazole in a Korean population. Pharmacogenetics. 1996;6:547–551. doi: 10.1097/00008571-199612000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Ieiri I, Kubota T, Urae A, et al. Pharmacokinetics of omeprazole (a substrate of CYP2C19) and comparison with two mutant alleles, CYP2C19m1 in exon 5 and CYP2C19m2 in exon 4, in Japanese subjects. Clin Pharmacol Ther. 1996;59:647–653. doi: 10.1016/S0009-9236(96)90004-1. [DOI] [PubMed] [Google Scholar]
- 9.de Morais SMF, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin in human. J Biol Chem. 1994;269:15419–15422. [PubMed] [Google Scholar]
- 10.de Morais SMF, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46:594–598. [PubMed] [Google Scholar]
- 11.Kimura M, Ieiri I, Mamiya K, Urae A, Higuchi S. Genetic polymorphism of cytochrome P450s, CYP2C19 and CYP2C9, in a Japanese population. Ther Drug Monit. 1998;20:243–247. doi: 10.1097/00007691-199806000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Xiao Z-S, Goldstein JA, Xie H-G, et al. Difference in the incidence of the CYP2C19 polymorphism affecting the S-mephenytoin phenotype in Chinese Han and Bai populations and identification of a new rare CYP2C19 mutant allele. J Pharmacol Exp Ther. 1997;281:604–609. [PubMed] [Google Scholar]
- 13.Ferguson RJ, de Morais SMF, Benhamou S, et al. A new genetic defect in human CYP2C19: Mutation of the initial codon is responsible for poor metabolism of S-mephenytoin. J Pharmacol Exp Ther. 1998;284:356–361. [PubMed] [Google Scholar]
- 14.Relling MV, Ayers D, Heideman RL. Mephenytoin phenotyping: lack of haematologic effect and timing of urine collections. Pharmacogenetics. 1991;1:42–49. doi: 10.1097/00008571-199110000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Xie H-G, Huang S-L, Xu Z-H, Xiao Z-S, He N, Zhou H-H. Evidence for the effect of gender on activity of (S)-mephenytoin 4′-hydroxylase (CYP2C19) in a Chinese population. Pharmacogenetics. 1997;7:115–119. doi: 10.1097/00008571-199704000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Andersson T. Omeprazole drug interaction studies. Clin Pharmacokinet. 1991;21:195–212. doi: 10.2165/00003088-199121030-00004. [DOI] [PubMed] [Google Scholar]
- 17.Andersson T. Pharmacokinetics, metabolism and interactions of acid pump inhibitors. Focus on omeprazole, lansoprazole and pantoprazole. Clin Pharmacokinet. 1996;31:9–28. doi: 10.2165/00003088-199631010-00002. [DOI] [PubMed] [Google Scholar]
- 18.Andersson T, Olsson R, Regardh C-G, Skanberg I. Pharmacokinetics of [14C]omeprazole in patients with liver cirrhosis. Clin Pharmacokinet. 1993;24:71–78. doi: 10.2165/00003088-199324010-00006. [DOI] [PubMed] [Google Scholar]
- 19.Landahl S, Andersson T, Larsson M, et al. Pharmacokinetic study of omeprazole in elderly healthy volunteers. Clin Pharmacokinet. 1992;23:469–476. doi: 10.2165/00003088-199223060-00006. [DOI] [PubMed] [Google Scholar]
- 20.Brockmoller J, Roots I. Assessment of liver metabolic function. Clinical implications. Clin Pharmacokinet. 1994;27:216–248. doi: 10.2165/00003088-199427030-00005. [DOI] [PubMed] [Google Scholar]


