Abstract
Essential hypertension is an escalating problem for industrialized populations. It is currently seen as a ‘complex’ genetic trait caused by multiple susceptibility genes the effects of which are modulated by gene‐environment and gene–gene interactions. Nevertheless, the success to date in identifying these susceptibility genes has been very limited. A number of candidates has been proposed, but demonstrating consistently the linkage or association with hypertension has been problematic. The data for angiotensinogen is undoubtedly the most extensive and meta‐analysis has confirmed a significant association overall, although the risk contributed by this gene appears to be modest (odds ratio of 1.2). Identifying further genes – probably conferring even smaller attributable risks – represents a major challenge for future developments in this area. This contrasts markedly with the success that has been achieved in the past 5 years in solving the molecular genetics of a number of rare familial hypertension syndromes. The true incidences of some of these disorders may be higher than first appreciated, but it is still unclear if the genes for these syndromes also play a part in essential hypertension. A more complete understanding of the genetic basis of essential hypertension should be possible in the coming years using new strategies that take advantage of the information provided by the human genome project. This knowledge will irrevocably change the way we approach this disease in terms of its diagnosis, risk assessment for end‐points such as stroke and heart disease, and the customised treatment that might be offered in the future.
Keywords: aldosteronism, angiotensinogen, genes, hypertension, monogenic, mutations, renin
Hypertension as a polygenic trait
Hypertension is a common and increasing public health problem that is estimated to cause as many premature deaths Worldwide as tobacco smoking [1]. The inheritance of hypertension has been debated for many years since Pickering and Platt clashed over the form of the blood pressure distribution. Pickering argued that blood pressure, like other traits such as height or weight, was continuously distributed [2] against Platt's belief that it was bimodal [3]. Their views implied rather different inheritance mechanisms: on the one hand multiple genes and on the other a single (or major) gene controlling blood pressure. With hindsight, the bimodal distribution was largely an artefact of observer bias, and the polygenic basis for blood pressure in the general population is now widely accepted. Indeed, the model of multiple susceptibility genes with environmental interaction is one that can be applied to many common diseases including asthma, diabetes and ischaemic heart disease. However, as a footnote to the Pickering‐Platt debate, we also appreciate that some familial forms of hypertension are inherited as single‐gene disorders, but they are a small proportion of the total cases.
Having settled on the likely model, an assessment is needed of the relative importance of genes vs environment. The genetic influence on a trait, such as blood pressure, is often expressed in terms of heritabilty, i.e. the fraction of the total interindividual variability (or variance) attributable to genes. This index requires comparison of blood pressures between pairs of subjects showing different familial connections and therefore differing degrees of genetic similarity; studies have variously employed extended families, twins or adoptees in an attempt to isolate environmental influences. It is no surprise perhaps that many pitfalls surround studies of heritabilty, but the range reported in the literature is 30–60% for blood pressure [4]. This of course should not be taken to imply that the remainder is environmental, as gene–environment interactions are essentially unresolvable from this figure. Another way of looking at the genetic contribution is in the relative risk for a sibling, i.e. the proportion of the siblings who are themselves hypertensive compared with the general population. This ratio, called the λs, is some 3.5 for hypertension [5]. This may seem a rather small value when compared with a single gene disorder such as cystic fibrosis (λs = 500), but it is within the range of other polygenic disorders such as ischaemic heart disease (λs≅2) and type I diabetes (λs = 15).
The task now facing hypertension genetics is to identify these multiple susceptibility genes and specifically in particular the molecular variants that modulate blood pressure. This short review highlights the important progress that has been so far towards this goal.
Animal models
Research into the genetics of hypertension has made extensive use of inbred animal strains since they enable a level of control over environment and breeding that would be impossible to achieve in human studies. During the 1960s, selecting and inbreeding individuals with high blood pressure established at least six major strains and a much larger number of substrains. Blood pressure is a polygenic trait in these models since cross‐breeding back onto their respective normotensive parental strains has usually produced a continuous distribution of blood pressures in the F1 offspring, and not the Mendelian pattern that Platt might have expected [6, 7]. Estimates of the number of genes operating in cross‐breeding studies have been put at between 2 and 6 [6, 8], although the assumptions in these models are necessarily crude and probably only represent genes with major contributions. The real power of cross‐breeding studies in these models is their ability to localize putative pressor genes by studying the cosegregation of the blood pressure trait with the alleles of known DNA markers. In such a segregation analysis, only markers on the same chromosome as a pressor gene will show nonrandom cosegregation and the tighter this statistical association the closer their proximity. The limited marker maps available for the rat genome initially hampered this approach, but now a number of chromosomal segments (dubbed quantitative trait loci or QTLs) carrying pressor genes have been identified using segregation analysis [9]. The regions covered by the QTLs are generally still too large to consider positionally cloning the causative pressor genes, but by further selective cross‐breeding these QTLs can be moved from one strain and expressed in another. The resulting congenic animals focus the search for the pressor gene to smaller chromosomal segments from where the isolation of pressor genes can begin. This is likely to be greatly helped in the next few years by the ability to rapidly identify differentially expressed genes within the QTLs. This strategy has recently succeeded in identifying a rodent susceptibility gene for another polygenic trait, insulin resistance [10].
There is, of course, no guarantee that identifying the causative genes for rodent QTLs will be relevant to human hypertension, but at least one rat QTL (BP10) is homologous with a region of human chromosome 17 containing both a human QTL [11], and the locus for Gordon's syndrome (see below). The other key finding of these rodent studies has been the realization that despite similar levels of high blood pressure susceptibility to end‐organ damage varies widely between different rodent strains. Hence, end‐organ damage is itself genetically determined and QTLs for renal failure, cardiac hypertrophy, and stroke have been identified that are distinct from pressor ones [12]. Identifying the causative genes could revolutionize our approach to risk assessment in human hypertensives.
Human studies
These have generally followed two main approaches; linkage studies based on allele sharing in affected relative pairs (usually sib‐pairs) or association studies comparing gene frequencies between cohorts of hypertensive and normal subjects. In both cases, either specific candidate genes (suggested from known pathophysiology of the disease) or anonymous DNA markers have been used. Successful sib‐pair studies using DNA markers have often employed them in large panels of several hundred DNA markers equally spaced over the entire genome. The resulting genome‐wide scans have become a standard tool for dissecting complex human traits as it allows detection of novel susceptibility genes that a candidate gene approach would inevitably miss. Evidence implicating a number of genes including angiotensinogen [13], α‐adducin [14], the β2‐adrenoceptor [15], G‐protein beta‐3 [16] and the β‐subunit of the epithelial sodium channel (β‐ENaC, see below) have been obtained using these methods. Nevertheless, the published data are conflicting and highlight some fundamental problems in establishing the cause‐effect relation between a putative susceptibility gene and hypertension. For example, angiotensinogen (AGT) was identified early on as a candidate gene because of the key role played by the renin‐angiotensin system in volume homeostasis; the levels of circulating AGT are also rate limiting for angiotensin II synthesis. The case was first supported by linkage in sib‐pair collections from Paris and Utah [13], and further verified in a smaller Caribbean collection [17]. Subsequently, two sequence variants were identified in the coding region of the AGT gene, T235M (a threonine to methionine substitution at residue 235) and T174M, which were particularly common, in complete linkage disequilibrium, and associated with hypertension. The increased frequency of the 235T variant in hypertensives could also be explained by its influence on AGT levels with 235T homozygotes having plasma levels 20% higher than 235 m homozygotes. Further work has clarified the relation of 235T and expressed levels of the AGT gene product by identifying a promoter variant that affects gene transcription [18]. It is likely that the effect of 235T on plasma AGT levels reflects its linkage disequilibrium with this promoter variant. Despite this apparently compelling evidence, it has not been possible to replicate the original linkage data in either a large European consortium of 690 sib‐pairs [19], or a somewhat smaller sib‐pair collection from Mainland China [20]. The association of the 235T variant with hypertension has also been impossible to establish in some populations, although a recent meta‐analysis using published data on some 5500 subjects reported a significant overall effect; the association is weak, however, with an odds ratio of just 1.2 [21].
These replication problems expose some inherent difficulties with the methods themselves. Firstly, both sib‐pair and association studies face the problem of genetic diversity with the risk attributable to any given gene variant liable to vary widely between different populations. Gene variants that were selected during evolution to conserve salt, for example, may play a much larger role in populations based on African hypertensives. The problem of replicating linkage results between hypertensive sib‐pair collections is not unprecedented and has become an increasing problem in the application of sib‐pairs to other complex diseases such as diabetes [22, 23]. Although the affected sib‐pair strategy is robust, it has low statistical power to detect even modest gene effects. It has been estimated that to replicate a significant linkage in a sib‐pair collection where n genes influence the complex trait would require a sample size (n‐1) times larger; for hypertension with an n of perhaps 6, very large sample sizes indeed will be required for replication [24]. The power of a sib‐pair collection is also dependent on matching within pairs. Affected sib‐pairs are often selected at random, yet selecting sib‐pairs whose blood pressure is highly concordant, or discordant, produces much more powerful collections [25]. A report using this approach has recently appeared and in a secondary analysis did suggested linkage to the region of the AGT gene [26]. By comparison, association studies are generally better powered to detect modest gene effects, although false‐positives are a hazard due to unmatched, and usually unrecognized, genetic differences between the hypertensive and control populations.
What is the future for the human studies? Technological advances in molecular biological tools will play an essential role. Genome–wide association studies are very attractive, but require the genotyping of many thousands of DNA markers in every subject. It is now possible to place all of the necessary probes for such a screen on a single high‐density microarray and these ‘gene‐chips’ should become a routine mapping tool in the near future. It may also be possible to rapidly map chromosomal segments containing susceptibility genes using the emergent method of genome mismatch scanning [27], which will again be aided by gene‐chip technology. But of equal importance in the future will probably be in the choice of study populations focusing in particular on intermediate phenotypes such as renin‐status.
Monogenic hypertension
Undoubtedly the greatest advances in recent years have been in the study of families in which the hypertension is inherited as a simple Mendelian character. The molecular bases for several of these disorders have now been established and are summarized in Table 1. Because of the subsequent impact on our understanding of the physiology of low‐renin hypertension they are worth considering in some detail.
Table 1.
Molecular basis for known familial (monogenic) hypertension syndromes.
| Monogenic syndrome | Mode of inheritance | Gene product | Chromosomal location |
|---|---|---|---|
| Apparent mineralocorticoid excess (AME) | Recessive | Non-functional 11-β-hydroxy steroid dehydrogenase (11β-HSD Type 2) enzyme | 16q |
| Glucocorticoid remediable aldosteronism (GRA) | Dominant | Hybrid aldosterone synthase (CYP11B2) enzyme expressed under ACTH control | 8p |
| Hypertensive forms of congenital adrenal hyperplasia | Recessive | Non-functional forms of the corresponding hydroxylases | 8q21 and 10q24 |
| (11-β and 17-α hydroxylase types) | (CYP11B1 and CYP17A) | ||
| Liddle's Syndrome (pseudo-aldosteronism) | Dominant | Constitutively expressed amiloride-sensitive sodium channel in distal nephron | 16p |
The index cases of these monogenic pedigrees have usually been highlighted because of other biochemical features, such as hypokalaemia or low plasma renin, which provides a useful intermediate phenotype to screen for other affected relatives. Further clues to the identity of the hypertensive gene have come from other unusual phenotypic features such as drug-sensitivity. For example, the affected members of Liddle's original 3-generation pedigree all showed hypertension and biochemical hyperaldosteronism, but the aldosterone levels were curiously low and did not respond to spironolactone [28]. Further investigation revealed that provided salt-intake was low the hypertension responded to amiloride or triamterene [29]. The cloning of the drug target for these diuretics, the amiloride-sensitive epithelial sodium channel (ENac), then provided an obvious candidate gene for the disorder [30]. Lifton and his group in Yale, subsequently showed that Liddle's original pedigree was linked to the locus on chromosome 16 containing the human β subunit for ENaC and identified truncation mutations in this and several other pedigrees [31]. The functionally active ENaC actually consists of three subunits (α, β and γ); the β and γ subunits are adjacent on the short arm of chromosome 16 and truncation mutations in either subunit can cause Liddle's syndrome [32, 33]. The truncations all involve a conserved motif within the C terminal region, which plays an essential role in channel internalization. When the truncated subunits are transfected into Xenopus oocytes there is a marked increase in surface expression of sodium channels and total sodium current [34]. A similar accumulation of apical sodium channels would explain the abnormal constitutive sodium flux in the distal nephron of Liddle's patients. This cannot be observed in vivo, but the increased epithelial sodium transport has been confirmed directly from measurements of potential difference across the nasal mucosa of these patients [35].
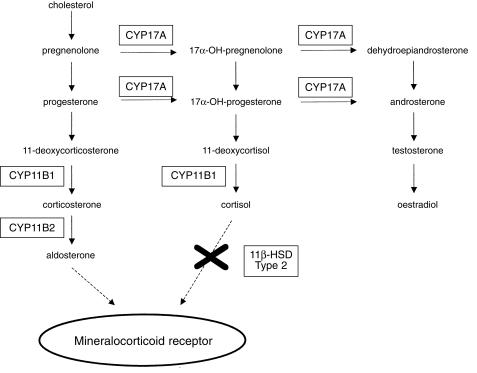
The syndrome of hyperaldosteronism responsive to exogenous glucocorticoids (Glucocorticoid-responsive Aldosteronism, GRA) was first described in 1966 in a father and son. Several large pedigrees have been subsequently described in Scotland, the USA and Australia all probably derived from Northern European founders. The elevated aldosterone levels in GRA are unusual in two respects: (1) they fall steadily during the day mirroring circadian changes in circulating ACTH; (2) they show the same postural response as aldosterone-producing adenomas (or strictly those that are ATII unresponsive) with a paradoxical fall. The molecular defect in GRA is the formation of a hybrid fusion gene of the antilepore type (after the haemoglobinopathy where this type of defect was first identified) [36]. The enzymes responsible for production of aldosterone (see Figure 1), aldosterone synthase (CYP11B2) and 11-β-hydroxylase (CYP11B1), are coded for by adjacent genes on chromosome 8, so the GRA fusion gene is likely to have arisen from nonhomologous pairing and unequal crossover between them. The product is of the form 5′-(promoter and exons 1–3 of CYP11B1)-(exons 5–9 of CYP11B2) with the breakpoints lying upstream of intron 4 in all cases studied to date. In vitro studies have supported this construct by showing that the first 3 exons of CYP11B1 can replace the homologous exons of CYP11B2 without loss of enzymatic function; substitution of > 3 exons causes inactivation of the hybrid gene product. By placing aldosterone production under ACTH regulation, large amounts of hybrid steroids (18-hydroxylase products) are produced from ectopically expressed CYP11B2 in the adrenal glomerulosa, and aldosterone production becomes glucocorticoid suppressible. One interesting feature of the condition is the broad phenotypic range. Some affected subjects present in early childhood while in one large Australian pedigree many affected subjects were normokalaemic and remained normotensive until late adult life.
Figure 1.
The adrenal steroid pathway.
Apparent mineralocorticoid excess (AME) was first reported in the 1970s amongst children with severe low-renin hypertension. Because of its sensitivity to spironolactone, the mineralocorticoid receptor was assumed to be involved. However, the levels of aldosterone were actually low in these subjects, as were the levels of other sodium-retaining corticosteroids. The first clue came from their abnormal conversion of cortisol to cortisone, but mutation scanning of the (type 1) 11β-hydroxysteroid dehydrogenase (HSD) gene was unrevealing. The breakthrough came with the identification of a second isoform of 11-β-HSD [37], which also provided an explanation for a longstanding puzzle: how could tissues such as the kidney respond selectively to aldosterone if the mineralocorticoid receptor could not distinguish cortisol from aldosterone? By locally metabolizing cortisol, the type 2 11-beta-HSD could ensure that tissues expressing this enzyme (kidney, parotid and placenta) only see and respond to circulating aldosterone [38] (Figure 1). This scheme was elegantly supported by the discovery of inactivating mutations within the type 2 11-βHSD gene in subjects with AME [39]. It is now clear that cortisol is a potent pressor held in check by the type 2 11-βHSD gene product. Loss of enzyme activity from either germ-line mutation or the liquorice metabolite, glycerrhetinic acid, exposes the mineralocorticoid receptor in the kidney to the paracrine effects of cortisol resulting in salt retention, hypokalaemia and hypertension.
A familial hypertension syndrome well recognized by paediatric endocrinologists is the 11-β-hydroxylase form of congenital adrenal hyperplasia (CAH). It is the second commonest form of CAH (after the 21-hydroxylase type) and index cases have been exclusively of Middle Eastern ancestry. It arises from inactivating mutations in the CYP11B1 enzyme (Figure 1) leading to accumulation of the potent mineralocorticoids, 11-deoxycortisol and DOC (11-deoxycorticosterone), which causes a low-renin hypertension. Since, the virilization in females can be very mild and does not correlate with the degree of mineralocorticoid excess, diagnosis may be delayed under adult life. In fact, it may present in adults as unexplained hypokalaemia or hypertension-related stroke. The rarer 17-α-hydroxylase form of CAH, inactivates the enzyme CYP17 causing low-renin hypertension (DOC accumulation) and concomitant oestrogen deficiency which is usually assumed to cause primary amenorrhoea in females and ambiguous genitalia in males. However, the effect on gonadal function may be less severe in some subjects, since a CYP17 mutation has been described in Japan associated with hypertension and premature ovarian failure. In fact, the index case was originally diagnosed on clinical grounds as having GRA [40].
There are two further dominantly inherited hypertension syndromes whose molecular basis remains unresolved. The first is the hypertension–bradydactyly syndrome, so-called because of a characteristic metacarpal shortening in affected subjects [41]. It has been linked to chromosome 12 and subjects have a normal renin/aldosterone axis. With the discovery of curious loops in the PICA arteries of these subjects, the suggestion has been made that they have ‘neurovascular’ hypertension secondary to brainstem compression. The second is Gordon's syndrome (pseudohypoaldosteronism type 2) characterized by hypertension and hyperkalaemia; the hypertension is of the low-renin type and affected subjects respond to either salt restriction or diuretics. Linkage studies have identified at least two loci for Gordon's syndrome on chromosomes 1 and 17 [42, 43], which excludes both the frusemide (Na,K,2 CI-) and thiazide-sensitive cotransporters as candidate genes. A novel ion channel or cotransporter gene may be involved in this syndrome.
It should be emphasized that published reports suggest that the monogenic syndromes are extremely rare even in tertiary referral populations. However, they are almost certainly underdiagnosed in the general hypertensive population: some mutations, for example, in AME [44] and GRA [45], confer a much milder phenotype with presentation in adult life; coinherited variants that lower blood pressure may also mask expression of the hypertensive mutation [45]; there is also sibling data suggesting that as a whole monogenic syndromes may be more frequent than is currently appreciated as a cause of low-renin hypertension [5]. This issue can be answered in the next few years when the chip technology becomes available to rapidly screen hypertensive populations for a large number of known mutations in these genes. Another unknown is whether the genes mutated in the monogenic disorders are susceptibility genes for the much commoner polygenic essential hypertension. The genes in Table 1 have been the subjects of extensive mutation scanning in an attempt to identify common functional gene variants in essential hypertensives. No fewer than seven common amino acid variants have been identified in the ENaC gene [46]. They appear to be particularly frequent in subjects of African origin and one group has reported positive association of one of the gene variants (T549M) [47]. However, expression of the mutant ENaCs in Xenopus oocytes failed to demonstrate any significant effect on function [46], although this may not be an appropriate test system for the T594M variant. Further work is clearly needed in this area.
The future
Major advancements can be expected in the next few years in the identification of the multiple genes that modify blood pressure. This will require the close interplay of animal and human approaches to the problem together with new molecular genetic techniques. The impact that this knowledge will have on the way we diagnose, treat and prevent this major health problem is likely to be profound.
References
- 1.Murray CJ, Lopez AD. Evidence-based health policy – lessons from the Global Burden of Disease Study. Science. 1996;274:740–743. doi: 10.1126/science.274.5288.740. 10.1126/science.274.5288.740. [DOI] [PubMed] [Google Scholar]
- 2.Pickering GW. The nature of essential hypertension. Lancet. 1959;ii:1027–1028. [PubMed] [Google Scholar]
- 3.Platt R. Heredity in hypertension. Q J Med. 1947;16:111–133. [PubMed] [Google Scholar]
- 4.Mongeau J-G. Heredity and blood pressure. Semin Nephrol. 1989;9:208–216. [PubMed] [Google Scholar]
- 5.Brown MJ. The causes of essential hypertension. Br J Clin Pharmacol. 1996;42:21–27. doi: 10.1046/j.1365-2125.1996.03785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrap SB. Genetic analysis of blood pressure and sodium balance in spontaneously hypertensive rats. Hypertension. 1986;8:572–582. doi: 10.1161/01.hyp.8.7.572. [DOI] [PubMed] [Google Scholar]
- 7.Tanase H, Suzuki Y, Ooshima A, Yamori Y, Okamoto K. Genetic analysis of blood pressure in spontaneously hypertensive rats. Jpn Circ J. 1970;34:1197–1212. doi: 10.1253/jcj.34.1197. [DOI] [PubMed] [Google Scholar]
- 8.Rapp JP. Genetics of Experimental and Human Hypertension. New York: McGraw-Hill; 1983. pp. 582–598. [Google Scholar]
- 9.Dominiczak AF, Jeffs B, Connell JM. New genetic concepts in hypertensive cardiovascular disease. Curr Opin Cardiol. 1998;13:304–311. doi: 10.1097/00001573-199809000-00003. [DOI] [PubMed] [Google Scholar]
- 10.Aitman TJ, Glazier AM, Wallace CA, et al. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 11.Julier C, Delepine M, Keavney B, et al. Genetic susceptibility for human familial essential hypertension in a region of homology with blood pressure linkage on rat chromosome 10. Hum Mol Genet. 1997;6:2077–2085. doi: 10.1093/hmg/6.12.2077. 10.1093/hmg/6.12.2077. [DOI] [PubMed] [Google Scholar]
- 12.Broeckel U, Shiozawa M, Kissebah AH, Provoost AP, Jacob HJ. Susceptibility genes for end-organ damage. New strategies to understand diabetic and hypertensive nephropathy. Nephrol Dial Transplant. 1998;13:840–842. doi: 10.1093/ndt/13.4.840. 10.1093/ndt/13.4.840. [DOI] [PubMed] [Google Scholar]
- 13.Jeunemaitre X, Soubrier F, Kotelevtsev YV, et al. Molecular basis of human hypertension: role of angiotensinogen. Cell. 1992;71:169–180. doi: 10.1016/0092-8674(92)90275-h. [DOI] [PubMed] [Google Scholar]
- 14.Glorioso N, Manunta P, Filigheddu F, et al. The role of alpha-adducin polymorphism in blood pressure and sodium handling regulation may not be excluded by a negative association study. Hypertension. 1999;34:649–654. doi: 10.1161/01.hyp.34.4.649. [DOI] [PubMed] [Google Scholar]
- 15.Svetkey LP, Chen YT, McKeown SP, Preis L, Wilson AF. Preliminary evidence of linkage of salt sensitivity in black Americans at the beta 2-adrenergic receptor locus. Hypertension. 1997;29:918–922. doi: 10.1161/01.hyp.29.4.918. [DOI] [PubMed] [Google Scholar]
- 16.Siffert W, Rosskopf D, Siffert G, et al. Association of a human G-protein beta3 subunit variant with hypertension. Nat Genet. 1998;18:45–48. doi: 10.1038/ng0198-45. [DOI] [PubMed] [Google Scholar]
- 17.Caulfield M, Lavender P, Farrall M, et al. Linkage of the angiotensinogen gene to essential hypertension. N Engl J Med. 1994;330:1629–1633. doi: 10.1056/NEJM199406093302301. [DOI] [PubMed] [Google Scholar]
- 18.Inoue I, Nakajima T, Williams CS, et al. A nucleotide substitution in the promoter of human angiotensinogen is associated with essential hypertension and affects basal transcription in vitro. J Clin Invest. 1997;99:1786–1797. doi: 10.1172/JCI119343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brand E, Chatelain N, Keavney B, et al. Evaluation of the angiotensinogen locus in human essential hypertension: a European study. Hypertension. 1998;31:725–729. doi: 10.1161/01.hyp.31.3.725. [DOI] [PubMed] [Google Scholar]
- 20.Niu T, Xu X, Cordell HJ, et al. Linkage analysis of candidate genes and gene–gene interactions in chinese hypertensive sib pairs. Hypertension. 1999;33:1332–1337. doi: 10.1161/01.hyp.33.6.1332. [DOI] [PubMed] [Google Scholar]
- 21.Kunz R, Kreutz R, Beige J, Distler A, Sharma AM. Association between the angiotensinogen 235T-variant and essential hypertension in whites: a systematic review and methodological appraisal. Hypertension. 1997;30:1331–1337. doi: 10.1161/01.hyp.30.6.1331. [DOI] [PubMed] [Google Scholar]
- 22.Mein CA, Esposito L, Dunn MG, et al. A search for type1 diabetes susceptibility genes in families from the United Kingdom. Nat Genet. 1998;19:297–300. doi: 10.1038/991. [DOI] [PubMed] [Google Scholar]
- 23.Concannon P, Gogolin-Ewens KJ, Hinds DA, et al. A second generation screen of the human genome for susceptibility to insulin-dependent diabetes mellitus. Nat Genet. 1998;19:292–296. doi: 10.1038/985. [DOI] [PubMed] [Google Scholar]
- 24.Suarez BK, Hampe CL. Linkage and association. Am J Hum Genet. 1994;54:554–559. [PMC free article] [PubMed] [Google Scholar]
- 25.Risch N, Zhang H. Extreme discordant sib pairs for mapping quantitative trait loci in humans. Science. 1995;268:1584–1589. doi: 10.1126/science.7777857. [DOI] [PubMed] [Google Scholar]
- 26.Xu X, Rogus JJ, Terwedow HA, et al. An extreme-sib-pair genome scan for genes regulating blood pressure. Am J Hum Genet. 1999;64:1694–1701. doi: 10.1086/302405. 10.1086/302405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung VG, Nelson SF. Genomic mismatch scanning identifies human genomic DNA shared identical by descent. Genomics. 1998;47:1–6. doi: 10.1006/geno.1997.5082. 10.1006/geno.1997.5082. [DOI] [PubMed] [Google Scholar]
- 28.Liddle GW, Bledsoe T, Coppage WS. A familial renal disorder simulating primary aldosteronism but with negligible aldosterone secretion. Trans Assoc Am Phys. 1963;76:199–213. [Google Scholar]
- 29.Wang C, Chan TK, Yeung RT, Coghlan JP, Scoggins BA, Stockigt JR. The effect of triamterene and sodium intake on renin, aldosterone, and erythrocyte sodium transport in Liddle's syndrome. J Clin Endocrinol Metab. 1981;52:1027–1032. doi: 10.1210/jcem-52-5-1027. [DOI] [PubMed] [Google Scholar]
- 30.Canessa CM, Horisberger JD, Rossier BC. Epithelial sodium channel related to proteins involved in neurodegeneration. Nature. 1993;361:467–470. doi: 10.1038/361467a0. [DOI] [PubMed] [Google Scholar]
- 31.Shimkets RA, Warnock DG, Bositis CM, et al. Liddle's syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 32.Hansson JH, Nelson Williams C, Suzuki H, et al. Hypertension caused by a truncated epithelial sodium channel gamma subunit: genetic heterogeneity of Liddle syndrome. Nat Genet. 1995;11:76–82. doi: 10.1038/ng0995-76. [DOI] [PubMed] [Google Scholar]
- 33.Hansson JH, Schild L, Lu Y, et al. A de novo missense mutation of the beta subunit of the epithelial sodium channel causes hypertension and Liddle syndrome, identifying a proline-rich segment critical for regulation of channel activity. Proc Natl Acad Sci U S A. 1995;92:11495–11499. doi: 10.1073/pnas.92.25.11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder PM, Price MP, McDonald FJ, et al. Mechanism by which Liddle's syndrome mutations increase activity of a human epithelial Na channel. Cell. 1995;83:969–978. doi: 10.1016/0092-8674(95)90212-0. [DOI] [PubMed] [Google Scholar]
- 35.Baker E, Jeunemaitre X, Portal AJ, et al. Abnormalities of nasal potential difference measurement in Liddle's syndrome. J Clin Invest. 1998;102:10–14. doi: 10.1172/JCI1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lifton RP, Dluhy RG, Powers M, et al. Hereditary hypertension caused by chimaeric gene duplications and ectopic expression of aldosterone synthase. Nat Genet. 1992;2:66–74. doi: 10.1038/ng0992-66. [DOI] [PubMed] [Google Scholar]
- 37.Agarwal AK, Rogerson FM, Mune T, White PC. Gene structure and chromosomal localization of the human HSD11K gene encoding the kidney (type 2) isozyme of 11 beta-hydroxysteroid dehydrogenase. Genomics. 1995;29:195–199. doi: 10.1006/geno.1995.1231. 10.1006/geno.1995.1231. [DOI] [PubMed] [Google Scholar]
- 38.Edwards CR, Stewart PM, Burt D, et al. Localisation of 11 beta-hydroxysteroid dehydrogenase – tissue specific protector of the mineralocorticoid receptor. Lancet. 1988;2:986–989. doi: 10.1016/s0140-6736(88)90742-8. [DOI] [PubMed] [Google Scholar]
- 39.Mune T, Rogerson FM, Nikkila H, Agarwal AK, White PC. Human hypertension caused by mutations in the kidney isozyme of 11 beta-hydroxysteroid dehydrogenase. Nat Genet. 1995;10:394–399. doi: 10.1038/ng0895-394. [DOI] [PubMed] [Google Scholar]
- 40.Miura K, Yasuda K, Yanase T, et al. Mutation of cytochrome P-45017 alpha gene (CYP17) in a Japanese patient previously reported as having glucocorticoid-responsive hyperaldosteronism: with a review of Japanese patients with mutations of CYP17. J Clin Endocrinol Metab. 1996;81:3797–3801. doi: 10.1210/jcem.81.10.8855840. [DOI] [PubMed] [Google Scholar]
- 41.Schuster H, Wienker TF, Toka HR, et al. Autosomal dominant hypertension and brachydactyly in a Turkish kindred resembles essential hypertension. Hypertension. 1996;28:1085–1092. doi: 10.1161/01.hyp.28.6.1085. [DOI] [PubMed] [Google Scholar]
- 42.Mansfield TA, Simon DB, Farfel Z, et al. Multilocus linkage of familial hyperkalaemia and hypertension, pseudohypoaldosteronism type II, to chromosomes 1q31–42 and 17p11- q21. Nat Genet. 1997;16:202–205. doi: 10.1038/ng0697-202. [DOI] [PubMed] [Google Scholar]
- 43.O'Shaughnessy KM, Fu B, Johnson A, Gordon RD. Linkage of Gordon's syndrome to the long arm of chromosome 17 in a region recently linked to familial essential hypertension. J Hum Hypertens. 1998;12:675–678. doi: 10.1038/sj.jhh.1000705. [DOI] [PubMed] [Google Scholar]
- 44.Li A, Tedde R, Krozowski ZS, et al. Molecular basis for hypertension in the ‘type II variant’ of apparent mineralocorticoid excess. Am J Hum Genet. 1998;63:370–379. doi: 10.1086/301955. 10.1086/301955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dluhy RG, Lifton RP. Glucocorticoid-remediable aldosteronism (GRA): diagnosis, variability of phenotype and regulation of potassium homeostasis. Steroids. 1995;60:48–51. doi: 10.1016/0039-128x(94)00010-a. 10.1016/0039-128x(94)00010-a. [DOI] [PubMed] [Google Scholar]
- 46.Persu A, Barbry P, Bassilana F, et al. Genetic analysis of the beta subunit of the epithelial Na+ channel in essential hypertension. Hypertension. 1998;32:129–137. doi: 10.1161/01.hyp.32.1.129. [DOI] [PubMed] [Google Scholar]
- 47.Baker EH, Dong YB, Sagnella GA, et al. Association of hypertension with T594M mutation in beta subunit of epithelial sodium channels in black people resident in London. Lancet. 1998;351:1388–1392. doi: 10.1016/s0140-6736(97)07306-6. 10.1016/s0140-6736(97)07306-6. [DOI] [PubMed] [Google Scholar]