Abstract
Aims
To determine the effects of verapamil and diltiazem on simvastatin metabolism in human liver microsomes and to compare their inhibitory potencies and CYP3A4 inactivation parameters with those reported previously for mibefradil.
Methods
Simvastatin metabolism was investigated in human liver microsomes in the presence and absence of verapamil or diltiazem (0.1–250 µm). Kinetics of CYP3A4 inactivation by verapamil and diltiazem were determined using testosterone as the substrate.
Results
When verapamil was coincubated with simvastatin, IC50 values ranged from 23 to 26 µm for all major metabolites. The IC50 values ranged from 4.8 to 5.6 µm on preincubation of verapamil for 30 min in the presence of an NADPH-generating system. Corresponding IC50 values for diltiazem ranged from110–127 µm and from 21–27 µm, respectively. Verapamil and diltiazem inhibited testosterone 6β-hydroxylation in a time- and concentration-dependent manner, key features of mechanism-based inactivation. Values for the inactivation parameters kinact and KI were 0.15 ± 0.04 min−1 (mean ± s.d.) and 2.9 ± 0.6 µm, respectively, for verapamil and 0.07 ± 0.01 min−1 and 3.3 ± 1.5 µm, respectively, for diltiazem.
Conclusions
The IC50 values for coincubation of verapamil and diltiazem were 46- and 220-fold higher, respectively, than those reported previously for mibefradil, and 16- and 71-fold higher, respectively, for preincubation. Thus, the results of this study suggest that verapamil and diltiazem are less likely than mibefradil to cause acute drug interactions with simvastatin in vivo. However, verapamil and diltiazem are moderate mechanism-based inhibitors of CYP3A4 and therefore may still cause significant inhibition of simvastatin metabolism in vivo during chronic therapy.
Keywords: CYP3A4, diltiazem, verapamil, simvastatin, interaction
Introduction
The HMG-CoA reductase inhibitor (statin) simvastatin is used extensively for treatment of hypercholesterolaemia and has a major impact in the prevention of coronary heart disease (CHD) [1]. Simvastatin is an inactive lactone pro-drug that is hydrolysed in vivo to simvastatin acid, a potent competitive inhibitor of HMG-CoA reductase the rate-limiting enzyme in cholesterol biosynthesis. The statin is generally well tolerated and causes few subjective side-effects during chronic treatment [1]. Myopathy, which may present as rhabdomyolysis, is a rare but serious side-effect of statin treatment and occurs in 0.1% of patients treated with standard doses of simvastatin or other HMG-CoA reductase inhibitors [2]. The risk of developing acute rhabdomyolysis appears to increase considerably on addition of drugs such as itraconazole [3] and mibefradil [4, 5] to the stable regimen of patients taking simvastatin. The onset of rhabdomyolysis has been associated with dramatic increases in plasma concentrations of HMG-CoA reductase inhibitory activity, due to inhibition of the cytochrome P4503A4 (CYP3A4)-mediated metabolism of simvastatin by the interacting drugs [6]. Results of an in vitro study have confirmed that at therapeutic concentrations, mibefradil inhibits the metabolism of simvastatin in human liver microsomes and acts as a powerful mechanism-based inhibitor of CYP3A4 activity [7].
Since the withdrawal of mibefradil from the market 2 years ago, there has been considerable debate regarding drug interactions with simvastatin, particularly with respect to the concomitant use of other calcium channel blockers [8, 9]. Many of these drugs are weak inhibitors of CYP3A4 [10], and diltazem and verapamil in particular, have been shown to increase the plasma levels of statins when used concurrently with simvastatin or the closely related lovastatin [11, 12]. Originally it was postulated that the mechanism of interaction by diltiazem was competitive in nature [13], but a recent in vitro study has demonstrated that diltiazem-mediated inhibition of midazolam metabolism by CYP3A4 occurs primarily by metabolite intermediate complex formation, which renders the enzyme inactive [14]. Thus, the mechanism of CYP3A4 inhibition by diltiazem appears to be similar to that observed with mibefradil [7]. It has also been reported that both diltiazem and verapamil inhibit testosterone 6β-hydroxylation due to metabolite-intermediate complexation with CYP3A [15]. The mechanism of inhibition was characterized for both inhibitors but there were limited data on the kinetics of inactivation.
In the present study the inhibitory effects of verapamil and diltiazem on simvastatin metabolism in human liver microsomes were investigated. In addition, the inactivation kinetics of verapamil and diltiazem were determined using incubation conditions described previously for the characterization of CYP3A4 inhibition by mibefradil [7]. Thus, it was possible to compare directly the inhibitory potencies and inactivation parameters of verapamil and diltiazem with those reported for mibefradil, a known potent inhibitor of CYP3A4 in vitro and in vivo [7].
Methods
Drugs and chemicals
Simvastatin was provided by the Merck Research Laboratories. Testosterone and its metabolite testosterone 6β-hydroxylase, dilitazem and verapamil were purchased from Sigma (London, UK). Glucose-6-phosphate (G6P) dehydrogenase (grade II) and the disodium salts of G6P and NADP were purchased from Boehringer Mannheim (Lewes, UK). Acetonitrile, ethyl acetate and methanol were obtained from Rathburn (Walkerburn, UK). All other reagents were of analytical grade and obtained from Merck (Poole, UK).
Human liver microsomes
Samples of human liver were obtained from ‘normal’ tissue taken from carcinoma patients with the approval of the Royal Hallamshire Hospital Ethics Committee and the local Coroner. The 10 samples of human liver were subsequently referred to as HL7, HL11, HL12, HL14, HL16, HL17, HL21, HL22, HL24 and HL25. There was no pathological evidence of hepatic disease in any of the liver samples used. The donors of HL14 and HL22 were female and the remaining donors were male. With the exception of the donor of HL25 who was a black Caribbean, the donors were Caucasian. None of the donors was a smoker. Drugs administered prior to or during organ removal were fentanyl, atracurium, thiopentone, morphine and droperidol.
Human liver microsomes were prepared as described previously [16] and stored at −80 °C as a suspension in 0.1 m potassium phosphate buffer (pH 7.4) containing 30% (v/v) glycerol. Microsomal protein was measured by the method of Lowry et al. [17] using bovine serum albumin as the standard.
Effects of verapamil and diltiazem on simvastatin metabolism
Incubation mixtures comprised 100 µl microsomal suspension (equivalent to 0.05–0.25 mg microsomal protein), inhibitors dissolved in 195 µl 1.15% KCl (w/v), 200 µl of a NADPH-generating system dissolved in 0.2 m potassium phosphate buffer (pH 7.4) and 5 µmol simvastatin added in 5 µl acetone. The NADPH-generating system consisted of 0.4 µmol NADP, 4 µmol G6P, 2 µmol MgCl2 and 0.4 units of G6P-dehydrogenase. Verapamil or diltiazem was either coincubated with simvastatin and microsomes before the reaction was initiated with the NADPH-generating system or preincubated with microsomes and the NADPH-generating system for 30 min at 37 °C before addition of simvastatin. Control experiments were also performed using preincubation and coincubation conditions to determine simvastatin metabolism in the absence of any inhibitor. Incubations were conducted at 37 °C and were terminated after 10 min by the addition of 0.5 ml acetone.
Preliminary experiments indicated that the final substrate concentration of 10 µm was comparable with the Km value of simvastatin and that the formation of metabolites at this concentration was linear up to 0.5 mg protein ml−1 and throughout the 10 min incubation period. A range of concentrations (0.5–250 µm) was used to investigate the effect of verapamil and diltiazem on the formation of simvastatin metabolites. Incubations were performed in triplicate at each inhibitor concentration using microsomes prepared from HL7. The experiment was repeated using microsomes from three different human livers, at single inhibitor concentrations of 5 and 20 µm for verapamil and diltiazem, respectively (approximate IC50 values).
Effects of verapamil and diltiazem on CYP3A4 activity
Testosterone 6β-hydroxylation was used as a marker of CYP3A4 activity [18]. Verapamil or diltiazem (0.5–250 µm) were either coincubated with testosterone and microsomes before the reaction was initiated with the NADPH-generating system or preincubated with microsomes and the NADPH-generating system for 30 min at 37 °C before the addition of testosterone (25 µmol in 5 µl acetone). Both sets of incubations were performed in the absence of inhibitor to determine control testosterone 6β-hydroxylase activity. The incubations were terminated after 10 min by the addition of 2 ml ethyl acetate. The substrate concentration used was comparable with the Km value of testosterone (50 µm). Incubations were performed in triplicate at each inhibitor concentration using microsomes prepared from HL7. To determine the time-dependent effect of CYP3A4 inactivation by the inhibitors, verapamil (2 µm) and diltiazem (10 µm) were preincubated in the presence of a NADPH-generating system for various times ranging from 0 to 45 min. The samples were then assayed for remaining CYP3A4 activity using testosterone (50 µm).
Microsomes prepared from 10 different human livers were preincubated with 1 µm verapamil for 30 min in the presence of an NADPH-generating system and were then assayed for remaining testosterone 6β-hydroxylase activity. Incubations were carried out in quadruplicate for each human liver. The experiment was repeated using 1 µm diltiazem.
Kinetics of CYP3A4 inactivation
Studies on the kinetics of CYP3A4 inactivation were performed using the testosterone 6β-hydroxylase assay at a substrate concentration of 250 µm, in order to minimize competitive inhibition by verapamil and diltiazem. Human liver microsomes were preincubated with 0.5, 1, 2 and 5 µm verapamil or 2, 5, 10 and 20 µm diltiazem in the presence of an NADPH-generating system for various times ranging from 0, 2, 5, 10, 15, 20 and 30 min at 37 °C. The incubation mixture was then assayed for remaining CYP3A4 activity by the addition of testosterone (125 µmol in 5 µl acetone). The reaction was terminated after 10 min by the addition of 2 ml ethyl acetate. Incubations were performed in duplicate and the experiment was carried out using microsomes prepared from three different human livers (HL7, HL12 and HL16).
In vitro metabolism of verapamil and diltiazem
The disappearance of verapamil and diltiazem in human liver microsomes was measured in order to determine a partition ratio for CYP3A4 inactivation by each of the inhibitors. Using the incubation conditions described previously for simvastatin, verapamil (0.5 nmol) and diltiazem (2.5 nmol) were incubated for various times ranging from 0 to 30 min. The reactions were terminated by the addition of 200 µl acetone. Incubations were carried out in duplicate using microsomes prepared from three different livers (HL7, HL12 and HL16).
Analytical procedures
Simvastatin and its metabolites were assayed using modifications of methods described previously [7, 19]. Ethyl acetate (2 ml) was added to the tubes containing the incubation mixture and the contents were shaken for 15 min. After centrifugation at 900 g for 5 min, the organic phase was transferred to conical glass tubes and evaporated to dryness under vacuum at 40 °C using a Buchler vortex-evaporator (Baired and Tatlock, Romford, UK). The residue was reconstituted in 200 µl mobile phase comprising water/acetonitrile (45 : 55, v/v) containing 1% triethylamine adjusted to pH 4 with orthophosphoric acid and subjected to h.p.l.c. analysis. The samples were injected onto a Waters RCM radial compression system containing a 0.8 × 10 cm NovaPak C18 cartridge, 4 µm particle size (Waters Chromatography, Watford, UK) fitted with a Guard-Pak guard column containing a C18 insert (Waters). A Model 510 chromatography pump (Waters) was used for solvent delivery at a flow rate of 2.5 ml min−1. The eluate was monitored using a SF769 variable wavelength u.v. detector (Kratos, Warrington, UK) set at 240 nm and 274 nm. Simvastatin is metabolized to several products, including the two major metabolites, 3′-hydroxy- and 6β-exomethylene-simvastatin [7, 18]. Identification of the 6β-exomethylene metabolite was made largely on the basis of its u.v. spectral characteristics, i.e. λmax = 274 nm, and also by comparison of its retention time with that reported previously [18]. The 3′-hydroxy-product was identified by its retention time and also the fact that it was the main metabolite observed at 240 nm using u.v. detection. The coefficient of variation for assay of both metabolites was less than 5% at 10 nmol ml−1 simvastatin.
The same h.p.l.c. system including the NovaPak C18 column was used to measure the formation of 6β-hydroxytestosterone and disappearance of diltiazem and verapamil. To assay 6β-hydroxytestosterone, 25 µl of a 10 µg ml−1 solution of 11β-hydroxytestosterone was added to the tubes containing the incubation mixture and 2 ml ethyl acetate. The samples were shaken for 15 min, prior to centrifugation at 900 g for 5 min. The organic phase was transferred to conical glass tubes and evaporated to dryness under vacuum at 40 °C using a Buchler vortex-evaporator. The samples were re-constituted in 200 µl mobile phase consisting of methanol/water (55 : 45, v/v) and injected onto the h.p.l.c. system. The samples were eluted from the column at a flow rate of 3 ml min−1 and were monitored by a u.v. detector set at 240 nm. The limit of determination of 6β-hydroxytestosterone was 30 pmol ml−1, and the coefficient of variation at 160 pmol ml−1 was less than 5%.
Verapamil and diltiazem were assayed by centrifugation of the incubation samples at 900 g for 5 min and injecting the supernatants directly onto the h.p.l.c. system. The samples were eluted from the column using a mobile phase consisting of acetonitrile/water (28%: 72%, v/v) containing 1% triethylamine (adjusted to pH 3.5 with orthophosphoric acid). The pump was set at a flow rate of 2.5 ml min−1 and the u.v. detector at 230 nm. The retention times of diltiazem and verapamil were 5.5 and 10.7 min, respectively. At 5 nmol ml−1 diltiazem and at 1 nmol ml−1 verapamil the coefficients of variation for assay were 6% and 5%, respectively.
Data analysis
The results were expressed as percentages of substrate consumed or metabolites formed in the presence of verapamil or diltiazem relative to the corresponding values obtained in the absence of any inhibitor (control). IC50 values were determined by a nonlinear regression analysis. The inactivation parameters kinact and KI were calculated according to the method of Silverman [20]. The initial-rate constant for inactivation (k, min−1) at each inhibitor concentration was estimated from the initial slope of the log-linear regression line of CYP3A4 activity remaining vs the preincubation time profile. Initial estimates of the inactivation parameters were then determined from the reciprocal plot of rate constants for inactivation (1/k) vs the respective inhibitor concentrations (1/I). These values were used as first estimates for iterative nonlinear regression analysis using the solver subprogram, which was implemented in Excel 97 to calculate values for kinact and KI according to the following equation:
where k is the initial rate constant for inactivation at the initial inhibitor concentration, Io; kinact is the maximum rate constant for inactivation; and KI is the inactivator concentration which produces half the maximal rate of inactivation, analogous to Km. The partition ratio was estimated from the initial disappearance rate constant of verapamil and diltiazem relative to the initial rate constant for inactivation of CYP3A4 activity at the same inhibitor concentrations.
Comparisons of CYP3A4 activities and percentage inhibition between control, verapamil and diltiazem groups were performed using a non-parametric (Kruskal–Wallis) one-way analysis of variance. Where this analysis indicated significant differences (P < 0.05) between groups, individual comparisons were made using the Mann–Whitney test.
Results
Inhibitory effects of verapamil and diltiazem on simvastatin metabolism
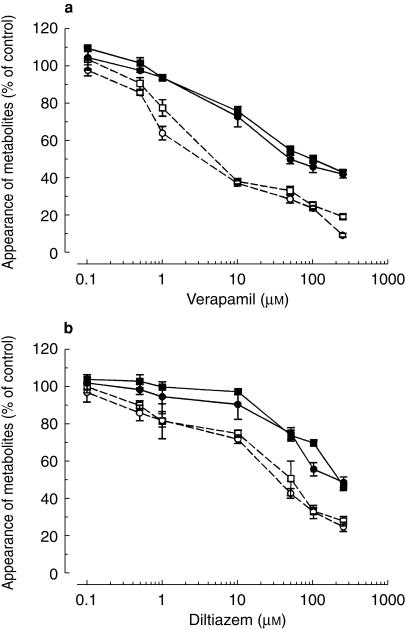
Verapamil and diltiazem inhibited simvastatin metabolism in a concentration-dependent manner (Figure 1a,b). In experiments where verapamil was coincubated with simvastatin, IC50 values ranged from 23 to 26 µm for the formation of all major metabolites (Figure 1a). When verapamil was preincubated for 30 min in the presence of an NADPH-generating system the degree of inhibition increased, the IC50 values ranging from 4.8 to 5.6 µm. Corresponding IC50 values for diltiazem ranged from110–127 µm and from 21 to 27 µm, respectively (Figure 1b).
Figure 1.
Concentration-dependent inhibitory effects of (a) verapamil and (b) diltiazem on formation of 3′-hydroxy-(▪) and 6β-exomethylene-simvastatin (•) in human liver microsomes. Verapamil or diltiazem were either coincubated with simvastatin and microsomes before the reaction was initiated with an NADPH-generating system (solid symbols) or preincubated with microsomes and an NADPH-generating system for 30 min at 37 °C before addition of simvastatin (open symbols). Values are expressed as mean (± s.d., n = 3 livers) percentage of the control in the absence of inhibitor.
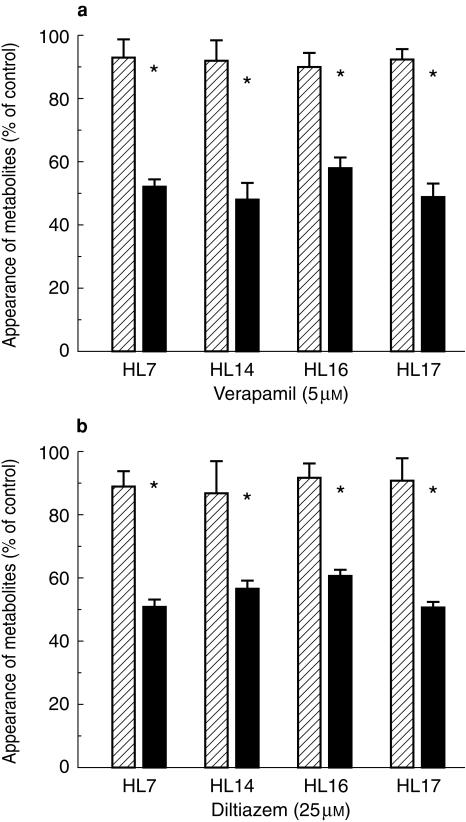
The experiment was repeated using microsomes from four different human livers and single inhibitor concentrations of 5 µm verapamil and 25 µm diltiazem. These concentrations were chosen because they approximated the IC50 values determined in the initial experiment. The degree of inhibition of 3′-hydroxy- and 6β-exomethylene-simvastatin formation was increased significantly in microsomes from each liver by preincubation of verapamil and dilitazem (P < 0.05) (Figure 2a,b).
Figure 2.
Inhibitory effects of (a) verapamil and (b) diltiazem on simvastatin 3′-hydroxylation in microsomes from four different human livers. Verapamil or diltiazem were either coincubated with simvastatin and microsomes before the reaction was initiated with an NADPH-generating system (hatched bars) or preincubated with microsomes and an NADPH-generating system for 30 min at 37 °C before addition of simvastatin (solid bars). Values are expressed as mean (± s.d., n = 4) percentage of the control in the absence of inhibitor.
Inhibition of testosterone 6β-hydroxylase activity by verapamil and diltiazem
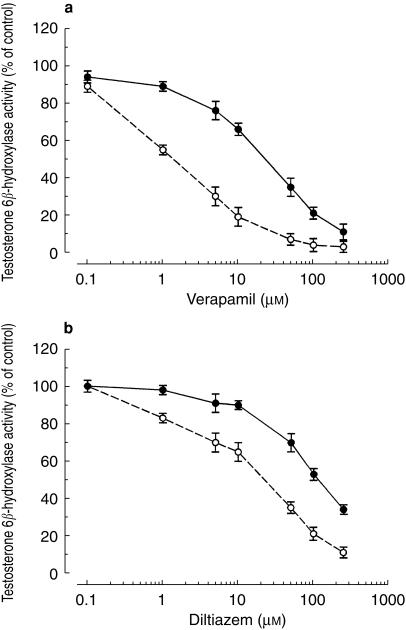
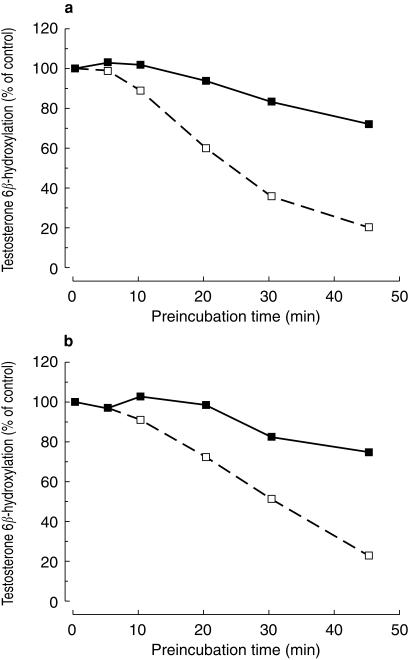
The inhibitory effects of verapamil and diltiazem on testosterone 6β-hydroxylation were also concentration dependent (Figure 3a,b). As was observed with simvastatin, IC50 values for preincubation were approximately 5-fold lower than those obtained for coincubation. IC50 values for inhibition of testosterone 6β-hydroxylation by preincubated verapamil and diltiazem were 27 and 122 µm, respectively. In coincubation experiments, the corresponding values were 3.1 and 26 µm, respectively. These values were comparable with those obtained when using simvastatin as the CYP3A4 substrate. The inhibitory effects of verapamil and diltiazem were also found to be time-dependent (Figure 4a,b).
Figure 3.
Concentration-dependent inhibitory effects of (a) verapamil and (b) diltiazem on testosterone 6β-hydroxylation in human liver microsomes. Verapamil or diltiazem were either coincubated with testosterone (50 µm) and microsomes before the reaction was initiated with an NADPH-generating system (•) or preincubated with microsomes and an NADPH-generating system for 30 min at 37 °C before addition of simvastatin (○). Values are expressed as mean (± s.d., n = 3) percentage of the control activity in the absence of inhibitor. Control activities for testosterone 6β-hydroxylase were 1.87 ± 0.12 nmol min−1 mg−1 protein for coincubation and 1.70 ± 0.10 nmol min−1 mg−1 protein for preincubation.
Figure 4.
Time-dependent loss of testosterone 6β-hydroxylase activity in human liver microsomes after preincubation with (a) verapamil and (b) dilltiazem. Microsomes were preincubated with 2 µm verapamil or 10 µm diltiazem for the designated time in the presence of an NADPH generating system (□) and then assayed for remaining testosterone 6β-hydroxylase activity. Values are expressed as mean (± s.d., n = 3) percentage of the control (i.e. activity at time zero in a sample that had not been preincubated with inhibitor, but that contained 2 µm verapamil or 10 µm diltiazem). Time courses of remaining testosterone 6β-hydroxylase activity in samples containing an NADPH-generating system but lacking either of the inhibitors are also shown for comparison (▪).
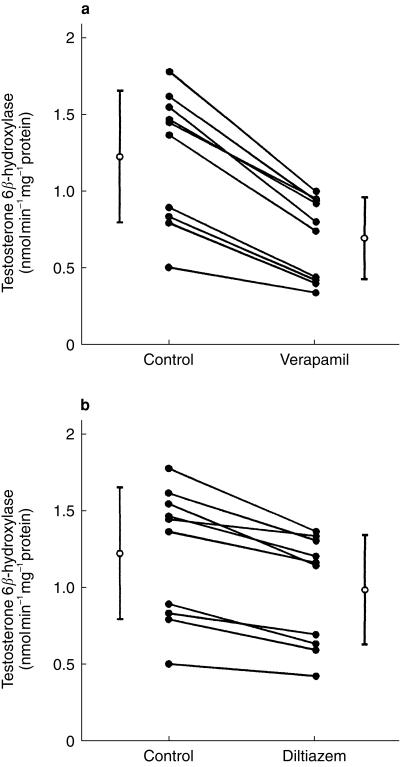
With preincubation in the presence of an NADPH-generating system, the inhibitory effects of 1 µm verapamil and 1 µm diltiazem on testosterone 6β-hydroxylation were investigated in microsomes prepared from 10 different human livers (Figure 5a,b). For each human liver, CYP3A4 activity was decreased significantly by preincubation with either verapamil or diltiazem (P < 0.05). Mean testosterone 6β-hydroxylase activity for all 10 human livers was reduced significantly from 1.22 ± 0.43–0.70 ± 0.27 nmol min−1 mg−1 protein (P < 0.05) after preincubation with verapamil. Testosterone 6β-hydroxylase activity after preincubation with diltiazem was reduced to 1.01 ± 0.39 nmol min−1 mg−1 protein. This was not significantly different from the control activity.
Figure 5.
Inhibition of testosterone 6β-hydroxylase activity in microsomes prepared from 10 different human livers after preincubation with (a) verapamil and (b) diltiazem. Microsomes were preincubated with an NADPH generating system in the absence (control) or presence of 1 µm verapamil or 1 µm diltiazem for 30 min, followed by assay of testosterone 6β-hydroxylase activity. Mean (± s.d., n = 10 livers) testosterone 6β-hydroxylase activities for the control and inhibitor groups are also shown (○).
Absolute reduction in testosterone 6β-hydroxylase activity after preincubation with verapamil was significantly correlated with baseline CYP3A4 activity (rs = 0.95; P < 0.001). Thus, more extensive inactivation was observed in microsomes with a higher baseline activity (Figure 5a). On preincubation with diltiazem, absolute reduction in CYP3A4 was not highly correlated with baseline activity (rs = 0.51).
Kinetics of CYP3A inactivation by verapamil and diltiazem
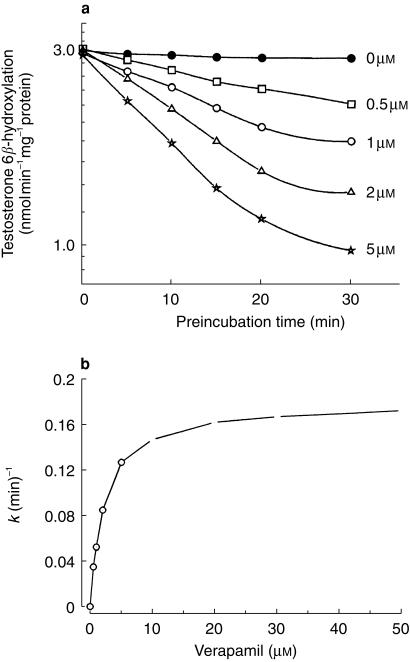
Mean (± s.d., n = 3 livers) kinact and KI values were 0.15 ± 0.04 min−1 and 2.9 ± 0.6 µm, respectively, for verapamil and 0.07 ± 0.01 min−1 and 3.3 ± 1.5 µm, respectively, for diltiazem (Table 1, Figure 6). The potency of CYP3A4 inactivation by verapamil and diltiazem was characterized further by calculation of the respective partition ratios, defined as moles inhibitor metabolized per moles enzyme inactivated. These values were found to be 10.3 ± 2.0 at 1 µm verapamil and 19.7 ± 1.1 at 5 µm diltiazem.
Table 1.
Inactivation parameters for verapamil and diltiazem.
| Verapamil | Diltiazem | |||||
|---|---|---|---|---|---|---|
| kinact (min−1) | KI (µm) | r | kinact (min−1) | KI (µm) | r | |
| HL7 | 0.16 | 2.9 | 7.6 | 0.07 | 2.2 | 18.1 |
| HL12 | 0.18 | 2.3 | 12.2 | 0.08 | 2.8 | 20.8 |
| HL16 | 0.10 | 3.4 | 11.1 | 0.06 | 5.0 | 20.1 |
| Mean ± s.d. | 0.15 ± 0.04 | 2.9 ± 0.6 | 10.3 ± 2.0 | 0.07 ± 0.01 | 3.3 ± 1.5 | 19.7 ± 1.1 |
kinact – maximum inactivation rate constant. KI – dissociation constant for the enzyme-inactivator complex. r – partition ratio.
Figure 6.
Time- and concentration-dependent inactivation of human liver microsomal testosterone 6β-hydroxylase activity by verapamil (a). Concentrations of verapamil present in the preincubation samples were 0 µm (NADPH only) (×), 0.5 µm (□), 1 µm (○), 2 µm (Δ) and 5 µm (K). Initial inactivation rate (k) constants were determined at each inhibitor concentration. The reciprocal of k was plotted against the reciprocal of the inhibitor concentration (I) to obtain initial estimates of kinact and KI. The inactivation parameters were calculated using a nonlinear regression analysis. A plot of inactivation rate constant vs increasing verapamil concentration is shown (b).
Discussion
The results of the present study demonstrate that verapamil and diltiazem inhibit simvastatin metabolism in human liver microsomes and that verapamil is the more potent inhibitor. The IC50 values determined from the coincubation experiments range from 23 to 26 µm, for verapamil and from 110 to 127 µm for diltiazem. The inhibitory potency of each drug was increased approximately fivefold on preincubation with human liver microsomes in the presence of an NADPH-generating system. Thus, the metabolites of verapamil and diltiazem are much more potent inhibitors of simvastatin metabolism than the parent drugs. The fact that the IC50 values were similar to those obtained when using the probe CYP3A4 substrate testosterone, confirms that verapamil and diltiazem inhibit the CYP3A4-mediated metabolism of simvastatin.
Testosterone 6β-hydroxylation was inhibited in a time-and concentration-dependent manner by verapamil and diltiazem showed a trend towards saturation of the inactivation rate. The loss of catalytic activity was also dependent on the presence of an NADPH generating system. The mean values of kinact and KI for verapamil were 0.15 ± 0.04 min−1 and 2.9 ± 0.6 µm, respectively. At 1 µm verapamil, the partition ratio was estimated to be 10.3 ± 20. The values of kinact, KI and partition ratio for other mechanism-based inhibitors of CYP3A4 that have been investigated are: 1.6 min−1, 7.5 µm and 1.35, respectively, for L-754 394 [21]; 0.4 min−1, 46 µm and 9.0, respectively, for gestodene [22]; and 0.4 min−1, 2.3 µm and 1.7, respectively, for mibedrafil [7]. Of the four drugs, verapamil has the highest partition ratio. This indicates that a higher number of molecules of verapamil have to be metabolized to inactivate the same amount of enzyme as the other three drugs. Furthermore, the kinact value of verapamil is 2.7-fold lower than that of mibefradil and gestodene, and 11-fold lower than that of L-754 394. This suggests that verapamil has a lower efficiency of inactivation and is a moderate mechanism-based inhibitor of CYP3A4 compared with other drugs that have been studied so far. In the present study, the mean kinact and KI values for diltiazem were 0.07 ± 0.01 min−1 and 3.3 ± 1.5 µm, respectively. At 5 µm diltiazem, the partition ratio was estimated to be 19.7 ± 1.1. These values indicate that diltiazem is less efficient than verapamil as an inactivator of CYP3A4. Jones et al. [14] reported kinact and KI values of 0.17 min−1 and 2.2 µm, respectively, for formation of a metabolite-intermediate complex by diltiazem in microsomes from human lymphoblastoid cells expressing CYP3A4(+ b5). The 2.4-fold lower kinact value obtained in our study may be due in part to the confounding presence of CYP3A5 in human liver microsomes. This enzyme can metabolize testosterone and also undergoes mechanism-based inactivation by diltiazem, but to a lesser degree than CYP3A4 [14]. It is also possible that other cytochrome P450s may have contributed to the metabolism of testosterone at the high substrate concentration of 250 µm, which was used to determine remaining CYP3A4 activity. The net result would be an underestimation of the inactivation rate of CYP3A4. Ma et al. [15] reported kinact and KI values of 0.03 min−1 and 0.5 µm for verapamil and 0.01 min−1 and 0.5 µm for diltiazem. The methodology was not described in detail and therefore it is not possible to explain the 5- to 10-fold differences observed between their values and those obtained in our study.
Myopathy, which can rarely present as rhabdomyolysis, is an uncommon but serious side-effect of statin treatment and occurs in 0.1% of patients treated with standard doses of simvastatin [1]. The onset of acute rhabdomyolysis has been associated with dramatic increases in plasma concentrations of HMG-CoA reductase inhibitory activity, due to inhibition of simvastatin metabolism by drugs such as itraconazole [3] or mibefradil [4] that have been introduced to the dosing regimen of simvastatin. The results of this study indicate that the degree of inhibition of simvastatin metabolism by verapamil and diltiazem is much lower than that observed with mibefradil, a known potent inhibitor of CYP3A4 in vitro and in vivo [4, 7]. The IC50 values for coincubation of verapamil and diltiazem are 46-and 220-fold higher, respectively, than those reported for mibefradil, and 16- and 71-fold higher, respectively, for preincubation. These data suggest that verapamil and diltiazem are much less likely than mibefradil to cause acute drug interactions with simvastatin in vivo. However, verapamil and diltiazem are moderate mechanism-based inhibitors of CYP3A4 and may still cause significant inhibition of CYP3A4-mediated metabolism of simvastatin in vivo during chronic therapy. The enzyme is continuously being regenerated in vivo and therefore the extent of CYP3A4 inactivation will be dependent on the turnover rate of the enzyme, as well as the inactivation parameters of the inhibitors. The fact that verapamil and diltiazem are less efficient CYP3A4 inactivators than mibefradil and other mechanism-based inhibitors studied so far, further suggests that they are less likely to cause severe drug interactions with simvastatin in vivo. To date, there have been no reports of rhabdomyolysis induced by a simvastatin–verapamil interaction and only one that was possibly induced by a simvastatin–diltiazem interaction [1].
The effects of verapamil and diltiazem on the pharmacokinetics of simvastatin and the closely related HMG-CoA reductase inhibitor, lovastatin have been investigated in healthy volunteers [11, 23]. Verapamil caused a 4.6-fold increase in the mean area under the concentration (AUC)-time curve of simvastatin [23] and diltiazem caused a fourfold increase in the mean AUC of lovastatin [11]. Verapamil (240 mg once daily) was administered for 2 days prior to simvastatin dosing, whereas the same dose of diltiazem was taken for 2 weeks prior to lovastatin dosing. The degree of interaction is dependent on CYP3A4 activity remaining after the period of administration of the mechanism-based inhibitors, which in turn is dependent on final inhibitor concentrations attained in vivo. Pretreatment with verapamil for 2 days, may not have been long enough for the inhibitor to evoke its maximum effect, thus resulting in an underestimation of the magnitude of the interaction with simvastatin. The increase in AUC of simvastatin caused by pretreatment with verapamil ranged from 1.5- to 17-fold among the 12 volunteers and the diltiazem-induced increase in lovastatin AUC ranged from 1.5- to 10.1-fold in the 10 subjects investigated. In both studies, the magnitude of interaction for each subject was dependent on their baseline hepatic CYP3A4 activity. Volunteers with high baseline activity experienced greater magnitudes of interaction whereas those with low baseline CYP3A4 activity experienced lower levels of interaction. The net result was a reduction in interindividual variability of simvastatin and lovastatin AUC after pretreatment with verapamil and diltiazem, respectively. The results of the present study reflect those obtained in vivo. Absolute reduction in testosterone 6β-hydroxylase activity after preincubation with verapamil was highly correlated with baseline CYP3A4 activity (rs = 0.95; P < 0.001). For diltiazem, the correlation was not significant but there was a definite trend. Greater inactivation was observed in human liver microsomes that had a higher baseline CYP3A4 activity. The net result was a reduction in variability of baseline CYP3A4 activity between livers (Figure 5a,b). This may have important clinical implications for the cholesterol-lowering response to simvastatin in individuals who are also taking verapamil or diltiazem. The results of a retrospective study reported previously by our group demonstrated that there was enhanced cholesterol reduction in patients who were taking simvastatin and diltiazem concurrently, and the interindividual variability in cholesterol-lowering response to simvastatin was significantly lower for patients taking diltiazem than for patients not taking diltiazem [24]. To date, no studies have been reported regarding the effect of verapamil on the cholesterol-lowering response to simvastatin.
Drug interactions with verapamil and diltiazem are likely to occur with other HMGCoA reductase inhibitors that are metabolized by CYP3A4, namely, atorvastatin and cerivastatin. However, the magnitude of interaction of these two statins with potent CYP3A4 inhibitors such as erythromycin, appears to be lower than those observed with simvastatin and lovastatin [25, 26]. Fluvastatin is metabolized primarily by CYP2C8/9 and is therefore unlikely to have a clinically significant interaction with either of the calcium channel blockers [27]. Similarly, metabolism is a minor route of pravastatin elimination, and CYP3A4 does not contribute significantly to the hepatic clearance of the drug [27].
According to the National Service Framework [28], approximately 8% of the UK population require statin treatment for secondary or primary prevention of coronary heart disease [29]. Co-prescription of simvastatin with one of the calcium channel blockers verapamil or diltiazem is likely to occur commonly. The findings of this study demonstrate that verapamil and diltiazem are not potent inhibitors of simvastatin metabolism in human liver microsomes. However, verapamil and diltiazem are moderate mechanism-based inhibitors of CYP3A4 and therefore may still cause significant inhibition of simvastatin metabolism in vivo during chronic therapy.
Acknowledgments
We thank Merck Research Laboratories for providing the simvastatin. We also wish to thank Professor G.T. Tucker for providing the human liver microsomes and laboratory facilities.
References
- 1.Scandinavian Simvastatin Survival Study Group. Randomised trial of cholesterol lowering in 4444 patients with heart disease: the Scandinavian Simvastatin Survival Study (4S) Lancet. 1994;344:1383–1389. [PubMed] [Google Scholar]
- 2.Blum CB. Comparison of four inhibitors of 3-hydroxy- 3-methylglutaryl-coenzyme A reductase. Am J Cardiol. 1994;73:3D–11D. doi: 10.1016/0002-9149(94)90626-2. [DOI] [PubMed] [Google Scholar]
- 3.Segaert MF, De Soete C, Vandewiele I, Verbanck J. Drug-interaction-induced rhabdomyolysis. Nephrol Dial Transplant. 1996;11:1846–1847. [PubMed] [Google Scholar]
- 4.Schmassmann-Suhijar D, Bullingham R, Gasser R, Schmutz J, Haefeli WE. Rhabdomyolysis due to interaction of simvastatin with mibefradil. Lancet. 1998;351:1929. doi: 10.1016/S0140-6736(05)78613-X. [DOI] [PubMed] [Google Scholar]
- 5.Roche. FDA announce new drug-interacting warnings for mibefradil. Am J Health-Syst Pharm. 1998;55:210. doi: 10.1093/ajhp/55.3.210a. [DOI] [PubMed] [Google Scholar]
- 6.Prueksaritanont T, Gorham LM, Bennett MA, et al. In vitro metabolism of simvastatin in humans – identification of metabolising enzymes and effect of the drug on hepatic P450s. Drug Metab Dispos. 1997;25:1191–1199. [PubMed] [Google Scholar]
- 7.Prueksaritanont T, Ma B, Tang C, et al. Metabolic interactions between mibefradil and HMG-CoA reductase inhibitors: an in vitro investigation with human liver preparations. Br J Clin Pharmacol. 1999;47:291–298. doi: 10.1046/j.1365-2125.1999.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruer PJK, Vega JM, Mercuri MF, Dobrinska MR, Tobert JA. Concomitant use of cytochrome P450 3A4 inhibitors and simvastatin. Am J Cardiol. 1999;84:811–815. doi: 10.1016/s0002-9149(99)00442-7. [DOI] [PubMed] [Google Scholar]
- 9.Neuvonen PJ, Kantola T, Kivisto KT. Calcium channel blocker–simvastatin interaction. Clin Pharmacol Ther. 1999;65:583. [PubMed] [Google Scholar]
- 10.Thummel KE, Wilkinson GR. In vitro and in vivo drug interactions involving human CYP3A. Ann Rev Pharmacol Toxicol. 1998;38:389–430. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- 11.Azie NE, Brater DC, Becker PA, Jones DR, Hall SD. The interaction of diltiazem with lovastatin and pravastatin. Clin Pharmacol Ther. 1998;64:369–377. doi: 10.1016/S0009-9236(98)90067-4. [DOI] [PubMed] [Google Scholar]
- 12.Kantola T, Kivisto K, Neuvonen P. Erythromycin and verapamil considerably increase simvastatin and simvastatin acid concentrations. Clin Pharmacol Ther. 1998;64:177–182. doi: 10.1016/S0009-9236(98)90151-5. [DOI] [PubMed] [Google Scholar]
- 13.Sutton D, Butler AM, Nadin L, Murray M. Role of CYP3A4 in human hepatic diltiazem N-demethylation: inhibition of CYP3A4 activity by oxidised diltiazem metabolites. J Pharmacol Exp Ther. 1997;282:294–300. [PubMed] [Google Scholar]
- 14.Jones DR, Gorski JC, Hamman MA, Mayhew BS, Rider S, Hall SD. Diltiazem inhibition of cytochrome P450, 3A activity is due to metabolite–intermediate complex formation. J Pharmacol Exp Ther. 1999;28:294–300. [PubMed] [Google Scholar]
- 15.Ma B, Prueksaritanont T, Lin JH. Drug interactions with calcium channel blockers: possible involvement of metabolite-intermediate complexation with CYP3A. Drug Metab Disp. 2000;28:125–130. [PubMed] [Google Scholar]
- 16.Otton SV, Crewe HK, Lennard MS, Tucker GT, Woods HF. Use of quinidine inhibition to define the role of the sparteine/debrisoquine cytochrome P450 in metoprolol oxidation in human liver microsomes. J Pharmacol Exp Ther. 1988;247:242–247. [PubMed] [Google Scholar]
- 17.Lowry OH, Roseburgh NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 18.He Kan Woolf T, Hollenberg PF. Mechanism-based inactivation of cytochrome P4503A4 by mifepristone (RU486) J Pharmacol Exp Ther. 1999;288:791–797. [PubMed] [Google Scholar]
- 19.Vickers S, Duncan CA, Vyas KP, et al. In vitro and in vivo biotransformation of simvastatin, an inhibitor. Of HMG-Coa Reductase Drug Metab Disp. 1990;18:138–145. [PubMed] [Google Scholar]
- 20.Silverman RB. Mechanism-Based Enzyme Inactivation: Chemistry and Enzymology Boca Raton. Vol. 1. Boca Raton: CRC Press; 1988. pp. 3–15. [Google Scholar]
- 21.Chiba M, Nishime JA, Lin JH. Potent and selective inactivation of human liver microsomal cytochrome P450 isoforms by L-754,394, an investigational human immune deficiency virus protease inhibitor. J Pharmacol Exp Ther. 1995;275:1527–1534. [PubMed] [Google Scholar]
- 22.Guengerich F. Mechanism-based inactivation of human liver microsomal cytochrome P450 III A4 by gestodene. Chem Res Toxicol. 1990;3:363–371. doi: 10.1021/tx00016a015. [DOI] [PubMed] [Google Scholar]
- 23.Kantola T, Kivisto KT, Neuvonen PJ. Erythromycin and verapamil considerably increase serum simvastatin and simvastatin acid concentrations. Clin Pharmacol Ther. 1998;64:177–182. doi: 10.1016/S0009-9236(98)90151-5. [DOI] [PubMed] [Google Scholar]
- 24.Rowland Yeo K, Yeo WW, Wallis EJ, Ramsay LE. Enhanced cholesterol reduction by simvastatin in diltiazem-treated patients. Br J Clin Pharmacol. 1999;48:610–615. doi: 10.1046/j.1365-2125.1999.00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang BB, Siedlek PH, Smithers JA, Sedman AJ, Stern RH. Atorvastatin pharmacokinetic interactions with other CYP3A4 substrates: erythromycin and ethinyl estradiol. Pharmacol Res. 1996;13(Suppl 9):S437. [Google Scholar]
- 26.Ochmann K, Mueck W, Unger S, Kuhlmann J. Influence of erythromycin pre- and co-treatment on the single dose pharmacokinetics of cerivastatin. Eur J Clin Pharmacol. 1997;52(Suppl):A139. doi: 10.1007/s002280050408. [DOI] [PubMed] [Google Scholar]
- 27.Desager JP, Horsmans Y. Clinical pharmacokinetics of 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase inhibitors. Clin Pharmacokin. 1996;31:348–371. doi: 10.2165/00003088-199631050-00003. [DOI] [PubMed] [Google Scholar]
- 28.Department of Health. London: DoH; 2000. National service framework for coronary heart disease. Modern standards and servic models. [Google Scholar]
- 29.Haq IUL, Ramsay LE, Pickin DM, Yeo WW, Jackson PR, Payne JN. Lipid-lowering for prevention of coronary heart disease: what policy now? Clin Sci. 1996;91:399–413. doi: 10.1042/cs0910399. [DOI] [PubMed] [Google Scholar]