Abstract
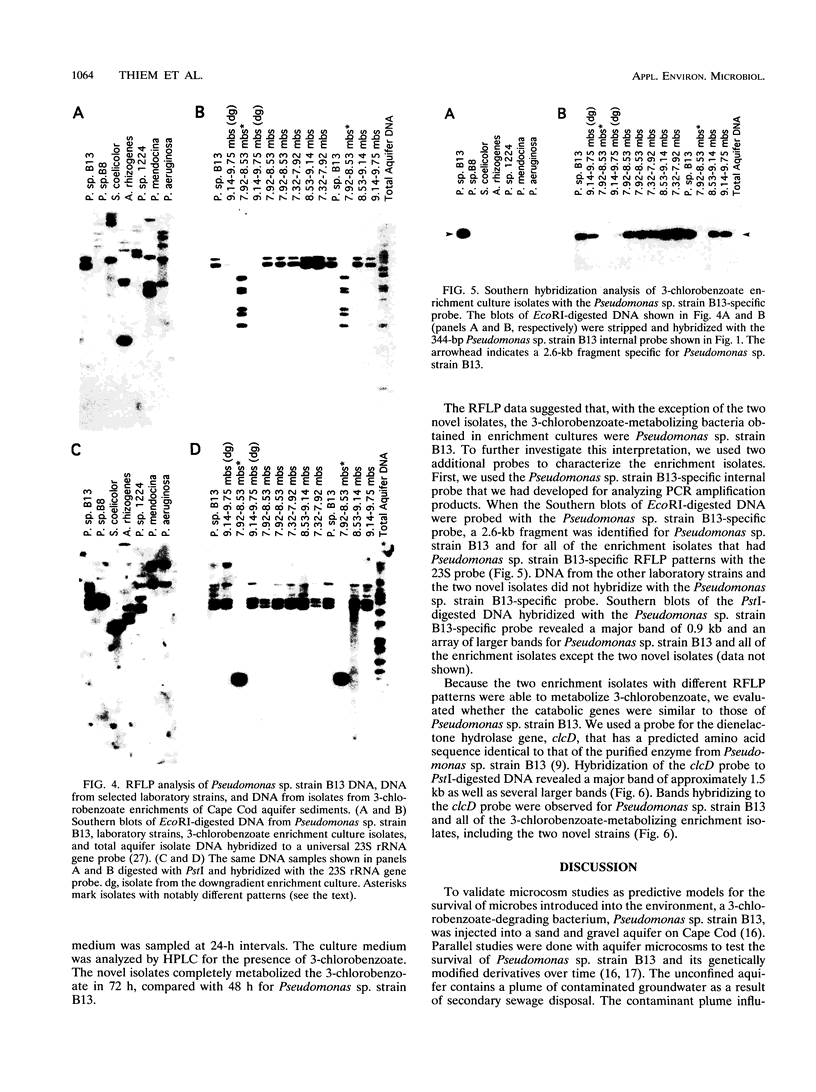
A PCR primer set and an internal probe that are specific for Pseudomonas sp. strain B13, a 3-chlorobenzoate-metabolizing strain, were developed. Using this primer set and probe, we were able to detect Pseudomonas sp. strain B13 DNA sequences in DNA extracted from aquifer samples 14.5 months after Pseudomonas sp. strain B13 had been injected into a sand and gravel aquifer. This primer set and probe were also used to analyze isolates from 3-chlorobenzoate enrichments of the aquifer samples by Southern blot analysis. Hybridization of Southern blots with the Pseudomonas sp. strain B13-specific probe and a catabolic probe in conjunction with restriction fragment length polymorphism (RFLP) analysis of ribosome genes was used to determine that viable Pseudomonas sp. strain B13 persisted in this environment. We isolated a new 3-chlorobenzoate-degrading strain from one of these enrichment cultures. The B13-specific probe does not hybridize to DNA from this isolate. The new strain could be the result of gene exchange between Pseudomonas sp. strain B13 and an indigenous bacterium. This speculation is based on an RFLP pattern of ribosome genes that differs from that of Pseudomonas sp. strain B13, the fact that identically sized restriction fragments hybridized to the catabolic gene probe, and the absence of any enrichable 3-chlorobenzoate-degrading strains in the aquifer prior to inoculation.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bej A. K., Mahbubani M. H., Atlas R. M. Detection of viable Legionella pneumophila in water by polymerase chain reaction and gene probe methods. Appl Environ Microbiol. 1991 Feb;57(2):597–600. doi: 10.1128/aem.57.2.597-600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej A. K., Steffan R. J., DiCesare J., Haff L., Atlas R. M. Detection of coliform bacteria in water by polymerase chain reaction and gene probes. Appl Environ Microbiol. 1990 Feb;56(2):307–314. doi: 10.1128/aem.56.2.307-314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H. J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99(1):61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- Frantz B., Ngai K. L., Chatterjee D. K., Ornston L. N., Chakrabarty A. M. Nucleotide sequence and expression of clcD, a plasmid-borne dienelactone hydrolase gene from Pseudomonas sp. strain B13. J Bacteriol. 1987 Feb;169(2):704–709. doi: 10.1128/jb.169.2.704-709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLLOWAY B. W. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955 Dec;13(3):572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- Harvey R. W., Smith R. L., George L. Effect of organic contamination upon microbial distributions and heterotrophic uptake in a Cape Cod, Mass., aquifer. Appl Environ Microbiol. 1984 Dec;48(6):1197–1202. doi: 10.1128/aem.48.6.1197-1202.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesack W., Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992 Aug;174(15):5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Natural genetic transformation of Pseudomonas stutzeri by sand-adsorbed DNA. Arch Microbiol. 1990;154(4):380–385. doi: 10.1007/BF00276535. [DOI] [PubMed] [Google Scholar]
- Regensburger A., Ludwig W., Schleifer K. H. DNA probes with different specificities from a cloned 23S rRNA gene of Micrococcus luteus. J Gen Microbiol. 1988 May;134(5):1197–1204. doi: 10.1099/00221287-134-5-1197. [DOI] [PubMed] [Google Scholar]
- Rojo F., Pieper D. H., Engesser K. H., Knackmuss H. J., Timmis K. N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987 Dec 4;238(4832):1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayler G. S., Layton A. C. Environmental application of nucleic acid hybridization. Annu Rev Microbiol. 1990;44:625–648. doi: 10.1146/annurev.mi.44.100190.003205. [DOI] [PubMed] [Google Scholar]
- Smith G. B., Tiedje J. M. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl Environ Microbiol. 1992 Jan;58(1):376–384. doi: 10.1128/aem.58.1.376-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay R. E., Loeber R., Gagnon C., Charlebois P., Larivée S., LeBlanc M. Disruptive boys with stable and unstable high fighting behavior patterns during junior elementary school. J Abnorm Child Psychol. 1991 Jun;19(3):285–300. doi: 10.1007/BF00911232. [DOI] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992 Feb;58(2):754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]