Abstract
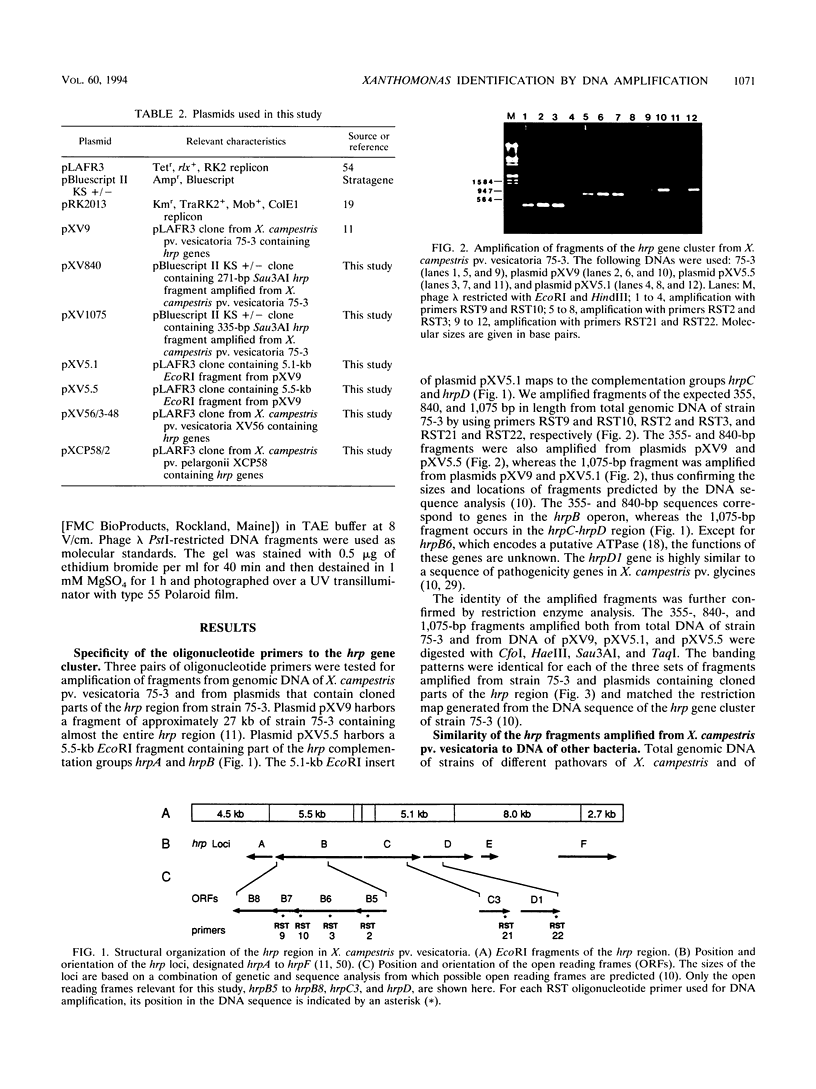
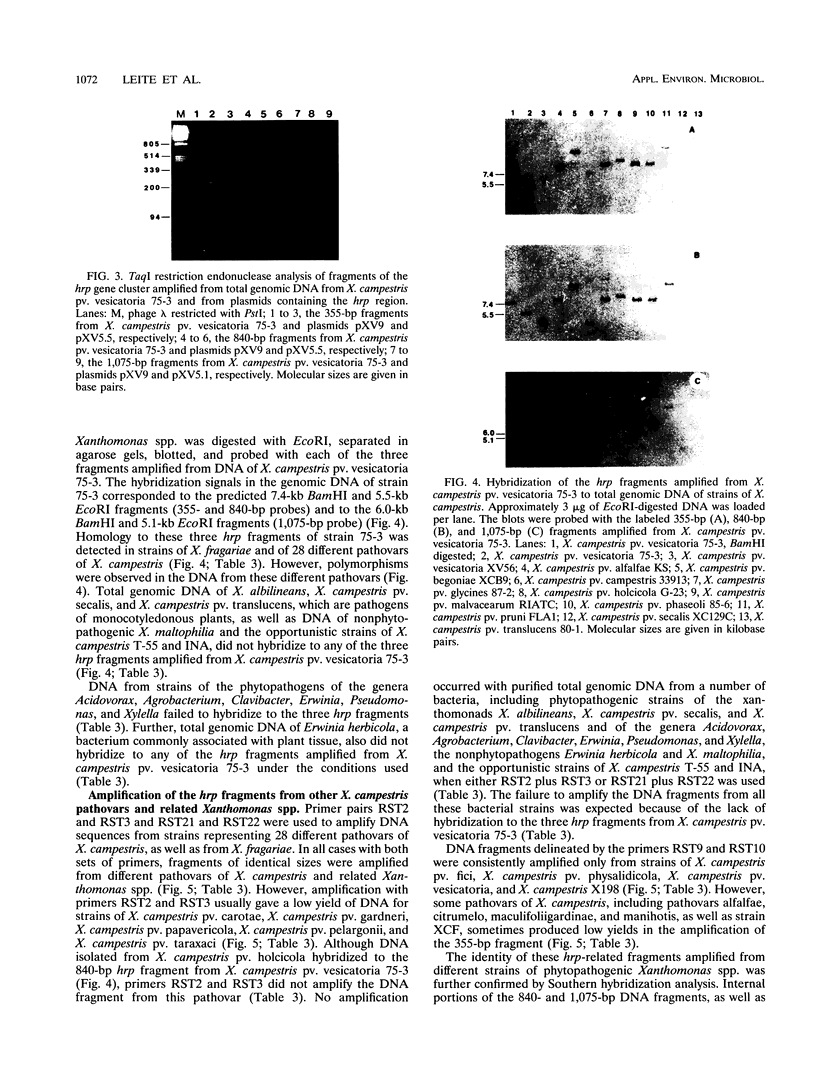
Three pairs of oligonucleotide primers specific for different regions of the hrp gene (hypersensitive reaction and pathogenicity) cluster of Xanthomonas campestris pv. vesicatoria were designed and tested for amplification of DNA isolated from a large number of different bacteria. DNA sequences related to the hrp genes were successfully amplified from X. fragariae and from 28 pathovars of X. campestris. No DNA amplification occurred with genomic DNA from phytopathogenic strains of X. campestris pv. secalis, X. campestris pv. translucens, and X. albilineans or from nonpathogenic opportunistic xanthomonads and phytopathogenic strains of the genera Acidovorax, Agrobacterium, Clavibacter, Erwinia, Pseudomonas, and Xylella. The DNA from those bacteria also failed to hybridize to hrp-specific fragments in Southern blot analysis. DNA fragments amplified with a particular primer pair were of identical size from each of the different phytopathogenic xanthomonads. However, restriction analysis of these fragments by using frequently cutting endonucleases revealed variation in the pattern for these hrp-related fragments amplified from the different Xanthomonas strains. The restriction patterns generated for the different fragments allowed distinction of the strains representing a pathovar or species of phytopathogenic xanthomonads. We believe that DNA amplification with hrp-specific oligonucleotide primers is a highly sensitive and specific method that can be applied for detection and identification of phytopathogenic xanthomonads.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benedict A. A., Alvarez A. M., Pollard L. W. Pathovar-Specific Antigens of Xanthomonas campestris pv. begoniae and X. campestris pv. pelargonii Detected with Monoclonal Antibodies. Appl Environ Microbiol. 1990 Feb;56(2):572–574. doi: 10.1128/aem.56.2.572-574.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereswill S., Pahl A., Bellemann P., Zeller W., Geider K. Sensitive and species-specific detection of Erwinia amylovora by polymerase chain reaction analysis. Appl Environ Microbiol. 1992 Nov;58(11):3522–3526. doi: 10.1128/aem.58.11.3522-3526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher C. A., Van Gijsegem F., Barberis P. A., Arlat M., Zischek C. Pseudomonas solanacearum genes controlling both pathogenicity on tomato and hypersensitivity on tobacco are clustered. J Bacteriol. 1987 Dec;169(12):5626–5632. doi: 10.1128/jb.169.12.5626-5632.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. S. Pressure-jump studies on the length-regulation kinetics of the self-assembly of myosin from vertebrate skeletal muscle into thick filament. Biochem J. 1981 Aug 1;197(2):309–314. doi: 10.1042/bj1970309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fenselau S., Balbo I., Bonas U. Determinants of pathogenicity in Xanthomonas campestris pv. vesicatoria are related to proteins involved in secretion in bacterial pathogens of animals. Mol Plant Microbe Interact. 1992 Sep-Oct;5(5):390–396. doi: 10.1094/mpmi-5-390. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garde S., Bender C. L. DNA probes for detection of copper resistance genes in Xanthomonas campestris pv. vesicatoria. Appl Environ Microbiol. 1991 Aug;57(8):2435–2439. doi: 10.1128/aem.57.8.2435-2439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough C. L., Genin S., Zischek C., Boucher C. A. hrp genes of Pseudomonas solanacearum are homologous to pathogenicity determinants of animal pathogenic bacteria and are conserved among plant pathogenic bacteria. Mol Plant Microbe Interact. 1992 Sep-Oct;5(5):384–389. doi: 10.1094/mpmi-5-384. [DOI] [PubMed] [Google Scholar]
- Hartung J. S., Daniel J. F., Pruvost O. P. Detection of Xanthomonas campestris pv. citri by the polymerase chain reaction method. Appl Environ Microbiol. 1993 Apr;59(4):1143–1148. doi: 10.1128/aem.59.4.1143-1148.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Lim S. M., Shaw P. D. Cloning and characterization of pathogenicity genes from Xanthomonas campestris pv. glycines. J Bacteriol. 1992 Mar;174(6):1923–1931. doi: 10.1128/jb.174.6.1923-1931.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite R. P., Jr, Egel D. S., Stall R. E. Genetic analysis of hrp-related DNA sequences of Xanthomonas campestris strains causing diseases of citrus. Appl Environ Microbiol. 1994 Apr;60(4):1078–1086. doi: 10.1128/aem.60.4.1078-1086.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren P. B., Peet R. C., Panopoulos N. J. Gene cluster of Pseudomonas syringae pv. "phaseolicola" controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J Bacteriol. 1986 Nov;168(2):512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte R., Bonas U. Expression of the Xanthomonas campestris pv. vesicatoria hrp gene cluster, which determines pathogenicity and hypersensitivity on pepper and tomato, is plant inducible. J Bacteriol. 1992 Feb;174(3):815–823. doi: 10.1128/jb.174.3.815-823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal S. E., Jackson L. A., Daniels M. J. Isolation of a Pseudomonas solanacearum-specific DNA probe by subtraction hybridization and construction of species-specific oligonucleotide primers for sensitive detection by the polymerase chain reaction. Appl Environ Microbiol. 1992 Nov;58(11):3751–3758. doi: 10.1128/aem.58.11.3751-3758.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- Van den Mooter M., Swings J. Numerical analysis of 295 phenotypic features of 266 Xanthomonas strains and related strains and an improved taxonomy of the genus. Int J Syst Bacteriol. 1990 Oct;40(4):348–369. doi: 10.1099/00207713-40-4-348. [DOI] [PubMed] [Google Scholar]