Abstract
Aims
Many substrates of cytochrome P450 (CYP) 3A4 are used for in vitro investigations of drug metabolism and potential drug–drug interactions. The aim of the present study was to determine the relationship between 10 commonly used CYP3A4 probes using modifiers with a range of inhibitory potency.
Methods
The effects of 34 compounds on CYP3A4-mediated metabolism were investigated in a recombinant CYP3A4 expression system. Inhibition of erythromycin, dextromethorphan and diazepam N-demethylation, testosterone 6β-hydroxylation, midazolam 1-hydroxylation, triazolam 4-hydroxylation, nifedipine oxidation, cyclosporin oxidation, terfenadine C-hydroxylation and N-dealkylation and benzyloxyresorufin O-dealkylation was evaluated at the apparent Km or S50 (for substrates showing sigmoidicity) value for each substrate and at an inhibitor concentration of 30 μm.
Results
While all CYP3A4 probe substrates demonstrate some degree of similarity, examination of the coefficients of determination, together with difference and cluster analysis highlighted that seven substrates can be categorized into two distinct substrate groups. Erythromycin, cyclosporin and testosterone form the most closely related group and dextromethorphan, diazepam, midazolam and triazolam form a second group. Terfenadine can be equally well placed in either group, while nifedipine shows a distinctly different relationship. Benzyloxyresorufin shows the weakest correlation with all the other CYP3A4 probes. Modifiers that caused negligible inhibition or potent inhibition are generally comparable in all assays, however, the greatest variability is apparent with compounds causing, on average, intermediate inhibition. Modifiers of this type may cause substantial inhibition, no effect or even activation depending on the substrate employed.
Conclusions
It is recommended that multiple CYP3A4 probes, representing each substrate group, are used for the in vitro assessment of CYP3A4-mediated drug interactions.
Keywords: allostery, CYP3A4, cytochrome P450 inhibition, in vitro probes
Introduction
Cytochrome P450 (CYP) is responsible for the metabolism of many drugs and is the target for a number of drug interactions of significant clinical concern. As a consequence in vitro studies using human liver tissue or recombinant enzymes are widely carried out to predict potential drug–drug interactions involving CYP prior to studies in vivo [1]. An effective in vitro screen must be a reliable predictor for the enzyme in question. Generally one probe substrate is used to characterize the inhibition of each of the various CYPs. It is becoming increasingly apparent that CYP3A4 demonstrates a number of phenomena that make this simple approach unreliable. Many in vitro probes for CYP3A activity have been established, including erythromycin (ER) [2], nifedipine (NF) [3], midazolam (MZ) [4], diazepam (DZ) [5], steroids [6], terfenadine (TF) [7] and cyclosporin (CY) [8].
Several groups have attempted to correlate the activities of various in vivo probes of CYP3A4 activity [9, 10]. In some instances a weak correlation has been found between two CYP3A substrates, e.g. ER and CY [11, 12]. However more commonly, the findings with one in vivo probe fails to accurately predict the metabolism of other CYP3A substrates [13–19]. The reasons for the lack of correlation have not been defined, however, they are likely to include extrahepatic metabolism [20] as well as other differences in the pharmacokinetic properties of the various probes.
The substrates of CYP3A4 are structurally diverse and exhibit a wide range of sizes and affinities, some also show atypical kinetic profiles including positive cooperativity [21] and substrate inhibition [4]. The interactions between CYP3A4 and its substrates and inhibitors are thought to be complex, and may result in inhibition of a competitive, noncompetitive or uncompetitive nature [10], partial inhibition [22], irreversible inactivation by mechanism-based inhibition [23] or activation [21] depending on the combination investigated. It has been suggested that the complex effects observed with substrates of CYP3A4 are attributable to the binding of multiple substrates within the active site of the enzyme [21, 24, 25]. As a consequence, the interactions observed with one CYP3A4 probe may not be representative of those observed with other CYP3A4 substrates. This may impact significantly on the extrapolation of drug interactions from the in vitro to in vivosituation or from one CYP3A4 substrate to another in vitro or in vivo.
The objectives of the present studies were to compare the effects of a set of chemical modifiers (n = 34), known to cause a range of inhibition values with at least one CYP3A4 assay (dextromethorphan (DX)), on the metabolism of 10 CYP3A4 substrates. The substrates chosen (ER, CY, DX, DZ, TS, MZ, triazolam (TZ), TF, NF and benzyloxyresorufin (BR)) have all been used as in vitro probes of CYP3A4 activity [2–8, 26–28], and well-established, validated assays are available for each. The results from each CYP3A4 assay were statistically compared to ascertain interrelationships between the various CYP3A4 substrates, and to aid the choice of the most representative assay(s) for predicting the modification of CYP3A4 activity.
Methods
Chemicals
[14C]-N-methyl ER was purchased from Dupont (Stevenage, UK), [14C]-N-methyl DZ and [3H]-CY were purchased from Amersham Life Science Ltd (Little Chalfont, Bucks, UK) and [14C]-N-methyl DX was supplied by SmithKline Beecham Pharmaceuticals (Welwyn, UK). Unlabelled substrates (ER, DZ, DX, TS, TF, MZ, CY, NF, TZ, BR and ethoxyresorufin), authentic metabolite standards (1-hydroxy MZ, 4-hydroxy MZ, 6β-hydroxy TS, TF alcohol (TFA), azacyclonol (TFZ) and oxidized (NF) and inhibitors (see Table 1) were obtained from Sigma Chemical Company (Poole, UK) with the exception of oxidized NF (UltraFine Chemicals, Manchester, UK), azacyclonol, cisapride, itraconazole (Janssen Chimica, Beerse, Belgium), metoclopramide (Wako Chemical Industries, Richmond, USA), nimodipine, nitrendipine (Tocris Cookson, Bristol, UK), prazepam (Warner Company, Pontypool, UK), propofol (Aldrich Chemicals Ltd, Gillingham, UK) and TF alcohol (a gift from Pfizer Ltd, Sandwich, Kent). All other reagents were of at least analytical grade. Microsomes from a human β-lymphoblastoid cell line engineered to express a recombinant human CYP3A4 and CYP reductase (CPR) were obtained from the Gentest Corporation (Woburn, MA, USA).
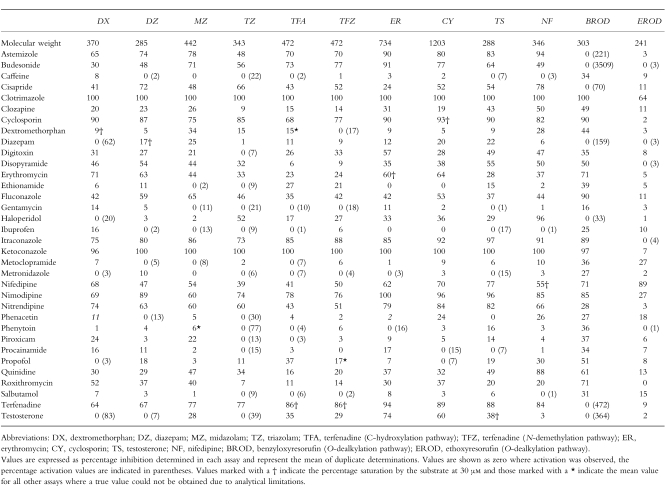
Table 1.
Modification of CYP3A4-mediated metabolism (10 substrates and EROD as a negative control) by 34 pharmaceutical agents.
Incubation conditions
The incubation times and protein concentrations employed were within the linear range for each assay. Experiments were carried out using a 0.25 ml reaction volume (duplicate incubations) containing 25–62.5 pmol CYP ml−1 of 0.05 m potassium phosphate buffer, pH 7.4 in the presence of an NADPH regenerating system (final concentration in each incubation; 1.2 mm NADP+, 5.5 mm glucose-6-phosphate, 0.4% (w/v) sodium bicarbonate and approximately 0.3 units glucose-6-phosphate dehydrogenase). The substrate (concentration defined below) and inhibitor (final concentration, 30 μm) were added to each incubation in acetonitrile or phosphate buffer depending upon the solubility of each, with the exception of the BR and ethoxyresorufin assays, where they were added in dimethylsulphoxide (DMSO). The final solvent concentration was 1% (v/v) in all cases. All samples were preincubated at 37° C for 5 min in a shaking water bath and each reaction was initiated by the addition of substrate (ER) or regenerating system (all others). Each reaction was terminated at the end of the incubation period and the metabolic products quantified as described below. Solvent control samples without inhibitor were also included for each assay. The analytical assays used, under the conditions described below, gave a coefficient of variation (n = 6) below 10%.
Analytical details for CYP3A4 probes
(a) Erythromycin
Experiments with [12C]/ [14C]-N-methyl ER (2.04 GBq/mmol, 13 KBq/incubation) were conducted at a protein concentration of 0.4 mg ml−1 at a final concentration of 20 μm (Km = 52 μm). Each reaction was terminated after 6 min by the addition of 10% (w/v) trichloroacetic acid. Following sedimentation of the precipitated proteins, aliquots (200 μl) were analysed by solid phase extraction (SPE) of the metabolic products, [14C]-formaldehyde and [14C]-formic acid using a Gilson ASPEC XL SPE Robot and EnviCARB™ SPE columns (size 3cc, Supelclean, Supelco, Poole, UK). The metabolic products were eluted with 2 ml h.p.l.c. grade water and the eluate was radioassayed and the amount of product was calculated from the d min−1 value after correction for the background d min−1 in blank incubates. The control activity was 0.6 pmol min−1 pmol−1 CYP.
(b) Dextromethorphan
Experiments with [12C]/ [14C]-N-methyl DX (2.01 GBq/mmol, 15.5 KBq/incubation) were conducted at a protein concentration of 0.4 mg ml−1 at a final concentration of 300 μm (Km = 660 μm). Each reaction was terminated after 30 min by the addition of 10% (w/v) trichloroacetic acid and the metabolic products, [14C]-formaldehyde and [14C]-formic acid, were analysed by SPE as for ER. The control activity was 1.07 pmol min−1 pmol−1 CYP.
(c) Diazepam
Experiments with [14C]-N-methyl DZ (2.11 GBq/mmol, 10 KBq/incubation) were conducted at a protein concentration of 1 mg ml−1 at a final concentration of 100 μm (S50 = 152 μm). Each reaction was terminated after 30 min by the addition of 10% (w/v) trichloroacetic acid and the metabolic products, [14C]-formaldehyde and [14C]-formic acid, were analysed by SPE as for ER. The control activity was 0.74 pmol min−1 pmol−1 CYP.
(d) Cyclosporin
Experiments with [3H]-cyclosporin (296 GBq/mmol, 74 KBq/incubation) were conducted at a protein concentration of 0.75 mg ml−1 at a final concentration of 1 μm (Km = 1 μm). Each reaction was terminated after 15 min by the addition of 100 μl ice-cold acetonitrile. Following sedimentation of the precipitated proteins, aliquots (200 μl) were analysed by h.p.l.c. with radiochemical detection using a 5 μm Ultrasphere ODS column (24 cm×4.6 mm: Beckman, High Wycombe, UK) at a temperature of 70° C. Initial chromatographic conditions were Water: Acetonitrile (45:55, flow rate 1.2 ml min−1), followed by a linear increase to 65% acetonitrile between 5 and 15 min, remaining at 65% until 18 min, followed by a linear increase to 83% between 18 and 30 min, remaining at 83% until 35 min, then returning to original run conditions at 36 min, followed by a 5 min re-equilibration period. Oxidized CY was quantified as a percentage of total radioactivity. The control activity was 0.11 pmol min−1 pmol−1 CYP.
(e) Testosterone
Experiments with [14C]-TS (2.02 GBq/ mmol, 4 KBq/incubation) were conducted at a protein concentration of 0.5 mg ml−1 at a final concentration of 50 μm (S50 = 51 μm). Each reaction was terminated after 15 min by the addition of 100 μl ice-cold acetonitrile. Following sedimentation of the precipitated proteins, aliquots (200 μl) were analysed by h.p.l.c. with radiochemical detection using a 4 μm C18 Novapak column (15 cm ×3.9 mm: Waters, Watford, UK) at a temperature of 50° C. The isocratic chromatographic conditions were methanol/water (25:75): methanol/water/acetonitrile (64:30:6) in a ratio of 20:80 delivered at a flow rate of 1 ml min−1. 6β-hydroxy TS was quantified as a percentage of the total radioactivity; its retention time was verified with that of an authentic standard. The control activity was 7.11 pmol min−1 pmol−1 CYP.
(f) Nifedipine
Experiments with NF were conducted at a protein concentration of 0.5 mg ml−1 at a final concentration of 25 μm (Km = 13 μm). Each reaction was terminated after 10 min by the addition of 150 μl ice-cold methanol. Following sedimentation of the precipitated proteins, aliquots (200 μl) were analysed by h.p.l.c. with u.v. detection (270 nm) using a 5 μm Prodigy ODS2 column (25 cm×4.6 mm: Phenomenex, Macclesfield, UK) at a temperature of 40° C. The isocratic chromatographic conditions were 0.1% trifluoroacetic acid in water: methanol (45:55, flow rate 1 ml min−1). Oxidized NF was quantified by comparison of peak areas with those of a calibration curve. The control activity was 0.96 pmol min−1 pmol−1 CYP.
(g) Terfenadine
Experiments with TF were conducted at a protein concentration of 0.5 mg ml−1 at a final concentration of 5 μm (Km = 7 μm for TFA and 4 μm for TFZ). Each reaction was terminated after 15 min by the addition of 150 μl ice-cold methanol. Following sedimentation of the precipitated proteins, aliquots (200 μl) were analysed by h.p.l.c. with fluorescence detection (excitation wavelength 230 nm and emission wavelength 280 nm) using a 5 μm Techsphere CN column (25 cm×4.6 mm: HPLC Technology, Macclesfield, UK) at ambient temperature. The isocratic chromatographic conditions were ammonium acetate (0.1 m): acetonitrile: methanol (64:23:13 adjusted to pH 5.7 with glacial acetic acid, flow rate 1 ml min−1). TFA and TFZ were quantified by comparison of peak areas with those of a calibration curve. The control activities were 0.70 and 0.32 pmol min−1 pmol−1 CYP for TFA and TFZ, respectively.
(h) Midazolam
Experiments with MZ were conducted at a protein concentration of 0.5 mg ml−1 at a final concentration of 5 μm (Km = 5 μm). Each reaction was terminated after 5 min by the addition of 125 μl ice-cold methanol. Following sedimentation of the precipitated proteins, aliquots (200 μl) were analysed by h.p.l.c. with u.v. detection (240 nm) using a 5 μm Supelcosil LC-ABZ column (15 cm×4.6 mm: Supelco, Poole, UK) at ambient temperature. The isocratic chromatographic conditions were 0.025 m ammonium acetate (pH 5.0): acetonitrile (65:35, flow rate 1.2 ml min−1). 1-hydroxy MZ was quantified by comparison of peak areas with those of a calibration curve. The levels of 4-hydroxy MZ were too low to quantify in the presence of the inhibitors. The effects of phenytoin could not be determined due to interference with the quantification of 1-hydroxy MZ. The control activity was 1.54 pmol min−1 pmol−1 CYP.
(i) Triazolam
Experiments with TZ were conducted at a protein concentration of 1 mg ml−1 at a final concentration of 200 μm (S50 = 200 μm). Each reaction was terminated after 30 min by the addition of 1 ml acetonitrile containing 300 ng ml−1 alprazolam as an internal standard. Samples were extracted following the addition of 50 μl ethanol and 1 ml ammonium acetate, pH 2.5. TZ 4-hydroxylation was measured by LC/MS/MS with positive ion electrospray ionization. Concentrations were expressed relative to a TZ standard curve, due to the unavailability of an authentic metabolite standard, and were expressed as arbitrary units.
(j) Benzyloxyresorufin and ethoxyresorufin
The O-dealkylation of benzyloxyresorufin (BROD) by CYP3A4 and the O-dealkylation of ethoxyresorufin (EROD) by CYP1 A2 (included as a negative control) were measured using methods based on those of Crespi et al. [28]. Experiments (duplicate incubations) were carried out in 96-well plates containing 0.12 pmol CYP/ml (CYP3A4) and 0.16 pmol CYP/ml (CYP1 A2). BROD and EROD were added in DMSO to achieve final concentrations of 3 μm (BROD, Km = 3 μm) or 0.5 μm (EROD, Km = 0.5 μm). The multiwell plates were preincubated at 37° C in a Cytofluor Multiwell Plate Reader (Series 4000, Perseptive Biosystems, Framingham, USA) for 5 min. Prewarmed cofactor solution (25 μl) was added to each well and the fluorescence was measured with an excitation wavelength of 530 nm and an emission wavelength of 590 nm. The plates were scanned for 10 cycles at 1 min intervals. Percentage inhibition values were obtained by comparison with the rates in control wells. BROD activity was determined in baculovirus Supersomes™ expressing CYP3A4 and CPR (also obtained from the Gentest Corporation, Woburn, MA, USA) due to the low metabolic turnover and large variability between controls and duplicate incubations in β-lymphoblastoid microsomes.
Data analysis
Results were expressed as percentage inhibition of the control activity. Mean control values were determined for each experiment. Compounds that demonstrated activation were expressed as 0% inhibition for the purposes of the correlation and cluster analyses. The data from two assays were compared by linear regression and the coefficient of determination (r 2) calculated. Differences between the percentage inhibition observed for each compound within each assay were calculated. All data sets were statistically compared and grouped by cluster analysis using STATISTICA (v 5.1) for Windows (StatSoft Inc, Tulsa, USA). The algorithms for joining data (tree clustering) were used to form successively larger clusters, using Euclidian distances as a measure of their similarity. Clustering of the data sets was determined by single linkage, and the results were presented as a hierarchical tree, where the horizontal axis denotes the linkage distance. Cluster analysis requires that all data sets are complete (i.e. 12 data sets containing all 34 data points, n = 408). In the three cases where data points were missing due to analytical interference, hence a mean value from all other determinations with that particular substrate was included. Where a modifier was also studied as a probe substrate (n = 8), the value for percentage inhibition was taken as the percentage saturation of the enzyme at the concentration used (30 μm), where Vmax is equivalent to 100% of the control activity. In both cases the numbers generated little deviation from the values obtained in the remaining assays.
Results
The compounds studied demonstrated a wide range of inhibition of CYP3A4-mediated metabolism and in some cases activation rather than inhibition was seen (Table 1). The data sets for the inhibition of each CYP3A4 substrate were compared by correlation analysis. Selected graphs showing the correlation of the inhibition data for the 34 compounds are shown in Figure 1. Each graph shows the line of identity. The coefficient of determination (r2) was calculated for each pair of CYP3A4 assays using values for percentage inhibition (Table 2). The values for modifiers which were also used as substrates and data missing due to analytical interference are not shown on the correlation graphs and were not included in the determination of ther2values. Activation values were included as zero inhibition on the correlation graphs and for the determination ofr2values. The inhibition of EROD in CYP1A2 β-lymphoblastoid microsomes by the 34 compounds was included as a negative control. All CYP3A4 probe substrates showed a poor correlation with this assay.
Figure 1.
Selected correlation graphs for the inhibition of CYP3A4-mediated metabolism. Values represent the mean of duplicate determinations.
Table 2.
Calculated r 2 values between 10 CYP3A4 assays.
Regression analysis was used to calculate percentage inhibition values for one probe substrate based on the inhibitory values of another probe. Examination of the differences between the inhibitory values observed for each compound allowed an alternative view of the degree of correlation between two probe substrates. Figure 2 illustrates some examples of these difference plots.
Figure 2.
Analysis of the differences between percentage inhibition values for the correlation between selected CYP3A4 substrates. Arbitrary cut off regions of ±10% are shown on each graph.
The degree of similarity between the 10 CYP3A4 probes was further evaluated by performing a cluster analysis; the resulting hierarchical tree is shown in Figure 3. The probe substrates can be categorized into two major groups, comprising of DX, DZ, MZ and TZ in one group and ER, CY and TS in another. TF, NF and BROD do not fall into either of these discrete groups, and do not correlate with each other. The range of inhibition and/or activation values observed for selected modifiers are shown in Figure 4. As the values are expressed as the percentage change from control, the top and bottom sectors of each graph illustrate that both activation and inhibition of metabolism, respectively, can occur with the same modifier depending on the probe substrate selected.
Figure 3.
Cluster analysis for 10 CYP3A4 substrates determined by single linkage using Euclidian distances to denote the degree of similarity between each variable.
Figure 4.
Modification of CYP3A4 metabolism by selected inhibitors. Substrates are shaded according to the groups defined by the cluster analysis. Percentage activation values are shown at the top of the bar where activation exceeded 100%.
Discussion
Several different CYP3A4 substrates have been used as probes of CYP3A4, however, existing reports in the literature are difficult to compare due to the varying experimental conditions employed. Furthermore many investigations utilize human liver microsomes, where contributions by other CYPs may confound the interpretation of the data. Heterologous expression systems have a distinct advantage in containing a single CYP isoform, and hence the effects on an individual isoform can be more clearly seen than with human liver microsomes.
Inhibition studies often focus on the determination of IC50 and Ki values, however, with a large bank of chemical modifiers and many CYP3A4 assays, both analysis time and resource are major issues when quantifying a wide range of substrate and inhibitor concentrations. For this reason, a single substrate—single inhibitor strategy was selected for the present study. The use of a single substrate concentration near or at the apparent Km or S50 (for substrates showing sigmoidal kinetics) was rationalized on the basis of the relationship between the IC50 and the Ki as described by the Cheng-Prusoff equation [29]. At the apparent Km, the IC50 value is equal to or twice the value of the Ki depending on whether the inhibition mechanism can be described by a noncompetitive or competitive mechanism, respectively. For substrates showing sigmoidicity the relationship is more complex [30], at the S50 when the Hill coefficient is equal to 2, the IC50 value is 1.4-fold lower than the Ki. Therefore at these substrate concentrations each assay will be sensitive to inhibition regardless of the mechanism of inhibition. The concentrations selected for ER, DZ and DX were below the Km to optimize the detection sensitivity for the inhibition of [14C]-formaldehyde and [14C]-formic acid production. A single inhibitor concentration of 30 μm was selected as previous experience of inhibition experiments suggests that this concentration will provide the most dynamic range for determination of inhibition with CYP3A4.
The data from the present study show that while each of the 10 CYP3A4 probe substrates investigated do show a degree of similarity there are a number of marked differences. As a consequence the probes can be categorized into distinct substrate groups based on correlation, difference and cluster analysis. This situation is summarized by the cluster diagram (Figure 3). All assays are within 50 linkage units of each other, with the exception of BR, yet distinct groupings are apparent. As might be anticipated, the groups defined by these analyses relate to the same chemical class, for example there is a benzodiazepine group (which includes DX as well as DZ, MZ and TZ), and also a large molecular weight group (which includes TS as well as ER and CY). TF is linked to both of these discrete groups, while NF and BROD are clearly not representative of any of the other CYP3A4 assays, and they also do not correlate with each other.
Visual inspection of the correlation graphs demonstrate that substrates within the same group clearly show a strong relationship, for example ER vs CY (Figure 1a) and DX vs DZ (Figure 1b). The difference plots for substrates within the same group (Figure 2a and 2b) show relatively little scatter of the data points about the zero line, with most points falling within the ±10% limit. The two TF metabolic routes are highly correlated (Figures 1c and 2c), suggesting that inhibition of TF metabolism is not regioselective. The correlation across the two defined groups is markedly poorer, for example DZ vs ER, where there is more scatter about the line of unity (Figure 1d) and a greater number of points show larger deviations from the ±10% limit on the difference plot (Figure 2d). Generally, the difference plots for TF (both pathways) show a larger number of outlying points falling below the zero line (for example CY and DZ in Figure 2e,f), indicating that inhibition by the modifiers was often under-predicted in the TF assay compared with substrates in the other defined groups.
All substrates showed a marked lack of correlation with NF, for example NF vs ER (Figure 1e) and NF vs DX (Figures 1f and 2g) demonstrating that NF did not fall into either of the other substrate categories. The lack of correlation with NF results in a general scatter of the data points at all levels of inhibition and a few marked outliers that have widely differing values in the different assays. The poorest correlations are observed with the BROD assay, where there is large scatter about the line of unity (for example ER vs BROD shown in Figure 1g) and very few points falling within the ±10% limit on the difference plot (Figure 2h). Very lowr2values were apparent between BROD and all other assays (0.11–0.36). As expected the negative control EROD showed no correlation with any of the CYP3A4 assay data, an example plot is shown in Figure 1h (EROD vs TS).
Ther2determinations (Table 2) and the cluster analysis (Figure 3) exclude the modifiers showing extensive activation, however, even without these data points BR is the substrate showing the greatest deviations from all other CYP3A4 assays. Interpretation of the data by cluster analysis has some limitations, since the values for activation have not been included and this method is sensitive to the number of data points used. However our interpretation of the cluster analysis is supported by the conclusions from both the correlation and difference analyses.
A comparison of the effects of each modifier across the range of CYP3A4 assays show how the extent of modification can vary depending on the assay being used. Compounds that show little or no inhibition of CYP3A4 are fairly consistent for all CYP3A4 assays studied, for example salbutamol (Figure 4a). Likewise, chemicals that show extensive inhibition of CYP3A4 metabolism at the concentration studied, also show similar results with each CYP3A4 assay, these include the azole antifungals, itraconazole (Figure 4b) and ketoconazole. Only a few exceptions are seen from these trends, and they occur most frequently with the BROD assay, which is clearly not representative of the other CYP3A4 substrates. The degree of inhibition seen for several CYP3A4 substrates is in agreement with the comparisons made by Bourrie et al. [31]. The Ki value determined for KZ inhibition of NF metabolism was 0.015 μm, consistent with the low Ki values reported for KZ inhibition of TS (0.2 μm), CY (0.2–0.3 μm), ER (0.5 μm) and amiodarone (0.3 μm). KZ is routinely used [32] as a potent selective CYP3A4 inhibitor for predicting drug metabolism in human liver microsomes. Our inhibition results for KZ against 10 different CYP3A4 substrates support this and indicate that KZ is a good choice regardless of the substrate utilized. The azole antifungals provide good examples of in vitro inhibition that are consistent with in vivo observations. Significant in vivo drug interactions were observed with CY [33] and with MZ [34], both examples from our two distinct groups of substrates.
The greatest variation in response is observed with the modifiers that show an intermediate level of inhibition. Excluding the activation occurring in the BROD assay, the chemical showing the largest range of modification of CYP3A4 mediated metabolism was haloperidol (Figure 4c), which caused 20% activation in the DX assay, and 96% inhibition in the NF assay. A full characterization of the inhibition of NF oxidation by haloperidol, confirmed that haloperidol has an IC50 value of approximately 0.1 μm, however, its effects in the other CYP3A4 assays were much less marked, with the IC50 in the DX assay being greater than 100 μm. It has been suggested [21, 24] that the large active site of CYP3A4 may accommodate more than one molecule simultaneously resulting in atypical kinetic and inhibition profiles. It is possible that haloperidol has the ability to inhibit the enzyme at more than one site, with the effect being dependent on the substrate present. Another possibility is that the site for inhibition by haloperidol is not available for occupation with certain substrate binding conformations.
Substrates showing activation in some assays, but potent inhibition in others include budesonide and astemizole (Figure 4d,e). These two compounds extensively activated the BROD assay only (3509% and 221% activation, respectively), the reason for such high activation of BROD by these modifiers is unknown. Even though these results were obtained in baculovirus expressed CYP3A4 (selected due to the low turnover of BROD in β-lymphoblastoid microsomes and the associated difficulty in measuring inhibited rates), comparable results (data not shown) were observed for these activators in the β-lymphoblastoid system, where the activity far exceeded the limit of detection of the assay. Thus the activation of the CYP3A4-mediated metabolism of this fluorescent probe is not specific to the baculovirus expression system. The greater activation observed for BROD than with other substrates may be due to the small molecular size of this fluorescent probe, allowing enhanced substrate binding in the presence of other CYP3A4 substrates and hence activation of the reaction. This highlights the need for caution when selecting a single fluorescent probe for use in determining CYP3A4 drug interactions in screening programs, as this isoform seems more prone to activation than other P450 isoforms [21, 24]. Compounds which are also substrates of CYP3A4 for example TS, ER and DZ (Figure 4f,g,h) also show a wide range of inhibition and/or activation effects in the different assays. This may also be a consequence of multiple substrate and modifier molecules binding to the enzyme, resulting in a variety of different effects depending on the exact binding conformations within the active site.
While these experiments have identified two distinct groups of CYP3A4 substrates, NF is seen to be quite distinct from these groups. Further investigations with less frequently used in vitro assays for CYP3A4, for example other dihydropyridines like felodipine, are required to confirm whether more than two distinct groups for CYP3A4 substrates do exist. A report by Soons et al. [35] demonstrated that the pharmacokinetics of three dihydropyridines, felodipine, nitrendipine and NF, were highly correlated in healthy subjects, presumably due to all three being metabolized primarily by CYP3A4. Differences in NF metabolism to that of other CYP3A4 substrates has also been noted in a bank of human liver samples, where relatively lowr2values were obtained for the comparison of NF oxidation with TF metabolism in contrast to other substrates [36].
In conclusion, the selection of a single substrate to investigate the effects of chemical modifiers on CYP3A4 should be approached with some caution. The results presented show that while all CYP3A4 assays do show a degree of similarity, there are some chemical modifiers which show a large range of effects between different CYP3A4 assays. It is therefore possible that the extensive inhibition of CYP3A4 caused by some drugs may under other conditions be either overlooked or underestimated leading to errors in predicting important drug–drug interactions. One solution would be to study all potential in vivo combinations in vitro. However this would be very onerous and on the basis of our analysis would be unnecessary. The current recommendation for investigators screening for inhibition of CYP3A4-mediated metabolism to highlight potential drug–drug interactions is to use three CYP3A4 assays. Selection of a substrate from each group, for example MZ and ER cover the benzodiazepine and macrolide substrate groups, and also give an adequate indication of the effects on TF. Additionally there appears little alternative to using NF per se to accurately detect potential interactions with this particular drug. This would enable a comprehensive characterization of inhibition of CYP3A4 metabolism whilst still maintaining the effectiveness of the in vitro approach to predicting drug interactions.
Acknowledgments
KEK was financially supported by a SmithKline Beecham studentship.
References
- 1.Wrighton SA, Vandenbranden M, Stevens JC, Shipley LA, Ring BJ. In vitro methods for assessing human hepatic drug metabolism: Their use in drug development. Drug Metab Rev. 1993;25:453–484. doi: 10.3109/03602539308993982. [DOI] [PubMed] [Google Scholar]
- 2.Watkins PB, Wrighton SA, Maurel P, et al. Identification of an inducible form of cytochrome P450 in human liver. Proc Natl Acad Sci. 1985;82:6310–6314. doi: 10.1073/pnas.82.18.6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guengerich FP, Martin MV, Beaune PH, Kremers P, Wolff T, Waxman DJ. Characterisation of rat and human liver microsomal cytochrome P450 forms involved in nifedipine oxidation, a prototype genetic polymorphism in oxidative drug metabolism. J Biol Chem. 1986;261:5051–5060. [PubMed] [Google Scholar]
- 4.Kronbach T, Mathys D, Umeno M, Gonzalez FJ, Meyer UA. Oxidation of midazolam and triazolam by human liver cytochrome P450IIIA4. Mol Pharmacol. 1989;36:89–96. [PubMed] [Google Scholar]
- 5.Andersson T, Miners JO, Veronese ME, Birkett DJ. Diazepam metabolism by human liver microsomes is mediated by both S-mephenytoin hydroxylase and CYP3A isoforms. Br J Clin Pharmacol. 1994;38:131–137. doi: 10.1111/j.1365-2125.1994.tb04336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waxman DJ, Attisano C, Guengerich FP, Lapenson DP. Human liver microsomal steroid metabolism: Identification of the major microsomal steroid hormone 6β-hydroxylase cytochrome P450 enzyme. Arch Biochem Biophys. 1988;263:424–436. doi: 10.1016/0003-9861(88)90655-8. [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues AD, Mulford DJ, Lee RD, et al. In vitro metabolism of terfenadine by a purified recombinant fusion protein containing cytochrome P4503A4 and NADPH-P450 reductase. Drug Metab Dispos. 1995;23:765–775. [PubMed] [Google Scholar]
- 8.Pichard L, Fabre I, Fabre G, et al. Cyclosporin A drug interactions: Screening for inducers and inhibitors of cytochrome P450 (cyclosporin A oxidase) in primary cultures of human hepatocytes and in liver microsomes. Drug Metab Dispos. 1990;18:595–606. [PubMed] [Google Scholar]
- 9.Watkins PB. Noninvasive tests of CYP3A enzymes. Pharmacogenetics. 1994;4:171–184. doi: 10.1097/00008571-199408000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Thummel KE, Wilkinson GR. In vitro and in vivo drug interaction involving human CYP3A. Ann Rev Pharmacol Toxicol. 1998;38:389–430. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- 11.Watkins PG, Hamilton TA, Annesley TM, Ellis CN, Kolars JC, Voorhees JJ. The erythromycin breath test as a predictor of cyclosporin blood levels. Clin Pharmacol Ther. 1990;48:120–129. doi: 10.1038/clpt.1990.126. [DOI] [PubMed] [Google Scholar]
- 12.Turgeon DK, Normolle DP, Leichtman AB, Annesley TM, Smith DE, Watkins PB. Erythromycin breath test predicts oral clearance of cyclosporin in kidney transplant recipients. Clin Pharmacol Ther. 1992;52:471–478. doi: 10.1038/clpt.1992.174. [DOI] [PubMed] [Google Scholar]
- 13.Hunt CM, Watkins PG, Saenger P, Stave GM, Barlascini N. Heterogeneity of CYP3A isoforms metabolising erythromycin and cortisol. Clin Pharmacol Ther. 1992;51:18–23. doi: 10.1038/clpt.1992.3. [DOI] [PubMed] [Google Scholar]
- 14.Watkins PB, Turgeon DK, Saenger P, et al. Comparison of urinary 6β-cortisol and the erythromycin breath test as measures of hepatic P450IIIA (CYP3A) activity. Clin Pharmacol Ther. 1992;52:265–273. doi: 10.1038/clpt.1992.140. [DOI] [PubMed] [Google Scholar]
- 15.Kinirons MT, O’Shea D, Downing TE, et al. Absence of correlations among three putative in vivo probes of human cytochrome P4503A activity in young healthy men. Clin Pharmacol Ther. 1993;54:621–629. doi: 10.1038/clpt.1993.199. [DOI] [PubMed] [Google Scholar]
- 16.Kinirons MT, O’Shea D, Groopman JD, Thummel KE, Wood AJJ, Wilkinson GR. Route of administration does not explain the lack of correlation between putative in vivo probes of cytochrome P4503A. Br J Clin Pharmacol. 1994;37:501P. [Google Scholar]
- 17.Krivoruk Y, Kinirons MT, Wood AJJ, Wood M. Metabolism of cytochrome P4503A substrates in vivo administered by the same route: Lack of correlation between alfentanil clearance and erythromycin breath test. Clin Pharmacol Ther. 1994;56:608–614. doi: 10.1038/clpt.1994.185. [DOI] [PubMed] [Google Scholar]
- 18.Lown KS, Thummel KE, Benedict PE, Shen DD, Turgeon DK. The erythromycin breath test predicts the clearance of midazolam. Clin Pharmacol Ther. 1995;57:16–24. doi: 10.1016/0009-9236(95)90261-9. [DOI] [PubMed] [Google Scholar]
- 19.Stein CM, Kinirons MT, Pincus T, Wilkinson GR, Wood AJJ. Comparison of the dapsone recovery ratio and the erythromycin breath test as in vivo probes of CYP3A activity in patients with rheumatoid arthritis receiving cyclosporin. Clin Pharmacol Ther. 1996;59:47–51. doi: 10.1016/S0009-9236(96)90023-5. [DOI] [PubMed] [Google Scholar]
- 20.Lown KS, Kolars JC, Thummel KE, Barnett JL, Kunze KL. Inter-patient heterogeneity in expression of CYP3A4 and CYP3A5 in small bowel: Lack of prediction by the erythromycin breath test. Drug Metab Dispos. 1994;22:947–955. [PubMed] [Google Scholar]
- 21.Ueng Y-F, Kuwabara T, Chun Y-J, Guengerich FP. Cooperativity in oxidations catalysed by cytochrome P450 3A4. Biochemistry. 1997;36:370–381. doi: 10.1021/bi962359z. [DOI] [PubMed] [Google Scholar]
- 22.Wang RW, Newton DJ, Scheri TD, Lu AYH. Human cytochrome P450 3A4-catalysed testosterone 6β-hydroxylation and erythromycin N-demethylation: Competition during catalysis. Drug Metab Dispos. 1997;25:502–507. [PubMed] [Google Scholar]
- 23.Bensoussan C, Delaforge M, Mansuy D. Particular ability of cytochrome P450, 3A to form inhibitory P450–iron–metabolite complexes upon metabolic oxidation of aminodrugs. Biochem Pharmacol. 1995;49:591–602. doi: 10.1016/0006-2952(94)00477-4. [DOI] [PubMed] [Google Scholar]
- 24.Shou M, Grogan J, Mancewicz JA, et al. Activation of CYP3A4: Evidence for the simultaneous binding of two substrates in a cytochrome P450 active site. Biochemistry. 1994;33:6450–6455. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- 25.Korzekwa KR, Krishnamachary N, Shou M, et al. Evaluation of atypical cytochrome P450 kinetics with two-substrate models: Evidence that multiple substrates can simultaneously bind to cytochrome P450 active sites. Biochemistry. 1998;37:4137–4147. doi: 10.1021/bi9715627. [DOI] [PubMed] [Google Scholar]
- 26.Gorski JC, Jones DR, Wrighton SA, Hall SD. Characterisation of dextromethorphan N-demethylation by human liver microsomes: Contribution of the cytochrome P450, 3A subfamily. Biochem Pharmacol. 1994a;48:173–182. doi: 10.1016/0006-2952(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 27.Von Moltke LL, Greenblatt DJ, Harmatz JS, et al. Triazolam biotransformation by human liver microsomes in vitro: Effects of metabolic inhibitors and clinical conformation of a predicted interaction with ketoconazole. J Pharmacol Exp Ther. 1996;276:370–379. [PubMed] [Google Scholar]
- 28.Crespi CL, Miller VP, Penman BW. Microtiter plate assays for inhibition of human drug metabolising cytochromes P450. Anal Biochem. 1997;248:188–190. doi: 10.1006/abio.1997.2145. [DOI] [PubMed] [Google Scholar]
- 29.Craig DA. The Cheng-Prusoff relationship: Something lost in the translation. Trends Pharmacol Sci. 1993;14:89–91. doi: 10.1016/0165-6147(93)90070-z. [DOI] [PubMed] [Google Scholar]
- 30.Leff P, Dougall IG. Further concerns over Cheng-Prusoff analysis. Trends Pharm Sci. 1993;14:110–112. doi: 10.1016/0165-6147(93)90080-4. [DOI] [PubMed] [Google Scholar]
- 31.Bourrie M, Meunier V, Berger Y, Fabre G. Cytochrome P450 inhibitors as a tool for the investigation of metabolic reactions catalysed by human liver microsomes. J Pharmacol Exp Ther. 1996;277:321–332. [PubMed] [Google Scholar]
- 32.Newton DJ, Wang RW, Lu AYH. Cytochrome P450 inhibitors: Evaluation of specificities in the in vitro metabolism of therapeutic agents by human liver microsomes. Drug Metab Dispos. 1995;23:154–158. [PubMed] [Google Scholar]
- 33.Gomez DY, Wacher VJ, Tomlanovich SJ, Herbert MF, Benet LZ. The effects of ketoconazole on the intestinal metabolism and bioavailability of cyclosporin. Clin Pharmacol Ther. 1995;58:15–29. doi: 10.1016/0009-9236(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 34.Olkkola KT, Ahonen J, Neuvonen PJ. The effect of the systemic antimycotics, itraconazole and fluconazole on the pharmacokinetics and pharmacodynamics of intravenous and oral midazolam. Anesth Analg. 1996;82:511–516. doi: 10.1097/00000539-199603000-00015. [DOI] [PubMed] [Google Scholar]
- 35.Soons PA, Mulders TM, Uchida E, Schoemaker HC, Cohen AF, Breimer DD. Stereoselective pharmacokinetics of oral felodipine and nitrendipine in healthy subjects: Correlation with nifedipine pharmacokinetics. Eur J Clin Pharmacol. 1993;44:163–169. doi: 10.1007/BF00315475. [DOI] [PubMed] [Google Scholar]
- 36.Yun CH, Okerholm RA, Guengerich FP. Oxidation of the antihistaminic drug terfenadine in human liver microsomes: Role of cytochrome P450, 3A (4) in N–dealkylation and C–hydroxylation. Drug Metab Dispos. 1993;21:403–409. [PubMed] [Google Scholar]